Abstract
The multiclass determination of antibiotic residues in the soil is challenging because of its complex physicochemical properties. In this study, a simple analytical method was developed to simultaneously extract and determine 58 antibiotics from the soil. A novel acidity-regulated extraction-partition-concentration protocol was established for the simultaneous extraction of five classes (23 sulfonamides, 18 quinolones, five tetracyclines, eight macrolides, and four chloramphenicols) of antibiotics from the soil. Compared to traditional methods, the sample preparation efficiency was significantly improved by four times (45 min vs. 230 min) by optimizing the extraction method and omitting the time-consuming solid-phase extraction (SPE) procedure. The ultra-high-performance liquid chromatography–tandem mass spectrometry (UHPLC-MS/MS) method was optimized to determine the 58 antibiotics in a single run by applying positive/negative switching acquisition mode in less than 10 min with the baseline separation of sulfameter and sulfamethoxypyridazine. Suitable recoveries, ranging between 60 and 120%, were obtained for most antibiotics, with RSD <20%. The limits of quantification (LOQ) of the method were 2 μg/kg and 5 μg/kg. Thus, this study provides a simple, reliable, and economical method for accurately and rapidly determining a multiclass of antibiotics in the soil.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Antibiotics have been widely used worldwide for veterinary and human therapy and in the agricultural sector as feed additives [1, 2]. Livestock breeding usage reportedly accounts for over 70% of global antibiotic consumption [3]. After antibiotics are administered to animals, 30–90% of them are released into the environment via excretion of feces and urine, and a significant percentage of these excretions remain in unchanged and active forms [1, 4]. Antibiotics can enter farmland soil from the use of animal manure and manure-based fertilizers in agricultural practices and reclaimed wastewater for irrigation [5]. The presence of antibiotics in animal manure and the subsequent high residue levels in farmland soil have become major concerns [6, 7]. High residue levels of antibiotics, especially tetracyclines and quinolones, in farmland soil have been reported in numerous studies [8, 9]. Residues of antibiotics in the soil can pose a severe threat to ecological safety by spreading antibiotic resistance genes [10,11,12] and affecting the structure and function of soil microbial communities [3, 13]. In addition, residual antibiotics in the soil can be transported and accumulated in plants, leading to potential human exposure [2, 14]. Therefore, monitoring antibiotic residues in the soil is critical for evaluating their potential ecological risk.
Antibiotics are ionic, polar organic compounds containing many ionic functional groups and acid dissociation constants (pKa). The sulfonamides show characteristics of either weak alkali due to aniline nitrogen (pKa values 5–11) or weak acids due to the N-H bond of the sulfonamide group (pKa values 2–3) [15]. Quinolones have carboxylic acid groups (pKa values 4.3–6.3) and one or more amine functional groups (pKa values 7.6–9.3). Tetracyclines have three pKa values (~3.3, 7.7, and 9.3). Macrolides have one or two amine groups (pKa values 7.1–9.4). Chloramphenicols have one or two amine groups (approximate pKa value 11). Therefore, cationic, anionic, and zwitterionic forms will be present for these antibiotics in the medium depending on the pH values. These antibiotics can be adsorbed into the soil by cation exchange, surface complexation, cation bridging, and hydrogen bonding [16]. Strong adsorption is generally observed for antibiotics, especially quinolones and tetracyclines [17]. Therefore, multiple extractions or pressurized liquid extraction (PLE) is generally required to guarantee satisfactory extraction efficiency [15, 18,19,20]. In addition, some antibiotics are unstable under acidic or/and alkaline conditions, such as sulfonamides and macrolides. As a result, determining multiclass antibiotic residues in the soil has long remained a challenge. Traditionally, solid-liquid extraction is the most popular analytical method for determining antibiotic residues in soil and other solid samples [15, 18, 21,22,23]. This methodology involves multiple extractions of the target analytes in soil using a buffer or buffer/organic mixed solvent, followed by solid-phase extraction (SPE) clean-up and concentration. Another commonly used method is pressurized liquid extraction (PLE), followed by SPE concentration and clean-up because of the enhanced extraction efficiency for quinolones and tetracyclines compared to that by the solid-liquid extraction method [19, 24, 25]. As PLE extraction requires a specific instrument and the procedures are complicated and tedious, ultrasonic extraction is reportedly used to extract antibiotics from soil and sediment [22, 26]. Although relatively stable and acceptable recovery and accuracy can be obtained by these methods, the disadvantages are apparent. These methods are complicated and time-consuming, making sample preparation inefficient.
To overcome this disadvantage, a quick, easy, cheap, effective, rugged, and safe (QuEChERS) methodology has been developed in recent years to simplify the sample preparation by omitting the tedious SPE procedure [27,28,29,30,31]. This method generally involves acetonitrile extraction, followed by dispersive SPE clean-up. However, the method performance of this method has been unsatisfactory thus far, owing to the complexity of the physicochemical properties of multiclass antibiotics. Only a limited number of analytes, mainly sulfonamides, chloramphenicols, and several other antibiotics, were applicable. In most cases, low recoveries were observed for most quinolones and tetracyclines due to low partition and extraction efficiency.
The objective of this study was to develop a new analytical method to improve the performance and extend the coverage of analyte types for the multi-residue analysis of antibiotics in soil. A novel acidity-regulated extraction-partition-concentration protocol was proposed to realize the simultaneous extraction and determination of five classes of antibiotics in soil. The parameters of chromatographic separation, buffer selection, extraction time and method, acidity regulation, and concentration were thoroughly optimized. The method was validated using five different types of soils, showing suitable performance in recovery, precision, linearity, and limits of quantification (LOQs). Finally, the proposed method was applied to determine paired farmland soil–vegetable samples near livestock breeding farms. This study provides a novel and simple method for the determination and risk assessment of antibiotic residues in the soil.
Material and methods
Reagent and materials
The 58 targeted antibiotics, including 23 sulfonamides, 18 quinolones, five tetracyclines, eight macrolides, and four chloramphenicols (Table 1), were purchased from Dr. Ehrenstorfer (Augsburg, Germany). Stock solutions of the standards were prepared by dissolving each antibiotic in methanol at 1000 or 100 mg/L, according to the solubility of each compound. A standard working mixture solution was prepared in methanol (5 mg/L).
Methanol and acetonitrile were of HPLC grade and purchased from Fisher Scientific (Fair Lawn, NJ, USA). The water was purified using a Milli-Q system (Millipore, Billerica, MA, USA). Buffer salts and other chemicals including formic acid, H3PO4, KH2PO4, citric acid, Na2HPO4, anhydrous MgSO4, NaCl, disodium ethylenediamine tetraacetate dihydrate (Na2EDTA·2H2O), and ammonia solution (7 M in methanol) were purchased from Shanghai Macklin Biochemical Co., Ltd (Shanghai, China). Dispersive solid-phase extraction adsorbents (PSA and C18) and polypropylene centrifuge tubes (50 mL for extraction and 15 mL for dispersive solid-phase extraction) were purchased from Agilent Technologies (Santa Clara, CA, USA). Potassium phosphate buffer was prepared by dissolving 27.2 g KH2PO4 and 1.35 mL H3PO4 in 1 L water.
Sample preparation
An aliquot of the soil sample (2 g) was accurately weighed into a 50 mL centrifuge tube. Na2EDTA·2H2O (0.4 g) was added, followed by 10 mL of potassium phosphate buffer and 10 mL of acetonitrile (containing 5% formic acid). The tube was sealed, vortexed for 3 min using a multi-position vortexer, and then centrifuged for 5 min at 4000 rpm. All supernatants were transferred into another 50 mL centrifuge tube. The sample was extracted again with 10 mL of potassium phosphate buffer and 10 mL of acetonitrile (containing 5% formic acid) under the same conditions. The supernatants were combined in a tube, and 10 g of NaCl was added and vigorously shaken several times. The tube was centrifuged for 5 min at 4000 rpm to obtain phase separation. The supernatant (8 mL) was transferred into a 15 mL centrifuge tube containing dispersive solid-phase extraction adsorbents (anhydrous MgSO4 1200 mg and C18 400 mg). The extract was vortexed for 1 min and centrifuged for 5 min at 4000 rpm. The supernatant (5 mL) was transferred into a 100 mL heart-shaped bottle, and 250 μL of ammonia solution (7 M in methanol) was added. The supernatant was carefully evaporated using a vacuum rotary evaporator in a water bath at 40 °C. The residue was redissolved in 1 mL of methanol/water (containing 1% formic acid) (1/1, v/v) and filtered through a 0.22 μm polytetrafluoroethylene (PTFE) filter for ultra-high-performance liquid chromatography–tandem mass spectrometry (UHPLC-MS/MS) analysis.
Instrument
An Exion UHPLC coupled to a quadrupole linear ion trap mass spectrometer (QTRAP 5500+, SCIEX) was used for antibiotic determination. A C18 (ACQUITY HSS T3 column, 2.1 × 100 mm, 1.7 μm, Waters Corporation) column was used for chromatographic separation. The column temperature was set at 40 °C, and the flow rate was 0.3 mL/min. The injection volume was 2 μL. Mobile phase A was water containing 0.1% formic acid, and mobile phase B was methanol. The gradient was programmed as follows: 0–1.5 min: 10% B, 1.5–5 min: gradient increase to 40 % B, 5–7.1 min: gradient increase to 90 % B, 7.1–8 min: hold 90 % B, 8–8.1 min: 10 % B; 8.1–10: hold 10 % B.
The mass spectrometry was operated in positive/negative switching mode with a DuoSpray ion source, and the source parameters were as follows: ion spray voltage floating (ISVF), 5500 V; temperature, 500 °C; nebulizing gas (GS1), 50 psi; heater gas (GS2), 50 psi; and curtain gas, 30 psi. Flow injection was performed for each antibiotic to optimize the MRM transitions and the corresponding MS parameters, including decluttering potential (DP) and collision energy (CE). The detailed chromatographic and MS parameters of the antibiotics are listed in Table 1.
Method validation
The analytical performance of the selected antibiotics was evaluated in terms of recovery, repeatability, LOQ, linearity, and matrix effects according to EU guideline SANTE/11312/2021 [32]. Validation was conducted using five different soil types (black soil, fluvo-aquic soil, drab earth, red soil, and paddy soil), free of the selected antibiotics. All recovery experiments were performed with five replicates for each soil type. The LOQ for each antibiotic was determined based on the recovery results and defined as the lowest concentration that could be quantified with acceptable accuracy (mean recovery within the range of 70–120%, in exceptional cases, 30–70% and 120–140%) and precision (relative standard deviation [RSD] ≤20%). Matrix-matched and solvent-based calibration curves (1, 2, 5, 10, 25, 50, 100, and 200 μg/L) were used to evaluate linearity and matrix effects.
Results and discussion
Optimization of the chromatographic and MS conditions
Acetonitrile has generally been used as the organic mobile phase in previous studies. However, under these mobile conditions, some antibiotic isomers with the same MRM transitions could not be chromatographically separated, leading to misannotation. In this study, sulfameter and sulfamethoxypyridazine, two structural isomers, could not be separated, even under optimized elution conditions using acetonitrile as the organic mobile phase (Fig. S1). Therefore, methanol and methanol/acetonitrile (1/1, v/v) with and without formic acid (0.1%) were tested and compared for chromatographic separation and ionization efficiency. Baseline separation between sulfameter and sulfamethoxypyridazine was obtained using methanol and methanol/acetonitrile (1/1, v/v) as the organic mobile phase (Fig. S2). A slight reduction in ionization efficiency was observed for several compounds such as sulfonamides (sulfaguanidine and sulfameter), quinolones (ciprofloxacin), macrolides (spiramycin, erythromycin, tilmicosin, josamycin, and tylosin), and chloramphenicols (florfenicol amine). A relatively strong ionization suppression was observed for sulfanitran, a rarely used sulfonamide, leading to a low mass response. However, for most antibiotics, the ionization efficiency was enhanced using methanol and methanol/acetonitrile (1/1, v/v) as the organic phase, and better performance was observed for methanol (Fig. S3). As the formic acid had been added to the water phase, no significant increase in ionization efficiency was observed with the addition of formic acid in the organic phase (Fig. S3). Methanol was used as the organic phase, and the MRM chromatogram of the selected antibiotics is shown in Fig. 1.
For the final injection solvent, acidified methanol/water (containing 0.1% formic acid) was generally used to obtain a more optimal peak shape and sensitivity [33]. In this study, different formic acid contents (0.1%, 0.5%, 1%, and 2%) were evaluated. As shown in Fig. S4, comparable responses were obtained for sulfonamides, macrolides, and chloramphenicols with different formic acid contents. However, higher responses for most quinolones and tetracyclines and better peak shapes for quinolones were obtained when higher contents of formic acid were used. Therefore, methanol/water (containing 1% formic acid) was used to dissolve the final extract.
Sample extraction
Traditional sample extraction procedures for antibiotic determination in soil and other solid samples involve extraction with buffer/organic solvent, followed by SPE concentration and clean-up with HLB cartridges, which are extremely complicated and time-consuming. The SPE procedure must be replaced by simple phase separation to increase sample preparation efficiency. Multiclass antibiotics with various polarity, solubility, pKa, and stability values under acidic and basic conditions may severely affect simultaneous extraction and determination. Three critical problems should be solved to guarantee satisfactory method performance: I) sufficient extraction efficiency, II) suitable recoveries during phase separation, and III) stability of the antibiotics during the final concentration process.
The extraction buffer is critical for the optimal extraction of a multiclass of antibiotics, and should be selected according to the physicochemical properties of the compounds. McIlvaine and potassium phosphate buffers combined with acetonitrile have been commonly used for extraction in previous studies [15, 18, 22]. Therefore, eight extract solvent combinations of McIlvaine buffers (pH 3, 4, 7) and potassium phosphate buffer (pH 3) with acetonitrile and acetonitrile containing 5% formic acid were investigated and compared in this study (Fig. S5). For sulfonamides, macrolides (except for azithromycin), and chloramphenicols, similar recoveries were obtained using different buffers with and without formic acid addition. However, for quinolones and azithromycin, most of the recoveries were relatively low (<30%) when McIlvaine buffers (pH 3, 4, 7) were used. A significant increase in the recovery was obtained when potassium phosphate buffer (pH3) was used, and a further increase was observed when formic acid was added. The addition of formic acid did not increase the extractability, altering the partition coefficient of antibiotics during phase separation. The increased effect of formic acid addition was observed for the partitioning of tetracyclines, with much higher recoveries (73–102%) obtained compared to the recoveries (25–52%) without formic acid. Subsequently, the recoveries of quinolones and tetracyclines with different contents of formic acid (0, 1, 2, 5, 10%) were determined and are shown in Fig. S6. The recoveries increased with an increase in the content of formic acid, and 5% was sufficient for satisfactory recoveries. Therefore, potassium phosphate buffer (pH 3) with 5% formic acid acetonitrile (1/1, v/v) was used as the final extract solvent.
The extraction method and time were optimized to obtain the best recoveries. Nine extraction combinations (Table S1) with different extraction solvent volumes, extraction times (2, 3, 4, 5, 7, and 10 min), and extraction methods (vortex and ultrasonic extraction) were evaluated, and the results are shown in Fig. S7. The overall recoveries were significantly lower for single extraction (E) than for double (D) and triple extraction (F). In addition, no further enhancement of the extraction efficiency was observed with a triple and an additional 10 min ultrasonic extraction (I). For the extraction time, 3 min was sufficient for the optimal extraction, and a prolonged time did not further increase the recoveries. In contrast, a decrease in recovery was observed for some sulfonamides with prolonged extraction time, which may be due to degradation under highly acidic conditions. Finally, double vortex extraction with a mixed solvent (10 mL of buffer and 10 mL of acidified acetonitrile) for 3 min (combination B) was performed.
The solvent/sample ratio can affect extraction efficiency. The recoveries for different sample volumes (1, 2, and 5 g) were tested using two types of soils, and the results are shown in Fig. S8 and S9. As can be seen from the figures, when 5 g soil was used, a significant reduction in the recoveries was observed for most antibiotics. Finally, 2 g of soil was used to balance the sensitivity and recovery of the proposed method.
During the extraction, Na2EDTA·2H2O was added to the samples to chelate the metals to guarantee the recovery of tetracyclines. However, excess amounts of Na2EDTA·2H2O chelate organic compounds, leading to decreased extraction efficiency for tetracyclines [34]. Therefore, 0.4 g Na2EDTA·2H2O was used, according to previous studies [15, 28].
Clean-up
Blank soil extracts fortified with 50 μg/L of each antibiotic were cleaned using different amounts of PSA and C18, the two most commonly used dispersive solid-phase extraction adsorbents in the QuEChERS method for various analytes [35]. The recoveries after clean-up using different adsorbents are shown in Fig. S10. C18 did not exhibit any adsorption of the different classes of antibiotics. However, for PSA, strong adsorption was observed for tetracyclines, even at the lowest amount of 5 mg/mL. Moderate adsorption of spiramycin occurred, leading to low recovery of 79.6% at 10 mg/mL. Weak adsorption of sulfonamides and quinolones was observed when a high amount of PSA (100 mg/mL) was used. Therefore, 50 mg/mL C18 was used for d-SPE clean-up to guarantee the recovery of tetracyclines.
Concentration
A high solvent (acetonitrile, 20 mL)/sample (2 g) ratio of 10:1 was used during sample extraction. The concentration of the final supernatant, acetonitrile, was necessary to obtain high detection sensitivity. In the traditional SPE method, methanol is generally used to elute the HLB cartridge and evaporate it to dryness using nitrogen with good recovery. In this study, the relatively high formic acid content in acetonitrile led to low recoveries of sulfonamides and macrolides during concentration because of their instability in acids. In addition, it took more than 60 min to evaporate 5 mL of acetonitrile by nitrogen blowing in a 40 °C water bath because of its relatively high boiling point, which exacerbates the degradation of sulfonamides and macrolides. To overcome this problem, an ammonia solution in methanol was added to adjust the pH of the acetonitrile, and nitrogen blowing was replaced by vacuum rotary evaporation to shorten the concentration time. The effect of different volumes of ammonia solution on the recoveries of sulfonamides and macrolides using the fortified blank extracts was evaluated (Fig. S11). For sulfonamides, the recoveries increased with an increasing volume of ammonia solution, and 250 μL was sufficient to avoid their degradation. For macrolides, all the compounds were highly sensitive (degradation rate over 80% when no ammonia solution was added) to acid, except for tilmicosin. The addition of ammonia solution could significantly reduce their degradation and increase their recovery, and no significant difference between the recoveries was observed for different volumes of ammonia solution. Finally, 250 μL of ammonia solution in methanol was used to adjust the pH of the final extracts.
Method validation
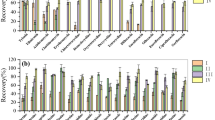
The proposed method was evaluated for 58 antibiotics in five soil types (black soil, fluvo-aquic soil, drab earth, red soil, and paddy soil). Detailed information on the soil, including soil type, mechanical composition, total organic carbon content, pH, and cation exchange capacity, is given in Table S2. Method parameters such as recovery, linearity, accuracy, matrix effects, and LOQ were calculated and evaluated according to the EU guideline SANTE/11312/2021[32]. A recovery study was performed to determine the method’s accuracy and precision at seven fortification levels of 2, 5, 10, 20, 50, 200, and 500 μg/kg in replicates (n=5). The recoveries and RSDs at different fortification levels in the five soil types are given in Tables S3–S9. Most recoveries of the selected antibiotics in the five soils ranged between 60 and 120%, with RSD < 20%. There were a few exceptions, with recoveries lower than 60%. Relatively low recoveries (38–73%) and poor repeatability were observed at different fortification levels and soil types for erythromycin, as it can easily degrade to anhydroerythromycin under acidic conditions [36]. Relatively low recoveries (40–60%) were obtained for sulfisomidine, sulfathiazole, sulfapyridine, sulfisoxazole, danofloxacin, enoxacin, norfloxacin, pipemidic acid, and oxytetracycline in some cases because of low acetonitrile/buffer partition coefficient during phase separation and/or low extraction efficiency. As suitable repeatability (RSD ≤20%) was obtained for these antibiotics, they are recognized as acceptable according to the EU guideline SANTE/11312/2021. However, low recoveries were obtained for sulfaguanidine and florfenicol amine because of the exceptionally low acetonitrile/buffer partition coefficient under the proposed extraction conditions.
The LOQs were determined based on the accuracy and precision data and were defined as the lowest fortification levels with acceptable recoveries and RSDs. The LOQs were 2 μg/kg for sulfonamides, quinolones, and macrolides, except sulfanitran. The low ionization efficiency owing to the use of methanol as the mobile phase led to poor mass responses for sulfanitran, and its LOQ was evaluated to be 10 μg/kg. For tetracyclines and chloramphenicols, the LOQ was 5 μg/kg.
Linearity was determined over a range of 1–200 μg/L in the solvent- and matrix-based standards. The coefficient of determination (r2), as presented in Tables S10–S15, was higher than 0.995 for most antibiotics in both solvent and matrix-based standards, which could guarantee accurate quantification.
Matrix effect is the suppression or enhancement of analyte responses due to the co-eluting matrix constitute. It is a major concern in small-molecule trace analysis by LC-MS and GC-MS [37]. As the matrix effect can severely compromise quantitative performance and reproducibility, it is an important performance parameter in the analytical method validation [38]. Soft matrix effects (suppression or enhancement of 0–20%) are negligible. However, certain approaches must be taken to compensate for matrix effects when they are medium (suppression or enhancement of 20–50%) or strong (suppression or enhancement > 50%). The matrix effects of the selected antibiotics were evaluated based on linear curves and were calculated using the following equation:
where, Smatrix and Ssolvent are the calibration curve slopes for each compound in the matrix-matched and solvent standards, respectively. The ME values for the five soil types are presented in Fig. S12. In general, matrix suppression was observed for sulfonamides, macrolides, and chloramphenicols, whereas, matrix enhancements were observed for quinolones and tetracyclines. For sulfonamides, 11 antibiotics exhibited weak matrix effects, 10 antibiotics exhibited moderate matrix effects, and only two (sulfadimethoxine and sulfanitran) exhibited strong matrix effects. For quinolones, matrix enhancements were observed for most antibiotics, except for sparfloxacin, orbifloxacin, and nalidixic acid. Strong matrix enhancement effects (>100%) were observed for danofloxacin, enoxacin, norfloxacin, cinoxacin, pefloxacin, and pipemidic acid. Therefore, matrix-matched standards must be applied for accurate quantification to compensate for strong matrix effects. As no significant differences were observed for different types of soil, any type of blank soil could be used to prepare matrix-matched standards.
Comparison with other methods
Many studies have reported the extraction and analysis of antibiotics in soils using SPE, PLE, and QuEChERS methods. We compared the analytical performance of the proposed method with other methods, as listed in Table S 16. Furthermore, the proposed method was compared with the SPE method (EPA 1694) in terms of recovery and RSDs; the results are shown in Fig. S13. For sample preparation efficiency, approximately 230 min (100 min for extraction, 15 min for rotary evaporation, 70 min for SPE sample loading and SPE cartridge drying, and 45 min for nitrogen blowing of the final extract) were required for the EPA 1694 method. However, only approximately 45 min (20 min for extraction, 15 min for phase separation and dispersive solid-phase extraction, 10 min for pH adjustment, and rotary evaporation) were required for the proposed method in this study. For the method performance, comparable or better recoveries and RSDs were obtained for most of the selected antibiotics using the proposed method, as shown in Table S16 and Fig. 13. In addition, the proposed method is more economical than the SPE method because of the omission of the SPE cartridge. These advantages render the proposed method superior to those of previous studies.
Method application
To further validate the feasibility of the proposed method, 15 paired soil and vegetable samples were collected from farmlands and greenhouses with long-term manure or manure-based fertilizer applications near livestock farms in Tianjin. The antibiotic residues in soil samples were determined using the methodology proposed in this study. The vegetable samples were analyzed using the QuEChERS method developed in our previous study, which was well validated to guarantee the accuracy of the results [39]. Notably, extracts containing high-level residues above the calibrated range must be diluted and re-injected, and the matrix-matched standards should also be diluted proportionately. Eight antibiotics, sulfadimidine, sulfamonomethoxine, ofloxacin, tilmicosin, chlortetracycline, oxytetracycline, tetracycline, and doxycycline, were detected in the samples. The concentrations of chlortetracycline and oxytetracycline in the paired soil and vegetable samples are shown in Fig. 2, and the concentrations of the other antibiotics in the soil samples are shown in Fig. 3. For the soil samples, high concentrations of oxytetracycline were detected, ranging between 12.7 and 1104 μg/kg (median 470.1 μg/kg, Fig. 2a). The concentrations of chlortetracycline ranged between 3.2 and 280 μg/kg (median 9.7 μg/kg, Fig. 2a). Relatively high concentrations of ofloxacin, ranging between 2.9 and 1124 μg/kg (median 16.0 μg/kg), were detected (Fig. 3). For the corresponding vegetable samples, two tetracycline antibiotics, chlortetracycline, and oxytetracycline, were detected at a detection rate of 100%, and no other antibiotics were detected (Fig. 2b). The concentrations were relatively low, with most samples being lower than 10 μg/kg, in agreement with a previous study. The concentrations of all five antibiotics (chlortetracycline, monensin, sulfamethazine, tylosin, and virginiamycin) in vegetables grown in soil fertilized with raw and composted turkey and hog manure were < 10 μg/kg [40]. High concentrations of fluoroquinolones ranging between 18.2 and 658.3 μg/kg were detected in an intensive vegetable cultivation area in Northern China [9]. In this study, no ofloxacin was detected in the green onion (sample 14), although a high concentration (1124.9 μg/kg) was detected in the corresponding soil. In addition, no positive correlation was observed between concentrations in the soil and vegetables. In most cases, variable results were obtained, even for the same class of antibiotics and plants, indicating the need for further studies.
Conclusions
A novel multi-residue analysis method for the simultaneous determination of sulfonamide, quinolone, tetracycline, macrolide, and chloramphenicol antibiotics in the soil is described. The traditional SPE procedure was replaced by an acidity-regulated extraction-partition-concentration protocol to improve sample preparation efficiency. Special attention was paid to extraction, phase separation, clean-up, and concentration. Satisfactory analytical performance was obtained for diverse types of soils, indicating the feasibility of the proposed method. Relatively high residue concentrations of oxytetracycline, chlortetracycline, and ofloxacin were observed in soils with long-term manure and manure-based fertilizer application.
References
Sarmah AK, Meyer MT, Boxall AB. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006;65(5):725–59.
Pan M, Chu L. Fate of antibiotics in soil and their uptake by edible crops. Sci Total Environ. 2017;599:500–12.
Jechalke S, Heuer H, Siemens J, Amelung W, Smalla K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014;22(9):536–45.
Zhang Q, Ying G, Pan C, Liu Y, Zhao J. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: source analysis, multimedia modeling, and linkage to bacterial resistance. Environ Sci Technol. 2015;49(11):6772–82.
Chen C, Li J, Chen P, Ding R, Zhang P, Li X. Occurrence of antibiotics and antibiotic resistances in soils from wastewater irrigation areas in Beijing and Tianjin, China. Environ Pollut. 2014;193:94–101.
Zhou X, Wang J, Lu C, Liao Q, Gudda FO, Ling W. Antibiotics in animal manure and manure-based fertilizers: Occurrence and ecological risk assessment. Chemosphere. 2020;255:127006.
Lyu J, Yang L, Zhang L, Ye B, Wang L. Antibiotics in soil and water in China–a systematic review and source analysis. Environ Pollut. 2020;266:115147.
Zhang H, Zhou Y, Huang Y, Wu L, Liu X, Luo Y. Residues and risks of veterinary antibiotics in protected vegetable soils following application of different manures. Chemosphere. 2016;152:229–37.
Li X, Xie Y, Li C, Zhao H, Zhao H, Wang N, et al. Investigation of residual fluoroquinolones in a soil–vegetable system in an intensive vegetable cultivation area in Northern China. Sci Total Environ. 2014;468:258–64.
Xie W, Shen Q, Zhao F. Antibiotics and antibiotic resistance from animal manures to soil: a review. Eur J Soil Sci. 2018;69(1):181–95.
Li J, Xin Z, Zhang Y, Chen J, Yan J, Li H, et al. Long-term manure application increased the levels of antibiotics and antibiotic resistance genes in a greenhouse soil. Appl Soil Ecol. 2017;121:193–200.
Wang F, Qiao M, Chen Z, Su J, Zhu Y. Antibiotic resistance genes in manure-amended soil and vegetables at harvest. J Hazard Mater. 2015;299:215–21.
Cycoń M, Mrozik A, Piotrowska-Seget Z. Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity. Front Microbiol. 2019;10:338.
Tasho RP, Cho JY. Veterinary antibiotics in animal waste, its distribution in soil and uptake by plants: A review. Sci Total Environ. 2016;563:366–76.
Hu W, Ma L, Guo C, Sha J, Zhu X, Wang Y. Simultaneous extraction and determination of fluoroquinolones, tetracyclines and sulfonamides antibiotics in soils using optimised solid phase extraction chromatography-tandem mass spectrometry. Int J Environ Anal Chem. 2012;92(6):698–713.
Zhi D, Yang D, Zheng Y, Yang Y, He Y, Luo L, et al. Current progress in the adsorption, transport and biodegradation of antibiotics in soil. J Environ Manag. 2019;251:109598.
Wang S, Wang H. Adsorption behavior of antibiotic in soil environment: a critical review. Front Environ Sci Eng. 2015;9(4):565–74.
Zhi S, Zhou J, Liu H, Wu H, Zhang Z, Ding Y, et al. Simultaneous extraction and determination of 45 veterinary antibiotics in swine manure by liquid chromatography-tandem mass spectrometry. J Chromatogr B. 2020;1154:122286.
Salvia M-V, Fieu M, Vulliet E. Determination of tetracycline and fluoroquinolone antibiotics at trace levels in sludge and soil. Appl Environ Soil Sci. 2015;2015:435741.
Białk-Bielińska A, Kumirska J, Borecka M, Caban M, Paszkiewicz M, Pazdro K, et al. Selected analytical challenges in the determination of pharmaceuticals in drinking/marine waters and soil/sediment samples. J Pharm Biomed Anal. 2016;121:271–96.
Huang Y, Cheng M, Li W, Wu L, Chen Y, Luo Y, et al. Simultaneous extraction of four classes of antibiotics in soil, manure and sewage sludge and analysis by liquid chromatography-tandem mass spectrometry with the isotope-labelled internal standard method. Anal Methods. 2013;5(15):3721–31.
Ho Y, Zakaria MP, Latif PA, Saari N. Simultaneous determination of veterinary antibiotics and hormone in broiler manure, soil and manure compost by liquid chromatography–tandem mass spectrometry. J Chromatogr A. 2012;1262:160–8.
Tetzner NF, Maniero MG, Rodrigues-Silva C, Rath S. On-line solid phase extraction-ultra high performance liquid chromatography-tandem mass spectrometry as a powerful technique for the determination of sulfonamide residues in soils. J Chromatogr A. 2016;1452:89–97.
Ezzariai A, Riboul D, Lacroix MZ, Barret M, El Fels L, Merlina G, et al. A pressurized liquid extraction approach followed by standard addition method and UPLC-MS/MS for a fast multiclass determination of antibiotics in a complex matrix. Chemosphere. 2018;211:893–902.
Jacobsen AM, Halling-Sørensen B, Ingerslev F, Hansen SH. Simultaneous extraction of tetracycline, macrolide and sulfonamide antibiotics from agricultural soils using pressurised liquid extraction, followed by solid-phase extraction and liquid chromatography–tandem mass spectrometry. J Chromatogr A. 2004;1038(1-2):157–70.
Yang J, Ying G, Zhao J, Tao R, Su H, Chen F. Simultaneous determination of four classes of antibiotics in sediments of the Pearl Rivers using RRLC–MS/MS. Sci Total Environ. 2010;408(16):3424–32.
Xu M, Qian M, Zhang H, Ma J, Wang J, Wu H. Simultaneous determination of florfenicol with its metabolite based on modified quick, easy, cheap, effective, rugged, and safe sample pretreatment and evaluation of their degradation behavior in agricultural soils. J Sep Sci. 2015;38(2):211–7.
Meng M, He Z, Xu Y, Wang L, Peng Y, Liu X. Simultaneous extraction and determination of antibiotics in soils using a method based on quick, easy, cheap, effective, rugged, and safe extraction and liquid chromatography with tandem mass spectrometry. J Sep Sci. 2017;40(16):3214–20.
Lee YJ, Choi JH, Abd El-Aty A, Chung HS, Lee HS, Kim SW, et al. Development of a single-run analytical method for the detection of ten multiclass emerging contaminants in agricultural soil using an acetate-buffered QuEChERS method coupled with LC–MS/MS. J Sep Sci. 2017;40(2):415–23.
Zhang Y, Li Y, Liu X, Sun Y. Determination of multiple antibiotics in agricultural soil using a quick, easy, cheap, effective, rugged, and safe method coupled with ultra-high performance liquid chromatography-tandem mass spectrometry. J Sep Sci. 2022;45(2):602–13.
Rashid A, Mazhar SH, Zeng Q, Kiki C, Yu C-P, Sun Q. Simultaneous analysis of multiclass antibiotic residues in complex environmental matrices by liquid chromatography with tandem quadrupole mass spectrometry. J Chromatogr B. 2020;1145:122103.
SANTE. 11312/2021, Analytical quality control and method validation procedures for pesticides residues analysis in food and feed, 2021, EU.
Zhang Z, Li X, Ding S, Jiang H, Shen J, Xia X. Multiresidue analysis of sulfonamides, quinolones, and tetracyclines in animal tissues by ultra-high performance liquid chromatography–tandem mass spectrometry. Food Chem. 2016;204:252–62.
Kim S, Carlson K. Quantification of human and veterinary antibiotics in water and sediment using SPE/LC/MS/MS. Anal Bioanal Chem. 2007;387(4):1301–15.
Kim L, Lee D, Cho H, Choi S-D. Review of the QuEChERS method for the analysis of organic pollutants: Persistent organic pollutants, polycyclic aromatic hydrocarbons, and pharmaceuticals. Trends Environ Anal Chem. 2019;22:e00063.
Gómez-Pérez ML, Romero-González R, Vidal JLM, Frenich AG. Identification of transformation products of pesticides and veterinary drugs in food and related matrices: use of retrospective analysis. J Chromatogr A. 2015;1389:133–8.
Lara-Ortega FJ, Robles-Molina J, Brandt S, Schütz A, Gilbert-López B, Molina-Díaz A, et al. Use of dielectric barrier discharge ionization to minimize matrix effects and expand coverage in pesticide residue analysis by liquid chromatography-mass spectrometry. Anal Chim Acta. 2018;1020:76–85.
Raposo F, Barceló D. Challenges and strategies of matrix effects using chromatography-mass spectrometry: an overview from research versus regulatory viewpoints, TrAC. Trends Anal Chem. 2021;134:116068.
He Z, Wang Y, Xu Y, Liu X. Determination of antibiotics in vegetables using QuEChERS-based method and liquid chromatography-quadrupole linear ion trap mass spectrometry. Food Anal Methods. 2018;11(10):2857–64.
Kang DH, Gupta S, Rosen C, Fritz V, Singh A, Chander Y, et al. Antibiotic uptake by vegetable crops from manure-applied soils. J Agric Food Chem. 2013;61(42):9992–10001.
Acknowledgment
This work was supported by the National Key Research and Development Program of China (2019YFC1606501).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests or funding.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 329 kb)
Rights and permissions
About this article
Cite this article
Shi, X., Zhang, S., Zhang, Y. et al. Novel and simple analytical method for simultaneous determination of sulfonamide, quinolone, tetracycline, macrolide, and chloramphenicol antibiotics in soil. Anal Bioanal Chem 414, 6497–6506 (2022). https://doi.org/10.1007/s00216-022-04206-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04206-0