Abstract
We present a novel simple strategy for the detection of Pb2+ based on G-quadruplex DNA and gold nanoparticles. First, gold nanoparticles were chemically adsorbed onto the surface of a thiol-modified gold electrode. Subsequently, the substrate DNA1 was adsorbed onto the surfaces of the gold nanoparticles via thiol–gold bonds, so that the complementary guanine-rich DNA2 could be hybridized to the gold electrode in sequence. [Ru(NH3)6]3+ (RuHex), which can be electrostatically adsorbed onto the anionic phosphate of DNA, served as an electrochemical probe. The presence of Pb2+ can induce DNA2 to form a stable G-quadruplex and fall off the gold electrode. The amount of RuHex remaining on the electrode surface was determined by electrochemical chronocoulometry (CC). The prepared biosensor showed high sensitivity for Pb2+ with a linear range with respect to ln(cPb2+) from 0.01 to 200 nM and a low detection limit of 0.0042 nM under optimal conditions. Because of the high selectivity of the Pb2+-specific DNA2, the designed biosensor also showed low false-positive signal rates with other metal ions in real-world examples. Therefore, this strategy has the potential for practical application in environmental monitoring.

ᅟ
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
With the development of modern agriculture and industry, environmental pollution has become increasingly serious. Contamination with heavy metals has aroused considerable public concern because they present a significant hazard to human health due to their bioaccumulation, toxicity, and low rate of clearance. Among the most toxic of the heavy metal ions is lead, which can cause serious damage to the central nervous system, kidneys, liver, skin, and other organs, even at concentrations measured in parts per million [1,2,3,4,5,6]. According to the U.S. Environmental Protection Agency (EPA), the maximum allowable contamination level of lead ions in drinking water is about 72 nM. Recently, lead contamination of soil, food, and drinking water has generated widespread concern. Natural soil is still the most widely used medium for plant cultivation, and lead ion pollution of cultivated land can be transferred into plants, which can affect food safety and animal health [7, 8]. The detection of trace amounts of lead ions in water and soil is consequently very important.
Traditionally, many detection methods have been used to measure lead ions, including atomic absorption spectrometry, mass spectrometry, atomic emission spectrometry, and inductively coupled plasma mass spectrometry (ICP-MS) [9,10,11,12,13]. These techniques have been successfully used in the laboratory but are not suitable for in-situ determinations, as complex pretreatment steps as well as unwieldy and expensive instruments are required to perform these methods. By contrast, electrochemical determination permits the use of ultrasensitive, portable, and inexpensive electrode-based systems. Chronocoulometry (CC), which was initially employed by Tarlov and coworkers [14], is a more accurate electrochemical method than cyclic voltammetry (CV) for quantifying the density of nucleic acid (DNA) absorbed on the surface of an electrode. Unlike other electrochemical methods, one obvious advantage of CC is that the charge generated by the reaction of species adsorbed on the electrode surface and the electrical double layer charge can be distinguished from the charge generated by the reaction of redox molecules that diffuse to the electrode surface [15]. Therefore, chronocoulometry was selected here to study the amount of [Ru(NH3)6]3+ (RuHex) bound to the anionic phosphate of DNA adsorbed on the surface of the electrode, which changes depending on the lead ion concentration.
Several types of DNA probes have been developed to bind specifically with Pb2+, including “8–17” DNAzyme [16, 17], GR-5 DNAzyme [18], and Pb2+-induced G-quadruplex DNAs [19,20,21]. In recent years, G-quadruplex DNAs have attracted much attention as possible sensors for bioanalytical detection [22, 23]. G-quadruplexes are higher-order structures formed from guanine-rich oligonucleotides through the stacking of planar G-tetrads [24]. There are many π–π interactions in the G-quadruplex structure, which consequently has a strong affinity for many fluorescent dyes. Thus, many fluorescent probes based on G-quadruplex structures have been studied [21, 23]. Furthermore, the presence of stabilizing cations (Ag+, Hg2+, Pb2+, etc.) can induce the formation of stable G-quadruplexes in short guanine-rich DNA sequences [25]. Among these G-rich DNAs, the T30695 molecule, 5′-(GGGT)4-3′, was found to form a stable G-quadruplex in the presence of Pb2+ [19, 26]. According to previous reports [27, 28], compared with other metal ions, Pb2+ is more efficient at stabilizing G-quadruplexes as the resulting M–O and O–O bonds are relatively short [29]. This extra stability of the Pb2+ complexes enables the use of short guanine-rich DNA sequences (T30695) for Pb2+ sensing with high sensitivity and selectivity.
The use of appropriate nanomaterials to modify the surface of an electrode can increase its specific surface area, meaning that its loading capacity for DNA will increase. Moreover, incorporating nanomaterials can improve the detection sensitivities and reduce the detection limits of biosensors based on DNA [30, 31]. Gold nanoparticles were first synthesized in a controllable manner in 1973 [32]. Since then, the method of preparing gold nanoparticles has been developed further, and is now very simple [33, 34]. Gold nanoparticles have many desirable properties, such as good chemical stability, biocompatibility, and electrochemical activity. Thus, they have been widely used in electrochemical biosensors [35,36,37].
In the work reported in the present paper, we designed an electrochemical biosensor for Pb2+ detection based on G-quadruplex DNA and gold nanoparticles. First, we used gold nanoparticles to modify the electrode in order to increase its specific surface area and improve its electronic transport properties. Secondly, the oligonucleotide substrate was chemically absorbed on the gold nanoparticles via thiol–gold bonds. Finally, a short guanine-rich DNA sequence was hybridized with the complementary substrate oligonucleotide. [Ru(NH3)6]3+ (RuHex) was used as an electrochemical probe that can bind to the anionic phosphate of DNA through electrostatic interactions, even at very low ionic strengths [38]. The presence of Pb2+ can induce the formation of stable G-quadruplexes in the short guanine-rich DNA sequence so that it falls off the double-stranded DNA. Consequently, the electrochemical signal decreases with increasing lead ion concentration, which makes it possible to detect Pb2+.
Materials and methods
Reagents
Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), chloroauric acid (HAuCl4·4H2O), hexaammineruthenium(III) chloride (Ru(NH3)6·Cl3), trisodium citrate, 1,2-ethanedithiol, tris(hydroxymethyl) aminomethane (Tris), and metal salts such as Pb(NO3)2 and CuAc2 were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). All reagents were analytical grade and used without further purification. Deionized water (18 MΩ cm) was used throughout the experiments.
The DNA sequences were obtained from GENEWIZ Biotechnology Co. Ltd. (Suzhou, China). The following two oligomers were used in this work: DNA1: 5′-CACCCACCCAC-(CH2)6-SH-3′; DNA2: 5′-GGGTGGGTGGGTGGGT-3′.
Instrumentation
The morphology of the gold nanoparticles was characterized by transmission electron microscopy (TEM; JEM-200CX, JEOL, Tokyo, Japan). For comparison with this sensor, an atomic absorption spectrophotometer (AAS; SP-3803AA, Shanghai Spectrum Instruments, Shanghai, China) with an air–acetylene burner was used to determine Pb2+. The adsorption wavelength was 283.3 nm and the spectral bandwidth was 1.4 nm. All electrochemical measurements, including cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS), and chronocoulometry (CC), were performed using a CHI 660E electrochemical workstation (Shanghai Chenhua Instrument Co. Ltd., Shanghai, China). A conventional three-electrode system comprising a saturated calomel electrode reference electrode, a platinum wire auxiliary electrode, and a AuNPs/dsDNA-modified working electrode was used. Cyclic voltammetry (CV) analysis was conducted using an electrolyte comprising 5 mM Fe(CN)63− and 0.1 M KCl at a scan rate of 100 mV/s in the range from −0.2 V to 0.6 V. In the EIS experiments, the frequency range was 0.01–100 kHz, the signal amplitude was 5 mV, and the electrolyte contained 5 mM Fe(CN)63−/4- and 0.1 M KCl. Chronocoulometry (CC) was performed in 10 mM Tris-acetate (pH 7.4) containing 50 μM [Ru(NH3)6]3+ within the potential range from −0.2 to 0.6 V. The pulse period was 0.25 s for CC.
Preparation of gold nanoparticles
Gold nanoparticles were prepared using the conventional citrate reduction method described previously [33, 34], albeit with the following minor modifications. Before the experiment, all glassware was immersed in aqua regia for 2 h in order to remove reducing substances. First, 100 mL of ultrapure water were added to a round-bottom flask and heated to boiling under constant stirring. Second, 384 μL of HAuCl4 (0.08 M) were added to the solution, after which 3.3 mL of trisodium citrate (38.8 mM, 60 °C) were added rapidly into the boiling solution. The color of the solution changed from colorless to wine red after boiling for another 15 min with constant stirring. After stirring for another 15 min, the solution was cooled to room temperature. The solution was stored at 4 °C until further use. The concentration of the prepared gold nanoparticles was about 2.2 nM, as assessed by spectrophotometry utilizing the Beer–Lambert law.
Fabrication of the electrochemical biosensor
The gold electrode (dp = 2 mm) was first polished to a mirror in an aqueous slurry of alumina particles (1.0, 0.3, and 0.05 mm, sequentially), and was then ultrasonically cleaned in ethanol and deionized water for 3 min each. After that, the gold electrode was dipped into freshly prepared piranha solution (98% H2SO4:30% H2O2 = 3:1 (v/v)) for 10 min, followed by a thorough rinse with deionized water in order to remove organic contaminants. Finally, the gold electrode was activated in 0.5 M H2SO4 by scanning between 0 and 1.6 V at a scan rate of 100 mV/s until a reproducible cyclic voltammogram (CV) was obtained. The pretreated gold electrode was then rinsed with water and dried with nitrogen.
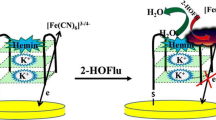
The fabrication steps are shown in Scheme 1. The self-assembled thiol monolayer (SAM) was formed on the surface of the pretreated gold electrode after dipping the electrode into 40 μM 1,2-ethanedithiol for 6 h at room temperature, after which it was rinsed with ethanol and subsequently with deionized water. The prepared SAM/Au electrode was then immersed in a 10 nM gold nanoparticle solution for 16 h at room temperature, during which the gold nanoparticles were immobilized on the surface of the Au electrode through chemical Au–S bonds.
The gold nanoparticle-modified SAM/Au electrode (AuNPs/SAM/Au electrode) was then incubated in a solution composed of 10 μM DNA1, 25 mM Tris-acetate buffer (pH 7.4), and 1 mM TCEP (included to reduce disulfide-bonded oligomers) for 10 h at room temperature. After this, the Au electrode with immobilized DNA1 on its surface (the DNA1/AuNPs/SAM/Au electrode) was hybridized with 15 μM DNA2 in 25 mM Tris-acetate, 0.3 M NaCl, pH 7.4 for 60 min at room temperature. The prepared electrochemical biosensor (the DNA2/DNA1/AuNPs/SAM/Au electrode) was stored in 25 mM Tris-acetate buffer (pH 7.4) at 4 °C until further use.
Results and discussion
Design of the strategy
The electrochemical biosensor for Pb2+ detection was based on G-quadruplex DNA and gold nanoparticles. The sensing mechanism applied in this work is shown in Scheme 1. First, the gold nanoparticles self-immobilized on the surface of a thiol-modified gold electrode. Then the gold nanoparticles were decorated with 3′-terminally thiol-modified DNA1 and its complementary DNA2 (T30695) in sequence. Ruthenium hexamine ([Ru(NH3)6]3+; RuHex for short) can bind to the anionic phosphate of DNA through electrostatic interactions [38]. Thus, RuHex served as an electrochemical probe in this strategy.
DNA2 can form a G-quadruplex structure in the presence of Pb2+ [19], which induces it to separate from double-stranded DNA. Concomitantly, the amount of RuHex adsorbed on the surface of the electrode will also decrease. Thus, the electrochemical signal depends on to the concentration of lead ions.
The results of the characterization of the gold nanoparticles used in this strategy are shown in Fig. 1. The average diameter of the prepared nanoparticles was about 15 nm, and they presented a fairly uniform morphology. Moreover, the characteristic absorption peak of gold nanoparticles at 520 nm was also visible.
Feasibility of the electrochemical biosensor
The feasibility of constructing an electrochemical biosensor based on G-quadruplex DNA utilizing a gold nanoparticle amplification strategy for Pb2+ detection was first verified (see Fig. 2). The cyclic voltammogram curves indicated that the CV signal of the thiol-modified Au electrode was significantly lower than that of the bare Au electrode. This confirmed that a self-assembled thiol monolayer had indeed formed on the surface of the Au electrode, as this monolayer hindered electron transfer. After AuNPs had assembled on the electrode surface, thus promoting electron transfer, the CV signal rose again. When DNA1 and DNA2 were chemically adsorbed on and hybridized to the electrode in sequence, the CV signal decreased again, which indicated that double-stranded DNA was successfully fabricated on the surface of the Au electrode.
The fabrication process of the biosensor was also monitored by electrochemical impedance spectroscopy (EIS). The electrical impedance-equivalent circuit, which is composed of the Warburg impedance (Zw), the charge-transfer resistance (Rct), the solution resistance (Rs), and the double-layer capacitance (Cdl), is shown in the inset of Fig. 3. The diameter of the semicircle is equal to the value of the Rct in the Nyquist plot of impedance spectroscopy. The bare Au electrode had the smallest Rct (Fig. 3a). After the Au electrode was modified with thiol groups, the Rct increased dramatically due to the low electron transfer rate (Fig. 3b). When AuNPs were assembled on the surface, the electron transfer rate increased, so the Rct was reduced (Fig. 3c). Subsequently, the Rct rose again due to the attachment of DNA1 to the Au electrode (Fig. 3d). After the formation of the duplex of DNA1 and DNA2 due to their hybridization on the surface of the biosensor, the Rct increased further (Fig. 3e). Importantly, the EIS measurements were in agreement with the results of the CV measurements. In general, the lower the electron transfer rate, the larger the Rct value.
Optimization of the detection conditions
The pH value, DNA1 concentration, incubation times of DNA1 and DNA2, and Pb2+ reaction time were optimized in order to obtain the best sensor fabrication and detection conditions. As shown in Fig. 4a, the chronocoulometric signals increased from pH 5.0 to 7.4 and then decreased. The maximum charge was observed at pH 7.4. This result was consistent with the theoretical expectations, since a pH of about 7.4 is optimal for many biological systems, such as human blood. We therefore chose pH 7.4 for both fabrication and Pb2+ detection.
In addition, the concentration of DNA1 attached to the electrode surface was also optimized. As shown in Fig. 4b, the chronocoulometric signal increased as the DNA1 concentration increased from 1 to 10 μM. However, there was no obvious change in the chronocoulometric signal when the DNA1 concentration was increased further. This indicated that the binding sites on the surfaces of the gold nanoparticles were saturated. Hence, 10 μM was selected as the optimal DNA1 concentration.
Furthermore, the hybridization time of DNA1 and DNA2 was also optimized. The chronocoulometric signal increased from 30 to 60 min, as shown in Fig. 4c. However, there was no change in the response signal when the incubation time was longer than 60 min due to the saturation of the hybridization sites. We consequently chose 60 min as the optimal incubation time.
Finally, the reaction time of the lead ions with DNA2 was also optimized. In the optimization experiment, the concentration of lead ions was 70 nM. As shown in Fig. 4d, the charge gradually decreased before 30 min, but almost no change was observed after that. At the beginning, Pb2+ induced the formation of the G-quadruplex structure in increasing amounts of DNA2, after which it was released into the electrolyte, taking the absorbed RuHex with it, which led to a decrease in the chronocoulometric signal. However, when the time was longer than 30 min, the reaction was saturated, and there was no change in the electrode’s total charge. Thus, we chose 30 min as the optimal Pb2+ reaction time.
Pb2+ detection sensitivity
Chronocoulometry is an analytical method that is well suited to quantifying the amount of RuHex bound to the anionic phosphate of DNA on the surface of a Au electrode [39]. The charge value (chronocoulometric signal) is proportional to the amount of RuHex adsorbed on the DNA. Increasing the concentration of Pb2+ causes more DNA2 to dissociate into the electrolyte. This in turn will lead to a decrease in the amount of RuHex adsorbed on the surface of the Au electrode. Hence, the charge decreases as the Pb2+ concentration increases (Fig. 5). According to the Cottrell equation, charge is linearly proportional to time raised to the power of 1/2. The intercept of the linear part of each curve in Fig. 5 represents the sum of the Faraday charge and the electrical double-layer charge, which we denoted Q.
This simple experimental process can be explored using the Cottrell equation as follows:
Chronocoulometry (CC) is the integrated form of chronoamperometry:
where n is the number of electrons transferred, F is the Faraday constant, A is the electrode’s surface area, DRuHex is the diffusion coefficient of RuHex in solution, C*RuHex is the concentration of RuHex in solution, Qdl is the electrical double-layer charge, ΓRuHex is the adsorption capacity of RuHex on the electrode surface, and nFAΓRuHex is the charge due to the reaction of RuHex adsorbed onto the electrode surface, that is, the Faraday charge.
Because the concentration of RuHex (50 μM) in the solution does not change, \( \frac{2{nFAD}_{\mathrm{RuHex}}^{1/2}{C}_{\mathrm{RuHex}}^{\ast }{t}^{1/2}}{\pi^{1/2}} \) and Qdl are constants.
Hence, ΔQc = Q0 − Qc = nFAΔΓRuHex(c = 0.01, 0.05, …, 200 nM; Q0 denotes the value of Q without Pb2+).
In Fig. 6, ΔQ equals the D value between Qc and Q0, which means that it represents the change in adsorption capacity of RuHex on the electrode due to the presence of Pb2+. As can be seen from Fig. 6, ΔQ is linearly proportional to ln(cPb2+) in the range 0.01–200 nM. The correlation equation is ΔQ = 0.106ln(cPb2+) + 0.5794, and R2 is 0.997. The detection limit (3σ) is about 0.0042 nM. This LOD value is comparable to those of most other reported Pb2+ sensors (Table 1). According to the EPA, the maximum allowable level of lead ions in drinking water is about 72 nM. Thus, the designed electrochemical sensor is sufficiently sensitive for Pb2+ detection in drinking water applications.
Detection selectivity for Pb2+
In order to determine the selectivity of the sensor, we tested the sensor for interference from a number of common metal ions: K+, Mg2+, Cd2+, Cu2+, Hg2+, Mn2+, and Ca2+. The concentration of Pb2+ was 70 nM while the concentrations of the other ions were all 0.5 μM. Moreover, the potential interference from common anions frequently encountered in environmental samples, i.e., CO32−, HCO3−, SO42−, NO3−, F−, and Cl− (1 μM), was also investigated. As shown in Fig. 7, no obvious ΔQ was observed in the presence of the other ions. This corroborates the findings that G-quadruplex DNA (T30695) can only be efficiently stabilized by Pb2+ due to the resulting relatively short M–O and O–O bonds [27, 28]. Thus, the designed biosensor will not be disturbed by other metal ions and showed good selectivity for Pb2+.
Analysis of water and soil samples
In order to test whether the proposed strategy could be applied in the outside environment, it was used to detect Pb2+ in real water and soil samples. Before detection, the water samples were filtered through a 0.2-μm membrane and then boiled for 10 min in order to remove chlorine.
The soil samples were collected from the top 0–20 cm of topsoil from orchards and farmland in the suburbs of Huai’an in China. Soil samples were collected from six points in each sampling area and pooled. Excess soil was discarded using a quarter-division method and the retained soil samples were heat-digested using the HCl–HNO3–HF–HClO4 four-acids method described previously [44] with minor modifications. After air-drying, grinding, and sifting (100 mesh), 0.300 g of the pretreated soil sample were placed in a polytetrafluoroethylene vessel. After moistening with water, 5 ml of hydrochloric acid (HCl) were added to the vessel, heated on a hot plate at 130 °C for 2 h, and then cooled. Subsequently, 5 mL of nitric acid (HNO3), 4 mL of hydrofluoric acid (HF), and 2 mL of perchloric acid (HClO4) were added and the vessel was capped and heated on a hot plate at 160 °C for 1 h, after which the cap was opened and heating was continued at 180 °C to remove silicon while continuously shaking the vessel. When white smoke was visible, the vessel was capped again and heating was continued at 180 °C for about 1 h until the black residue was fully digested. Finally, the cap was removed and the samples were heated until there was no white smoke and the viscous liquid was colorless. If the black residue was not digested sufficiently, 2 mL of HNO3, 2 mL of HF, and 1 mL of HClO4 were added to the vessel and the above process was repeated until there was no black residue. After cooling the soil digestion solution, the walls of the vessel were washed with distilled water and the solution was transferred into a 25-mL volumetric flask. The soil digestion solution was diluted 100 times with distilled water and the pH value was adjusted to 7.4 with Tris-acetate buffer before detecting lead ions.
For comparison purposes, the concentrations of lead ions in the samples were also measured by atomic absorption spectrophotometry (AAS) in accordance with the national standard method (GB/T7475–1987 and GB/T 23739–2009).
As can be seen in Table 2, the content of Pb2+ in tap water was too low to be detected. The recoveries (biosensor/AAS) from these samples were between 99.1 and 100.8%, and the relative standard deviation (RSD) was acceptable, as shown in Table 2. The results indicated that the proposed strategy has the potential for application to real-world water samples and soil monitoring.
Conclusions
In this work, we designed a novel electrochemical biosensor for Pb2+ detection. We increased the specific surface area and electrical conductivity of the sensor using gold nanoparticles and specific DNA oligonucleotides for the purpose of electrochemical signal amplification. A short guanine-rich DNA (DNA2) was used for the specific recognition of Pb2+. This sensing strategy was demonstrated to have a low detection limit (0.0042 nM) and good selectivity toward Pb2+. Good recoveries from water and soil samples compared to the national standard AAS method indicated that the biosensor has the potential to be utilized in actual environmental monitoring. This strategy could also be applied to construct electrochemical sensors based on ion-specific G-quadruplex DNAs for the detection of other metal ions, such as Hg2+, Ag+, and K+.
References
Lu Y, Li X, Wang G. Tang W. A highly sensitive and selective optical sensor for Pb2+ by using conjugated polymers and label-free oligonucleotides. Biosens Bioelectron. 2013;39(1):231–5.
Needleman H. Lead poisoning. Annu Rev Med. 2004;55(1):209–22.
Godwin HA. The biological chemistry of lead. Curr Opin Chem Biol. 2001;5(2):223–7.
Liu S, Wei W, Sun X, Wang L. Ultrasensitive electrochemical DNAzyme sensor for lead ion based on cleavage-induced template-independent polymerization and alkaline phosphatase amplification. Biosens Bioelectron. 2016;83:33–8.
Schneider E, Clark DS. Cytochrome P450 (CYP) enzymes and the development of CYP biosensors. Biosens Bioelectron. 2013;39(1):1–13.
Dalavoy TS, Wernette DP, Gong M, Sweedler JV, Lu Y, Flachsbart BR, et al. Immobilization of DNAzyme catalytic beacons on PMMA for Pb2+ detection. Lab Chip. 2008;8(5):786–93.
Kothe EA. Bio-geo interactions in metal-contaminated soils. Berlin: Springer; 2012.
Hou Q, Ma A, Wang T, Lin J, Wang H, Du B, et al. Detection of bioavailable cadmium, lead, and arsenic in polluted soil by tailored multiple Escherichia coli whole-cell sensor set. Anal Bioanal Chem. 2015;407(22):6865–71.
Yang L, Saavedra SS. Chemical sensing using sol-gel derived planar waveguides and Indicator phases. Anal Chem. 1995;67(8):1307–14.
Liu J, Chen H, Mao X, Jin X. Determination of trace copper, lead, cadmium, and Iron in environmental and biological samples by flame atomic absorption spectrometry coupled to flow injection on-line coprecipitation preconcentration using DDTC-nickel as coprecipitate carrier. Int J Environ Anal Chem. 2000;76(4):267–82.
Elfering H, Andersson JT, Poll KG. Determination of organic lead in soils and waters by hydride generation inductively coupled plasma atomic emission spectrometry. Analyst 1998;123(4):669–674.
Rebôcho J, Carvalho ML, Marques AF, Ferreira FR, Chettle DR. Lead post-mortem intake in human bones of ancient populations by 109Cd-based X-ray fluorescence and EDXRF. Talanta. 2006;70(5):957–61.
Butler OT, Cook JM, Harrington CF, Hill SJ, Rieuwerts J, Miles DL. Atomic spectrometry update. Environmental analysis. J Anal Atom Spectrrom. 2006;21(2):217–43.
Steel AB, Herne TM, Tarlov MJ. Electrochemical quantitation of DNA immobilized on gold. Anal Chem. 1998;70(22):4670–7.
Yao B, Liu Y, Tabata M, Zhu H, Miyahara Y. Sensitive detection of microRNA by chronocoulometry and rolling circle amplification on a gold electrode. Chem Commun. 2014;50(68):9704–6.
Wu Y-F, Cai Z-M, Wu G-H, Rong M-C, Jiang Y-q, Yang C-YJ, et al. A novel signal-on DNAzyme-based electrochemiluminescence, sensor for Pb2+. Sens Actuator B-Chem. 2014;191:60–6.
Tang S, Lu W, Gu F, Tong P, Yan Z, Zhang L. A novel electrochemical sensor for lead ion based on cascade DNA and quantum dots amplification. Electrochim Acta. 2014;134:1–7.
Zhao X-H, Kong R-M, Zhang X-B, Meng H-M, Liu W-N, Tan W, et al. Graphene–DNAzyme based biosensor for amplified fluorescence “turn-on” detection of Pb2+ with a high selectivity. Anal Chem. 2011;83(13):5062–6.
Zhu Y, Zeng G-M, Zhang Y, Tang L, Chen J, Cheng M, et al. Highly sensitive electrochemical sensor using a MWCNTs/GNPs-modified electrode for lead(II) detection based on Pb2+-induced G-rich DNA conformation. Analyst. 2014;139(19):5014–20.
Wang H, Wang DM, Huang CZ. Highly sensitive chemiluminescent detection of lead ion based on its displacement of potassium in G-quadruplex DNAzyme. Analyst. 2015;140(16):5742–7.
Bhasikuttan AC, Mohanty J. Targeting G-quadruplex structures with extrinsic fluorogenic dyes: promising fluorescence sensors. Chem Commun. 2015;51(36):7581–97.
Ge J, Li X-P, Jiang J-H, Yu R-Q. A highly sensitive label-free sensor for mercury ion (Hg2+) by inhibiting thioflavin T as DNA G-quadruplexes fluorescent inducer. Talanta. 2014;122:85–90.
Xu L, Shen X, Hong S, Wang J, Zhang Y, Wang H, et al. Turn-on and label-free fluorescence detection of lead ions based on target-induced G-quadruplex formation. Chem Commun. 2015;51(38):8165–8.
Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364.
Doluca O, Withers JM, Filichev VV. Molecular engineering of guanine-rich sequences: Z-DNA, DNA triplexes, and G-quadruplexes. Chem Rev. 2013;113(5):3044–83.
Liedl T, Simmel FC. Switching the conformation of a DNA molecule with a chemical oscillator. Nano Lett. 2005;5(10):1894–8.
Kotch FW, Fettinger JC, Davis JT. A lead-filled G-quadruplex: insight into the G-quartet's selectivity for Pb2+ over K+. Org Lett. 2000;2(21):3277–80.
Li T, Dong S, Wang EA. Lead(II)-driven DNA molecular device for turn-on fluorescence detection of lead(II) ion with high selectivity and sensitivity. J Am Chem Soc. 2010;132(38):13156–7.
Guo L, Nie D, Qiu C, Zheng Q, Wu H, Ye P, et al. Chen G. A G-quadruplex based label-free fluorescent biosensor for lead ion. Biosens Bioelectron. 2012;35(1):123–7.
Gumpu MB, Krishnan UM, Rayappan JBB. Design and development of amperometric biosensor for the detection of lead and mercury ions in water matrix—a permeability approach. Anal Bioanal Chem. 2017;409(17):4257–66.
Dönmez KB, Çetinkaya E, Deveci S, Karadağ S, Şahin Y, Doğu M. Preparation of electrochemically treated nanoporous pencil-graphite electrodes for the simultaneous determination of Pb and Cd in water samples. Anal Bioanal Chem. 2017;409(20):4827–37.
Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nature Phys Sci. 1973;241:20.
Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc. 1998;120(9):1959–64.
Wang J, Meng W, Zheng X, Liu S, Li G. Combination of aptamer with gold nanoparticles for electrochemical signal amplification: application to sensitive detection of platelet-derived growth factor. Biosens Bioelectron. 2009;24(6):1598–602.
Yang Y, Yuan Z, Liu X-P, Liu Q, Mao C-J, Niu H-L, et al. Electrochemical biosensor for Ni2+ detection based on a DNAzyme-CdSe nanocomposite. Biosens Bioelectron. 2016;77:13–8.
Ayesh AI, Karam Z, Awwad F, Meetani MA. Conductometric graphene sensors decorated with nanoclusters for selective detection of Hg2+ traces in water. Sens Actuator B-Chem. 2015;221:201–6.
Xu X, Duan G, Li Y, Liu G, Wang J, Zhang H, et al. Fabrication of gold nanoparticles by laser ablation in liquid and their application for simultaneous electrochemical detection of Cd2+, Pb2+, Cu2+, Hg2+. ACS Appl Mater Inter. 2014;6(1):65–71.
Ho PS, Frederick CA, Saal D, Wang AHJ, Rich A. The interactions of ruthenium hexaammine with Z-DNA: crystal structure of a Ru(NH3)6 3+ salt of d(CGCGCG) at 1.2 Å resolution. J Biomol Struct Dyn. 1987;4(4):521–34.
Yang X, Xu J, Tang X, Liu H. Tian D. A novel electrochemical DNAzyme sensor for the amplified detection of Pb2+ ions. Chem Commun. 2010;46(18):3107–9.
Zhan F, Gao F, Wang X, Xie L, Gao F, Wang Q. Determination of lead(II) by adsorptive stripping voltammetry using a glassy carbon electrode modified with β-cyclodextrin and chemically reduced graphene oxide composite. Microchim Acta. 2016;183(3):1169–76.
Yang D, Wang L, Chen Z, Megharaj M, Naidu R. Anodic stripping voltammetric determination of traces of Pb(II) and Cd(II) using a glassy carbon electrode modified with bismuth nanoparticles. Microchim Acta. 2014;181(11):1199–206.
Mehta VN, Solanki JN, Kailasa SK. Selective visual detection of Pb(II) ion via gold nanoparticles coated with a dithiocarbamate-modified 4′-aminobenzo-18-crown-6. Microchim Acta. 2014;181(15):1905–15.
Cai F, Wang Q, Chen X, Qiu W, Zhan F, Gao F, et al. Selective binding of Pb2+ with manganese-terephthalic acid MOF/SWCNTs: theoretical modeling, experimental study and electroanalytical application. Biosens Bioelectron. 2017;98:310–6.
Nogueira TAR, Jose de MW, Oliveira LR, Fonseca IM, Peruca de MGM, Marcussi SA, et al. Nickel in soil and maize plants grown on an oxisol treated over a long time with sewage sludge. Chem Spec Bioavailab. 2009;21(3):165–73.
Acknowledgements
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20131406, BK20130917), the Six Talent Peaks Project in Jiangsu Province (2015-JY-009), the State Key Laboratory of Bio-organic and Natural Products Chemistry, CAS (SKLBNPC15429), and the National Science Foundation for Young Scholars of China (21225626, 21506101).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Xu, S., Chen, X., Peng, G. et al. An electrochemical biosensor for the detection of Pb2+ based on G-quadruplex DNA and gold nanoparticles. Anal Bioanal Chem 410, 5879–5887 (2018). https://doi.org/10.1007/s00216-018-1204-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1204-6