Abstract
Due to the complex manufacturing process of therapeutic monoclonal antibodies, it is hardly possible to produce an identical copy of the original product (originator). Consequently, follow-on products (biosimilars) must demonstrate their efficacy being similar to the originator in terms of structure and function. During this process, a variety of analytical methods are required for this purpose. This study focuses on three particularly relevant analytical techniques: high-resolution mass spectrometry, fragment crystallisable (Fc) affinity chromatography, and two-dimensional peptide mapping. Each analytical method proved able to identify specific differences between originator and biosimilar. High-resolution mass spectrometry was used to characterize the glycan pattern. It was shown that a trastuzumab biosimilar did not have the G0:G0F sugar modification identified in the originator. The application of affinity chromatography to rituximab showed that originator and biosimilar interacted differently with the immobilized Fc receptor. Furthermore, 2D-HPLC peptide mapping demonstrated the influence of orthogonality of separation dimensions, leading to differentiation of a rituximab originator and biosimilar.
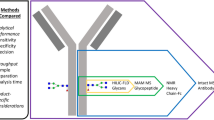
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Therapeutic monoclonal antibodies (mAbs) represent a group of drugs with significant market growth [1]. They are proteins with a molecular weight of about 150,000 Da. Each antibody is divided into two light (25,000 Da) and two heavy chains (50,000 Da) [2, 3]. Their production is usually carried out in animal cell cultures such as Chinese hamster ovary cells (CHO) or mouse myeloma cells (NS0 and SP2/0) [1, 3]. These mammalian expression systems lead to a characteristic glycosylation pattern ensuring therapeutic efficacy [1].
The highly complex production process also has an impact on the generation of post-translational modifications, which is the reason for it being impossible to produce an exact copy of the original substance (originator). The follow-on products are therefore called biosimilars and not generics. However, in order to ensure the efficacy and safety of the biosimilar, the regulatory authorities only accept minor modifications from the originator [3].
The European Medicines Agency (EMA) [4] has therefore published a guideline to assess the similarity of biological medicinal products containing monoclonal antibodies. Herein, a stepwise approach consisting of non-clinical and clinical studies is recommended. Step 1 of the non-clinical studies includes in vitro assays such as determination of binding to antigens or to the respective isoforms of the relevant fragment crystallisable (Fc) gamma receptors. In step 2, it is to be assessed whether additional in vivo studies are necessary. Among other things, the presence of additional quality attributes (e.g., new post-translational modifications) that were not present in the originator must be taken into account. On the basis of the analytical evaluation, a decision must be made whether in vivo studies are required or clinical trials should be initiated. The scope of the clinical studies needed will depend on the results of the non-clinical studies, and a stepwise approach will be taken for these as well [4].
However, the characterization of monoclonal antibodies does not end with the approval of the drug, as a continuous quality monitoring of the active pharmaceutical ingredient (API) has to be performed. This is because fluctuations in the production process or up-scaling can lead to modifications such as an altered glycosylation pattern [5,6,7].
Modern analytical methods are required for the detection of biosimilarity and subsequent analytical quality control. An elegant approach is the characterization of the glycosylation pattern by high-resolution mass spectrometry (HRMS). The glycosylation pattern of mAbs has an impact on safety and efficacy, so that its characterization is mandatory [8]. The determination of molecular weight (MW) by LC-HRMS is useful for this purpose, which has the advantage that no additional sample preparation steps are required [3, 9]. However, it should be noted that it is not the MW that is determined, but rather the most frequent isotopic mass, which nevertheless approximates the MW in the case of large macromolecules such as mAbs [10]. However, ionization in the mass spectrometer of intact monoclonal antibodies results in a charge pattern which is superimposed by the isotope pattern. Such superposition exists for all glycosylation species. These complex patterns can be converted by deconvolution algorithms into the most frequent isotope mass or, approximately, the MW [9].
Although an HRMS approach can provide information about the presence or absence of post-translational modifications, it is difficult to link them to a therapeutic effect. For the correlation, appropriate binding assays or affinity chromatography need to be used. For example, retention time in affinity chromatography can be correlated with antibody-dependent cell-mediated cytotoxicity (ADCC). Furthermore, affinity chromatography offers the possibility to use low-cost spectroscopic detection methods without the need for HRMS measurements. Despite the use of low-cost detection techniques, the robustness of the method is not compromised since other matrix components interact only slightly with the stationary phase and can therefore be chromatographically separated from the analyte [11].
In addition to the analysis of intact monoclonal antibodies, peptide mapping needs to be performed for complete characterization, as some modifications would otherwise remain undetected. An example of this is deamidation, which leads to a mass shift of only 1 Da [2]. However, one-dimensional liquid chromatography usually does not have the necessary peak capacity to allow complete sequence recovery [12]. To increase peak capacity, comprehensive two-dimensional liquid chromatography (LC × LC) can be used. Here, a sample mixture is first chromatographically separated and the eluate of the column is continuously transferred to a second separation phase with a different separation mechanism. How much the separation mechanisms differ with respect to the analyte can be indicated by calculation of the orthogonality [13,14,15,16]. In simplified terms, this describes how well the substances are distributed across the two-dimensional separation space.
The aim of this study therefore was to demonstrate that different analytical methods can be used for the characterization of the originator and biosimilar. Specifically, the advantages and disadvantages of glycan analysis by LC-HRMS, Fc chromatography, and 2D-HPLC will be discussed and demonstrated using analysis examples of originator and biosimilar.
Materials and methods
Chemicals and reagents
The monoclonal antibodies bevacizumab, cetuximab, daratumumab, omalizumab, rituximab, and trastuzumab were sourced as originator, which includes the drugs Avastin (Hoffmann-La Roche, Basel, Switzerland), Erbitux (Merck KGaA, Darmstadt, Germany), Darzalex (Janssen-Cilag GmbH, Neuss, Germany), Xolair (Novartis Pharma, Basel, Switzerland), MabThera (Hoffmann-La Roche, Basel, Switzerland), and Herceptin (Hoffmann-La Roche, Basel, Switzerland). Furthermore, the biosimilars Truxima (Celltrion Healthcare Hungary Kft., Budapest, Hungary), Rixathon (Sandoz, Holzkirchen, Germany), and Herzuma (Celltrion Healthcare Hungary Kft., Budapest, Hungary) were used. In addition, a rituximab research sample was provided by Tosoh Bioscience GmbH, Darmstadt, Germany. Water, acetonitrile, and sodium chloride were purchased from Merck KGaA (Darmstadt, Germany). Formic acid (FA), PBS buffer, sodium acetate, guanidine hydrochloride (GuHCl), dithiothreitol (DTT), ammonium bicarbonate (NH4HCO3), and tris(hydroxymethyl)aminomethane hydrochloride (TrisHCl) were purchased from Sigma-Aldrich (St. Louis, USA). Sequencing grade modified trypsin was purchased from Promega (Madison, USA). Isopropanol (IPA) was obtained from Th. Geyer (Renningen, Germany), hydrochloric acid from Fluka (St. Louis, USA), and sodium hydroxide and ammonia from AppliChem (Darmstadt, Germany).
Analysis conditions and detector settings for the determination of the glycan pattern by LC-HRMS
The analytical system consists of a 1260 Infinity II (Agilent Technologies, Waldbronn, Germany) liquid chromatography system coupled to a 6560 Ion Mobility LC/Q-TOF (Agilent Technologies, Waldbronn, Germany). The liquid chromatography system includes a pump, autosampler, column thermostat, and diode array detector. The whole system was controlled via Masshunter Workstation software (Build 9.0.9044.1 SP1) from Agilent Technologies (Waldbronn, Germany). Separation was performed at 80 °C. The mobile phase consisted of water + 0.1% formic acid (mobile phase A) and acetonitrile/isopropanol/water (70:20:10 v/v/v) + 0.1% formic acid (mobile phase B). A Zorbax 300 SB-C8 2.1 × 150 mm, 5 µm (Agilent Technologies, Waldbronn, Germany) stationary phase was installed. A Zorbax 300 SB-C8 2.1 × 12.5 mm, 5 µm (Agilent Technologies, Waldbronn, Germany) was used as precolumn. The injection volume was 0.1 µL and the flow rate was set to 0.5 mL min−1. The concentration of the API was 2.5 mg mL−1. The gradient program was as follows: 0.0–2.0 min 5–5% B; 2.0–3.0 min 5–50% B; 3.0–6.0 min 50–50% B; 6.0–6.5 min 50–90% B; 6.5–9.5 min 90–90% B; 9.5–10.5 min 90–5% B; 10.0–14.0 min 5–5% B. The mass spectrometer recorded all signals in the range of m/z 500–5000. ESI positive was selected as ionization mode. The other settings were as follows: 350 °C gas temperature, 12 L min−1 gas flow, 60 psig nebulizer gas, 400 °C sheath gas temperature, 11 L min−1 sheath gas flow, 5500 V capillary voltage (VCap), 2000 V nozzle voltage, 380 V fragmentor voltage, 750 V octopole rod repel voltage (OctopoleRPPeak).
The signals of the antibody were evaluated using Masshunter BioConfirm (Build 10.0.10136.0) from Agilent Technologies (Waldbronn, Germany). For deconvolution using the Maximum Entropy (MaxEnt) algorithm, the signals from m/z 2000 to 3500 with a retention time of 5.1–5.8 min are used, which have a signal-to-noise ratio of at least 30. The baseline is adjusted by a factor of 7. The mass range to be calculated is 140,000–160,000 Da with a mass resolution of 0.05 Da. The charge states are considered to be proton adducts.
2D-HPLC and the Fc affinity chromatographic method
A 1290 Infinity II 2D-LC system (Agilent Technologies, Waldbronn, Germany) was used. It consisted of two HPLC pumps, two column ovens, two diode array detectors, a multisampler, and three valve drives, yet only the flow path of the first dimension was used. The system was controlled via OpenLab CDS Chemstation Edition Rev. C.01.09 144 (Agilent Technologies, Waldbronn, Germany). A TSKgel FcR-3A-NPR 4.6 × 75 mm stationary phase (Tosoh Bioscience GmbH, Darmstadt, Germany) at 15 °C was selected for analysis. A 50 mM citrate buffer was used as mobile phase. Mobile phase A was adjusted to a pH of 5.9 and mobile phase B to 4.3. The flow rate was set to 0.5 mL min−1 and the injection volume to 5 µL. The concentration of the API was 5 mg mL−1. The gradient program was 0–20 min 0–62.5% B, 20–20.1 min 62.5–100% B, 20.1–40 min 100–100% B, 40–40.1 min 100–0% B, and 40.1–50 min 0–0% B. Detection was performed at 280 nm.
Selectivity screening for 2D-HPLC
Selectivity screening was performed using the 2D-HPLC system described above. However, the injection volume was increased to 20 µL. The preparation of the samples, as well as their concentration, is described below. The modulation time was 1 min for RPC methods (pH 2.7) in the second dimension and 1.5 min for HILIC methods. An active solvent modulation (ASM) factor of 3 was used. The importance and use of an ASM valve is explained in the Supplementary Part. The combinations of stationary phases and pH tested were RP(pH 10)/RP(pH 2.7), RP(pH 10)/HILIC, RP(pH 2.7)/HILIC, SCX/RP(pH 2.7), SEC/RP(pH 2.7), HIC/RP(pH 2.7), SCX/HILIC, SEC/HILIC, and HIC/HILIC. The detailed analysis conditions can be found in Table 1.
Tryptic digest
In order to generate the peptides for selectivity screening, a mixture of the monoclonal antibodies bevacizumab, cetuximab, daratumumab, omalizumab, rituximab, and trastuzumab at a concentration of 1 mg mL−1 was tryptically digested. For this purpose, 50 µL of sample was mixed with 50 µL of denaturation buffer. The denaturation buffer consisted of 3 M GuHCl, 100 mM TrisHCl, and 8 mM DTT. The mixture was heated to 95 °C for 20 min using a WiseTherm HB-48P heating block (Witteg Labortechnik GmbH, Wertheim, Germany). After cooling to room temperature, 575 µL of a 50 mM NH4HCO3 solution was added first, followed by 50 µL of trypsin solution (1 µg absolute). Tryptic digestion was performed after mixing for 21 h at 37 °C in an IPP55 Plus incubator (Memmert GmbH + Co. KG, Büchenbach, Germany). The reaction was quenched by adding 25 µL of formic acid.
Results and discussion
Intact analysis performed on mAbs: innovator and biosimilar
Before the evaluation of the mass spectrum of a mAb can be performed, the charge distribution obtained must be deconvoluted. The result of such a deconvolution is shown in Fig. 1 for trastuzumab, where the drug product Herceptin is the originator and Herzuma the biosimilar. Furthermore, results for the monoclonal antibody rituximab can be obtained from the Supplementary Part.
The data in Fig. 1 show the glycan pattern of trastuzumab, with the originator giving rise to five signals. The biosimilar, in contrast, had only four clearly prominent signals. This is because the G0:G0F modification could not be detected in the biosimilar. A more detailed evaluation of the signals can be found in Table 2.
The deconvolved masses of the individual signals are consistent with theory and analyses of other research groups [2, 17]. The masses for Herceptin and Herzuma were found identical, which was to be expected since the latter is an approved biosimilar. For the G0:G0F modification, it should be noted that it is the sugar modification with the lowest intensity. Although the G0:G0F modification was not detected, Lee et al. demonstrated that the G0 glycan is present in Herzuma to a similar extent as in Herceptin [18]. Since the proportion of the individual sugar modifications are known to vary with the production batch, these variations can be recognized using LC-HRMS [19]. This means that it is difficult to determine the cause of such differences, as molecular differences can be masked by batch variations. In order for the differences to be traceable to the analyte, various batches must be examined.
Regardless of the reason for the absence of the modification, these minor differences between originator and biosimilar could be successfully detected. Hence, LC-HRMS of intact mAbs is a suitable tool to characterize the glycosylation pattern.
Quantification of affinity for an Fc gamma receptor IIIA ligand
Although HRMS proved well suited for glycan analysis, it is difficult to predict the effects to the API from the sugar modifications detected. Thereby, the glycan structure has an influence on the binding affinity to the Fc receptor, which is involved in antibody-dependent cell-mediated cytotoxicity (ADCC). Using affinity chromatography with an immobilized Fc receptor, it is thus possible to generate a correlation to ADCC and at the same time make a statement about the sugar modifications responsible for this [7, 11].
Figure 2 shows the application of Fc affinity chromatography to different rituximab drugs, where MabThera is the originator and Truxima and Rixathon are the biosimilars. Furthermore, a rituximab biosimilar candidate currently in development (R&D sample) was investigated.
Chromatogram of the analysis of rituximab by FcR affinity chromatography. The originator MabThera (blue dashed) was compared with the biosimilars Truxima (green dotted) and Rixathon (orange dotted and dashed), as well as a research sample (red solid). All samples showed three signals, whereby increasing retention time is associated with increasing affinity for the Fc receptor and thus increased ADCC activity
The analysis gives three main signals for all samples, which is in agreement with the literature [11]. It should be noted that longer retention leads to increased ADCC activity. This relationship was confirmed by Chakrabarti et al. for rituximab by correlating FcR chromatography analysis results with an ADCC binding assay. They were able to show that ADCC activity was highest from the last eluting compound (peak 3) and lowest for the first eluting compound (peak 1). The activity from the unseparated monoclonal antibody represented a mixture and is intermediate between that of peak 1 and peak 2 [11]. The correlation between ADCC activity and retention time in FcR chromatography was also shown for other mAbs [7]. A quantitative evaluation of Fig. 2 can be found in Table 3.
The comparison of the R&D sample with MabThera shows comparable results for all three signals, making this also a promising biosimilar candidate.
Interestingly, the normalized area of peaks 1 and 3 of the biosimilar Rixathon differed significantly from the originator MabThera (50.4% vs. 38.4% and 9.2% vs. 18.1%, respectively), suggesting lower ADCC activity. However, studies demonstrated comparable ADCC activity between Rixathon and MabThera [20, 21]. Considering the influence of sugar modifications on the Fc affinity chromatography, it could be shown that more terminal galactose as well as a higher proportion of afucosylated glycans led to a higher retention [11]. Studies have also shown that the glycan structure of Rixathon was within the range of the batch variability of MabThera. Nevertheless, small differences were found in sugar modifications with a low intensity [20, 21]. The results of our own HRMS analysis are shown in Table 4.
The results of HRMS analysis show that the sugar modification G0F:G0F has a relative area of 27.6% ± 2.3% for the biosimilar Rixathon and 20.1% ± 3.5 for the originator MabThera. Since this modification elutes mainly at peak 1 in affinity chromatography, the increased area of peak 1 for Rixathon can thus be explained. Similarly, the difference of peak 3 in affinity chromatography can be explained. This can be attributed to the lower proportion of modifications with a higher proportion of terminal galactose (see Table 4).
Fc affinity chromatography can thus be used to characterize monoclonal antibodies and estimate their ADCC properties. Due to its sensitivity to changes in glycan composition, it is a suitable tool to detect differences in manufacturing batches and biosimilars. Furthermore, this technique can be used to quickly check whether a biosimilar candidate is promising and should be investigated further.
Comparison of biosimilar and originator by 2D-HPLC
The use of comprehensive 2D-HPLC, where the two separation dimensions are orthogonal, increases peak capacity and hence chromatographic separation. In this study, we applied a highly orthogonal method to samples from tryptic digest yielding results that proved equal to mass spectrometer utilization.
Figure 3 shows the determination of orthogonality for a tryptic digest of a mixture containing six monoclonal antibodies, using different separation mechanisms. Each spot represents at least one peptide.
The combinations of SEC/HILIC and HIC/HILIC cannot be recommended due to solvent incompatibilities. The combination of SEC/RP and HIC/RP did not provide good orthogonality, which can be seen from the fact that the substances were not distributed across the separation space, but rather formed clusters that were not well resolved. The other five combinations used led to a distribution of the substances across the separation space.
Although our analytical data show that there are potentially several suitable combinations (RP pH 10/RP pH 2.7; RP pH 10/HILIC; RP pH 2.7/HILIC; SCX/RP pH 2.7; SCX/HILIC), in practice it may not be appropriate to use the combination with the highest peak capacity. For example, Vanhoenacker et al. showed that a combination of RP/RP had a higher peak capacity, but some oxidative changes of trastuzumab could only be detected with the SCX/RP combination [12]. This means that regardless of the maximum peak capacity, diverse combinations of separation mechanisms should be used for complete characterization.
Figure 4 shows an example of the application of a possible combination (RP pH 2.7/HILIC) to distinguish originator and biosimilar monoclonal antibodies.
The enlarged areas of Fig. 4 show a different distribution of the substance spots. Although no mass spectrometer was used to identify the peptides, a different peak pattern of the biosimilar peptides indicates that they were peptides that differed from the originator, which means that biosimilar and originator can be distinguished by using a UV detector.
Therefore, our data show that it is possible to detect differences in mAb samples (innovator vs. biosimilar) with a UV detector due to the specific peak pattern across the two-dimensional separation space.
Conclusion
Characterization of monoclonal antibodies and evaluation of biosimilar and originator similarity make great demands on analytical techniques. A wide variety of instrumental analytical techniques was needed to obtain sufficient structural and functional information. However, by choosing and combining LC-HRMS, Fc affinity chromatography, and 2D-HPLC, we could show that analytical techniques are becoming sufficiently sophisticated to greatly reduce sample preparation, allow structural characterization, distinguish originator and biosimilar functionally, and offer the possibility to quickly identify new biosimilar candidates.
Although a large amount of information can be obtained by combining the above analytical methods, they also have certain limitations when considered individually. High-resolution mass spectrometry of intact mAbs is an excellent tool to characterize the glycan structure, but it does not provide information about its effects on ADCC. Fc affinity chromatography, on the other hand, can be used to determine the ADCC properties of a mAb, but cannot be used to accurately characterize the glycan structure. If additional information about the primary sequence is required, characterization should be performed at the peptide level. Here, 2D-HPLC coupled to UV detection offers the possibility to distinguish a biosimilar from its originator if the separation mechanisms have a high orthogonality.
Abbreviations
- mAbs:
-
Monoclonal antibodies
- CHO:
-
Chinese hamster ovary cells
- EMA:
-
European Medicines Agency
- API:
-
Active pharmaceutical ingredient
- HRMS:
-
High-resolution mass spectrometry
- 2D-HPLC:
-
Two-dimensional liquid chromatography
- ADCC:
-
Antibody-dependent cell-mediated cytotoxicity
- Fc:
-
Fragment crystallisable
References
Grilo AL, Mantalaris A. The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. 2019;37(1):9–16. https://doi.org/10.1016/j.tibtech.2018.05.014.
Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianferani S. Characterization of therapeutic antibodies and related products. Anal Chem. 2013;85(2):715–36. https://doi.org/10.1021/ac3032355.
Beck A, Sanglier-Cianferani S, Van Dorsselaer A. Biosimilar, biobetter, and next generation antibody characterization by mass spectrometry. Anal Chem. 2012;84(11):4637–46. https://doi.org/10.1021/ac3002885.
Agency EM (2012) Guideline on similar biological medicinal products containing monoclonal antibodies – non-clinical and clinical issues. London
Shukla AA, Thommes J. Recent advances in large-scale production of monoclonal antibodies and related proteins. Trends Biotechnol. 2010;28(5):253–61. https://doi.org/10.1016/j.tibtech.2010.02.001.
Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19(9):936–49. https://doi.org/10.1093/glycob/cwp079.
Xie LQ, Zhang EH, Xu YP, Gao WY, Wang LL, Xie MHW, Qin PL, Lu LH, Li SP, Shen PC, Jiang WD, Liu S. Demonstrating analytical similarity of trastuzumab biosimilar HLX02 to Herceptin (R) with a panel of sensitive and orthogonal methods including a novel Fc gamma RIIIa affinity chromatography technology. BioDrugs. 2020;34(3):363–79. https://doi.org/10.1007/s40259-020-00407-0.
Sandra K, Vandenheede I, Sandra P. Modern chromatographic and mass spectrometric techniques for protein biopharmaceutical characterization. J Chromatogr A. 2014;1335:81–103. https://doi.org/10.1016/j.chroma.2013.11.057.
Reinders LMH, Klassen MD, Jaeger M, Teutenberg T, Tuerk J. Development of an analytical method to assess the occupational health risk of therapeutic monoclonal antibodies using LC-HRMS. Anal Bioanal Chem. 2018;410(11):2829–36. https://doi.org/10.1007/s00216-018-0966-1.
Hesse M, Meier H, Zeeh B (2012) Spektroskopische Methoden in der organischen Chemie; 8. Edition, Georg Thieme Verlag KG, Stuttgart, ISBN 978-3-13-576108-4
Chakrabarti A, Kervinen J, Müller E, Tanaka T, Muranaka K (2020) Analytical characterization of monoclonal antibodies with novel Fc receptor-based chromatography technique, monoclonal antibodies. In: Monoclonal antibodies. https://doi.org/10.5772/intechopen.95356
Vanhoenacker G, Vandenheede I, David F, Sandra P, Sandra K. Comprehensive two-dimensional liquid chromatography of therapeutic monoclonal antibody digests. Anal Bioanal Chem. 2015;407(1):355–66. https://doi.org/10.1007/s00216-014-8299-1.
Semard G, Peulon-Agasse V, Bruchet A, Bouillon JP, Cardinael P. Convex hull: a new method to determine the separation space used and to optimize operating conditions for comprehensive two-dimensional gas chromatography. J Chromatogr A. 2010;1217(33):5449–54. https://doi.org/10.1016/j.chroma.2010.06.048.
Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of separation in two-dimensional liquid chromatography. Anal Chem. 2005;77(19):6426–34. https://doi.org/10.1021/ac050923i.
Camenzuli M, Schoenmakers PJ. A new measure of orthogonality for multi-dimensional chromatography. Anal Chim Acta. 2014;838:93–101. https://doi.org/10.1016/j.aca.2014.05.048.
Leonhardt J, Teutenberg T, Buschmann G, Gassner O, Schmidt TC. A new method for the determination of peak distribution across a two-dimensional separation space for the identification of optimal column combinations. Anal Bioanal Chem. 2016;408(28):8079–88. https://doi.org/10.1007/s00216-016-9911-3.
Delobel A, Cantais F, Catrain A, Dereux E, Van Vyncht G. Therapeutic antibody glycosylation analysis: a contract research organization perspective in the frame of batch release or comparability support. Methods Mol Biol. 2013;988:115–43. https://doi.org/10.1007/978-1-62703-327-5_8.
Lee J, Kang HA, Bae JS, Kim KD, Lee KH, Lim KH, Choo MJ, Chang SJ. Evaluation of analytical similarity between trastuzumab biosimilar CT-P6 and reference product using statistical analyses. mAbs. 2018;10(4):547–71. https://doi.org/10.1080/19420862.2018.1440170.
Damen CWN, Chen WB, Chakraborty AB, van Oosterhout M, Mazzeo JR, Gebler JC, Schellens JHM, Rosing H, Beijnen JH. Electrospray ionization quadrupole ion-mobility time-of-flight mass spectrometry as a tool to distinguish the lot-to-lot heterogeneity in N-glycosylation profile of the therapeutic monoclonal antibody trastuzumab. J Am Soc Mass Spectr. 2009;20(11):2021–33. https://doi.org/10.1016/j.jasms.2009.07.017.
Visser J, Feuerstein I, Stangler T, Schmiederer T, Fritsch C, Schiestl M. Physicochemical and functional comparability between the proposed biosimilar rituximab GP2013 and originator rituximab. BioDrugs. 2013;27(5):495–507. https://doi.org/10.1007/s40259-013-0036-3.
Jurczak W, Cohen S, Illidge TM, da Silva A, Amersdorffer J. Scientific rationale underpinning the development of biosimilar rituximab in hematological cancers and inflammatory diseases. Future Oncol. 2019;15(36):4223–34. https://doi.org/10.2217/fon-2019-0430.
Acknowledgements
We would like to thank the Federal Ministry for Economic Affairs and Energy for funding the INNO-KOM project “Sensitive method for the detection of airborne proteins” (49VF170039). Furthermore, we would like to thank Tosoh Bioscience for providing materials and samples.
Author information
Authors and Affiliations
Contributions
Lars M. H. Reinders: conceptualization, methodology, validation, formal analysis, investigation, data curation, writing—original draft, writing—review and editing, visualization, project administration, funding acquisition. Martin D. Klassen: conceptualization, methodology, writing—review and editing, supervision, funding acquisition. Thorsten Teutenberg: conceptualization, writing—review and editing, supervision, funding acquisition. Martin Jaeger: writing—review and editing, supervision. Torsten C. Schmidt: writing—review and editing, supervision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Reinders, L.M.H., Klassen, M.D., Teutenberg, T. et al. Comparison of originator and biosimilar monoclonal antibodies using HRMS, Fc affinity chromatography, and 2D-HPLC. Anal Bioanal Chem 414, 6761–6769 (2022). https://doi.org/10.1007/s00216-022-04236-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04236-8