Abstract
Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases. The main functions of these metalloproteinases are the degradation of the stromal connective tissue and basement membrane components. Recent data from model systems suggest that MMPs are involved in breast cancer (BC) initiation, invasion, and metastasis. Particularly, MMP-11 (stromelysin-3) is expressed in stromal fibroblasts adjacent to epithelial tumor cells, and high levels of this metalloproteinase were associated with tumor progression and poor prognosis of BC. Consequently, MMP-11 involved in these processes can be a candidate as a new potential prognostic biomarker in BC. Bioimaging techniques based on laser ablation/desorption and mass spectrometry are rapidly growing in biology and medicine for studies of biological systems to provide information of biomolecules (such as proteins, metabolites, and lipids) and metals with lateral resolution at the micrometer scale. In this study, matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) has been used for the first time to investigate the distribution of MMP-11 in human breast cancer tissues in order to show a possible correlation between cancerous and healthy samples, by differential proteomics and using such differences for possible cancer diagnosis and/or prognosis. Additionally, those human breast tissue samples were analyzed in parallel by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) in order to gather additional information about the elemental distribution of Zn and its possible associations with MMPs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is one of the leading causes of cancer death among women worldwide. Clinical parameters, such as the size of the primary tumor, the histological grade, classification, and regional lymph node involvement, are generally useful for prognosis [1]. The early detection of the disease is vital for a successful patient treatment and supervision. In this sense, the profile of biomarkers may provide a valuable insight into the underlying mechanisms of disease progression, thus aiding in understanding the prediction, cause, diagnosis, progression, regression, and outcome of disease treatments [2]. Thus, detection of cancer characteristic biomarkers in tissue sections is essential for the identification and classification of tumor cells and for the choice of the therapy. In the case of breast cancer, estrogen receptor (ER), progesterone receptor (PR), and oncogene Her-2/neu [3, 4] have been investigated as possible biomarkers.
The number of studies on application and better understanding of traditional and new breast cancer biomarkers, and prognostic factors, is continuously growing, and they might allow the early identification of individuals with a high risk of breast cancer [5].
During the last years, breast cancer research has been focused on the study of concentration levels of trace elements naturally present in breast tissues. Malignant breast tumor presents high levels of trace elements, so a change in trace element levels in tissue might be a useful and significant new tool for cancer prevention, diagnosis, and treatment [6]. In this vein, essential elements such as Ca, Fe, Cu, and Zn are believed to play key roles, since they are responsible for a great number of metabolic and biological processes. In fact, related to the increased metabolism rates on tumors, trace elements have been shown to appear in altered concentrations in cancer tissues [7]. Therefore, the aforementioned concentration levels of elements could be used as potential breast cancer biomarkers.
The role of trace elements in breast cancer is complex, since many types of molecules, cells, and tissues are affected [8]. Zinc, in particular, is a trace element which is vital for numerous cellular processes, particularly the cell growth, playing an important role in cancer etiology and outcome. Moreover, Zn is a cofactor of the extracellular matrix metalloproteinases (MMPs), a family of 28 zinc-dependent endopeptidases, which are involved in the degradation of the extracellular matrix, facilitating the breakdown of the basal membrane and/or matrix connective tissue components [9]. This mechanism is associated with a facilitated proliferation, and therefore, the stage of tumor progression is positively correlated with the expression of MMPs in many cases. Eiró et al. and Vizoso et al. identified subgroups of tumors showing a high molecular profile of MMP/TIMP expression by stromal cells, both at the tumor center and at the invasive front, which were strongly associated with a higher prevalence of distant metastasis [10, 11]. Particularly, MMP-11 expression by mononuclear inflammatory cells (MICs) coupled with the expression of pro-inflammatory proteins support tumor escape and invasion, hence promoting metastasis [12]. Thus, MMP-11 expression levels should be included into traditional classification schemes to provide new prognostic and predictive tools to clinicians [13]. Therefore, the combination of epidemiological, analytical, and biological studies becomes increasingly relevant for the investigation of the role of Zn and/or MMP-11 in the promotion and/or maintenance of the tumor.
On the other hand, the application of bioimaging techniques based on laser ablation or desorption coupled to MS is rapidly growing in biology and medicine to provide information of key biomolecules (such as proteins, metabolites, and lipids) [14,15,16,17] and metals [18,19,20] with lateral resolution at the micrometer scale. Such techniques constitute today an important potential tool to study the differential distribution of metals (e.g., Zn) and proteins (e.g., MMP-11) in healthy and breast cancer tissue samples. Therefore, valuable complementary information regarding the role, uptake, transport, and storage of Zn and MMP-11 and their associations with irregular protein abnormalities can be obtained using a combination of elemental and molecular MS-based imaging techniques.
In this work, two complementary molecular and elemental imaging methods, combining spatially resolved information from matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) and from laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS), were developed and applied to human breast tissue samples. The observed elemental and molecular spatial distributions are correlated, and they are compared with the histological aspects observed for the same breast tissues in an attempt to investigate possible biological implications derived from results coming from a combination of elemental and molecular imaging information.
Experimental
Materials and reagents
All chemicals were used in the highest quality available. Water was purified by an Aquatron Water Still purification system model A4000D (Barloworld Scientific, Nemours Cedex, France). Rhodium standard solution (1000 mg L−1) was purchased from SCP Science (Baie-D’Urfé, Canada). Trypsin used for tryptic digestion and α-cyano-4-hydroxycinnamic acid (CHCA) used as a matrix were purchased from Sigma-Aldrich (St. Louis, MO, USA). All gases used for ICP-MS and LA analysis (argon and helium, each 99.999% purity) were obtained from Westfalen AG (Münster, Germany).
Patients’ characteristics and tissue sectioning
This study comprised four women with a histologically confirmed diagnosis of early invasive breast cancer of a ductal type (histological grade 2). Figure 1 shows histological images from the healthy and tumor tissue samples. In the tumoral tissue (Fig. 1b), cancer cells can be distinguished from stromal cells. Cancer cells are large cells with large nuclei arranged forming either an acinar or trabecular pattern, whereas stromal cells are spread. In the normal tissue (Fig. 1a), no large cell with large nuclei compatible with cancer cells can be observed.
Frozen human breast tissue samples (non-tumoral and tumoral, from the same patients) were provided by the Biobanco Central de Asturias (Asturias, Spain) and were stored at −20 °C prior to sectioning. Tissue without tumor corresponds to surgical margins (at least 2 cm away from the tumor). Healthy and tumor sections (20 μm thickness) were prepared using a Cryotome (CryoStar NX70, Thermo Scientific, Bremen, Germany) held at −20 °C. Tissue slices were thaw mounted onto glass slides for LA-ICP-MS measurements, and for MALDI-MSI measurements, conductive indium tin oxide (ITO) glass slides (Sigma-Aldrich) were used. Parallel sections from the same tissue sample were analyzed by LA-ICP-MS and MALDI-MS techniques for eventual comparisons of the images observed.
In situ tryptic digestion
A tryptic digestion was done directly onto the tissue. As a result, the identification and distribution of this protein had to be achieved by its peptide fingerprint.
Different tryptic digestion protocols were investigated [21,22,23,24], and the parameters to be optimized were the following and are summarized in Table 1: enzyme concentration (0.1–1 g L−1 trypsin), solvent (ammonium bicarbonate or water), incubation time (overnight, 24 h), as well as the temperature at which the enzymatic reaction would take place (room temperature or 37 °C). Finally, the following conditions turned out to possess optimal properties, in terms of better protein fragmentation, getting the signals of corresponding peptides and higher intensities: 1 g L−1 trypsin (Sigma-Aldrich) in bi-distilled water was sprayed three times using an airbrush (Harder & Steenbeck, Germany) (20 psi) onto the tissue and then incubated at 37 °C for 24 h. Between the consecutive sprays, the tissue was completely dried.
MALDI matrix deposition
The MALDI matrix was applied to the digested samples by sublimation onto the sample. In this case, CHCA appeared to have optimal properties to ionize target peptides and was therefore selected in all experiments as a matrix. The matrix was applied using an iMLayer (Shimadzu, Kyoto, Japan) (600 mg CHCA, 20 min, 250 °C, 5 × 10−2 Pa). This matrix vapor deposition system facilities a controlled matrix deposition with adjusted homogeneous thickness for reproducible sample preparation.
Instrumentation
LA-ICP-MS experiments were carried out using a CETAC LSX-213 laser ablation system (Teledyne CETAC Technologies, Omaha, NE, USA) working at a wavelength of 213 nm and were hyphenated to an Agilent 7500ce ICP-MS (Agilent Technologies, Santa Clara, USA). Helium was used as the carrier gas. The LA-ICP-MS system was tuned for maximum sensitivity prior to each experiment using the reference material NIST 612 trace elements in glass. The ICP-MS was also tuned to minimize the formation of oxides by monitoring the oxide ratio (232Th16O+/232Th+, m/z 248:232) as well as the doubly charge ratio (137Ba++/137Ba+, m/z 68.5:137.0). A value of less than 1% for these ratios indicated that the derived signals from potential matrix-based polyatomic interferences were negligible. Furthermore, natural isotopic abundance ratios were monitored to confirm the absence of interfering polyatomic species.
Imaging measurements were performed by multiple line scan ablation of frozen breast tissue sections (line per line) using a spot diameter of 25 μm and no space distance between adjacent lines. A rhodium standard, introduced via liquid sample introduction and merged with the aerosol from the ablation chamber, was used as the internal standard in order to compensate plasma fluctuations and surveil plasma stability. Typical operational parameters are summarized in Table 2.
MALDI-MSI experiments were performed using an iMScope TRIO (Shimadzu, Kyoto, Japan), consisting of a mass microscope equipped with an atmospheric pressure ion-source chamber for matrix-assisted laser desorption/ionization (AP-MALDI) and a quadrupole ion trap time-of-flight (QIT-TOF) mass analyzer. The optical microscope combined with the mass spectrometer permitted the precise determination of relevant tissue region prior to performing mass spectrometry imaging (MSI). The instrument is also equipped with an ultraviolet laser (355 nm) that allows to achieve high spatial resolution down to 5 μm.
Tissue sections were analyzed in the positive ion mode. The optimized operative conditions of the instrument are listed below and were used for all subsequent measurements of the healthy as well as the tumor tissue samples: mass range = m/z 600–1600, shots = 100, accumulation = 1, sample voltage = 3.5 kV, detector voltage = 1.90 kV, laser spot size = 25 μm, and pixels = 50 μm × 50 μm. For 2D MSI analysis and overlay of images, data were visualized using Imaging MS Solution software (Shimadzu).
MALDI-MSI data were searched using the MASCOT software against the SwissProt database and were based on the MOWSE algorithm. Within the MASCOT search engine, the enzyme selected was trypsin and experimental ion peptide tolerance was set to ±1 Da. The criteria also included that one missed cleavage was allowed in order to account for incomplete digestion.
Results and discussion
Elemental distribution of Zn by LA-ICP-MS
Human breast tissue samples were analyzed by LA-ICP-MS in order to provide information about possible differential spatial distribution of Zn in healthy and tumor samples. Breast samples were prepared as described before and ablated line per line using a focused laser beam. Figure 2 shows the observed Zn distributions of healthy and cancer tissue samples of a selected patient. A rather homogeneous Zn distribution was observed in the healthy tissue (Fig. 2a). In contrast, Fig. 2b shows that Zn distribution in the tumor sample presents hot spots where the amount of Zn is higher than that in the rest of the analyzed tissue. The recent work done in our laboratory [25] demonstrated that not only do normal and malignant tissue types differ in zinc concentration but also within individual patients, and cancerous breast cells tend to accumulate four times more than non-cancerous breast cells (17.8 ± 3.4 and 4.7 ± 1.2 μg g−1, respectively). It should be stressed here that increased Zn levels were also found and reported in other studies using X-ray analytical techniques [26, 27]. Several comparative studies have provided evidence that altered zinc homeostasis occurs in breast cancer. In vivo studies showed that the growth of mammary carcinomas is suppressed following zinc depletion in humans [28]. Cellular zinc status profoundly affects cellular health and has numerous roles in cancer initiation, progression, termination, and potentially, prevention. The zinc acquisition of breast cancer suggests the potential for this mineral’s involvement, and perhaps requirement, for breast malignancy. The interaction between zinc status, zinc transporter expression, and immune function is interwoven with cellular signaling pathways providing multiple mechanisms for the role of zinc in cancer. Although the role of Zn is not yet well clarified in the literature, the observed increased Zn levels in tumor tissues may be associated with structural and metabolic aspects derived from tumor development. These changes in the Zn levels, associated to cancer, have the potential to provide a novel early biomarker for breast cancer.
Distribution of MMP-11 by means of MALDI-MSI
Using the optimized conditions described in the “Experimental” section, the differential protein analysis of healthy and tumor breast tissue samples was carried out by means of MALDI-MSI. Mass spectra from healthy and tumor tissue samples were obtained directly from the in situ tryptically digested breast tissue sections (see Fig. 3) under study. Figure 3a shows the obtained spectra of a healthy breast tissue sample. Since a tryptic digestion was performed, protein identification can be carried out through the peptide fingerprint using a database. A comparatively high number of MMP-11-specific peptides were experimentally observed, as compared with those theoretically expected (from the database) for that protein. Figure 3b shows the spectra obtained by MALDI-MSI for the tumor tissue (the analysis of the tumor sample was conducted under identical conditions). The peptides that belong to the protein of interest (MMP-11) are selected and then introduced in the MASCOT software to assign the found peptide fragments to the target protein.
Comparing the two obtained mass spectra from healthy and tumor samples, it can be seen in Fig. 3 that the tumor sample presents more fragmentation as well as higher signal intensities. This can be due to the higher abundances of the corresponding proteins in the tumor tissue. Table 3 shows the theoretical and the observed masses of the peptides belonging to the digested MMP-11, which were detected in both healthy and tumor tissue samples (21 and 25 peptides, respectively). These peptides covered the 18 and 20% of the total amino acid sequence of MMP-11 found in healthy and tumor tissue samples, respectively. As can be seen in the case of tumor tissue (third column in Table 3), higher correlations between observed and theoretical peptides (from the database) were achieved, as well as higher protein sequence coverage. The protein coverage visualizes the mapping of supporting peptides to the target protein.
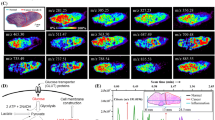
Two parallel sections (1 and 2) from the tumor tissue were then analyzed, and the results were converted into images. The found m/z and their respective intensities were measured over the entire sample and were assigned in a heat map (see Fig. 4). Figure 4a (1 and 2) shows the distribution of protonated [M+H]+ ion derived from the proteolytic fragments of MMP-11 at m/z 768.5. As can be seen, in both sections, the highly abundant MMP-11 peptides were detected predominantly in one specific area of the tumor sample. Moreover, the reproducibility of the sample treatment and the MALDI technique is demonstrated. Also that, although several parallel cuts are made in the tissue, the tumor preserves its structure.
MALDI images generated, from two parallel sections of a tumor tissue, from a found characteristic peptide of the MMP-11 (m/z 768.5 (a), 1 and 2). MALDI images from a tumor tissue, generated from three specific peptide signals belonging to MMP-11 (m/z 768.5, 786.6, and 822.5 (b), 1) and overlay signals of these peptides from MMP-11 (m/z 768.5 + 789.6 + 822.5 (b), 2)
The distributions of three protonated ions characteristic from the MMP-11 are shown in Fig. 4b as well as the corresponding overlapping image of these three MMP-11 peptides at m/z 768.5 + 789.6 + 822.5. As can be seen, all MMP-11 peptides appeared gathering in a specific zone, indicating an increased abundance of this protein in the tumor region. This study shows that the level of MMP-11 was significantly higher in breast cancer tissue than in healthy one. These results are consistent with reports indicating that MMPs are overexpressed in breast cancer and suggesting that the detection of concentrations of MMP proteins might be useful in distinguishing between benign and malignant breast diseases, as they could be associated with pathologic parameters in breast cancer patients [29, 30]. So, MMP-11 might become a potential prognostic marker as well as a therapeutic target for BC patients.
Comparative elemental and molecular results
The observed elevated Zn levels can be associated with an overexpression of the MMP-11 (see Fig. 5), but of course, they may be due to other Zn-containing enzymes/proteins in the investigated cancer tissue.
Comparative images obtained from the same frozen breast tumor tissue by LA-ICP-MS (Zn distribution) and MALDI-MSI (MMP-11 distribution) techniques and the overlay image obtained from the Zn and MMP-11 images (red signal corresponds to Zn distribution, green signal corresponds to MMP-11 distribution, and the correlation between them is shown in yellow)
In any case, the results from the two complementary (elemental and molecular) imaging techniques tend to confirm that the occurrence of Zn and MMP-11 is correlated: this can be observed in Fig. 5, where the distribution of Zn within the tumor section obtained from LA-ICP-MS matched well with the localization of the MMP-11 peptide signals obtained by MALDI-MS analysis for the two parallel sections of the analyzed breast cancer tissue. In other words, hot spots of Zn occur in the area where maximum intensity of MMP-11 is detected. Thus, complementary imaging information (obtained by elemental and molecular MS techniques of the same samples) has proved useful to correlate Zn levels in the tissue with overexpression of MMP-11, even if the selectivity of measuring MMP-11 via elemental Zn levels by ICP-MS is still debatable.
Conclusions
In this study, a methodology for the analysis of the potential tumor biomarker MMP-11 in tissue samples was developed (it has been shown that overexpression of this protein is associated with the occurrence of tumor). Initially, different in situ trypsin digestion protocols were applied and optimized (to select the better conditions) to be then applied to in situ digestion of the MMP-11 protein in healthy and tumor breast tissue samples. It has to be emphasized that the methodology used here of a spraying system for the in situ application of the digestion solution enables to maintain the integrity of the tissue as well as the native distribution of the proteins. The recorded m/z was finally compared with theoretical data for the targeted protein from the database. By this in situ bottom-up approach, it was possible to assign with spatial resolution the observed specific peptides to their original protein MMP-11. This proceeding is particularly interesting when certain proteins are not easily accessible by conventional MALDI-MSI (e.g., high mass proteins). It was demonstrated that MMP-11 was not homogeneously distributed and showed higher abundances in specific areas of the investigated tissue. Furthermore, higher MMP-11 levels were found in the tumor tissue compared to the healthy one, indicating an altered metabolism and so pointing to MMP-11 as a possible candidate biomarker for breast cancer. Since the protein MMP-11 contains Zn, elemental mapping of the metal by means of LA-ICP-MS was attempted in parallel and images results were compared eventually to those from MALDI-MSI. Images in Fig. 5 show that Zn distribution correlates with the observed abundance of the target protein, MMP-11, indicating that Zn measurement could offer an indirect, more robust, and sensitive method for MMP-11 determination [31].
Yet, it should be emphasized that elemental imaging via ICP-MS alone might not be sufficient to trace the MMP-11 distribution, since many different enzymes (and metabolic processes) are reliant upon Zn as a cofactor and the required specificity for the Zn-MMP-11 cannot be secured by direct LA-ICP-MS measurements. However, the observed correlation between LA-ICP-MS and MALDI-MSI images in Fig. 5 warrants further investigations in the search for in situ combined elemental and molecular information of breast cancer tissues. Moreover, by resorting to in situ labeling of MMP-11 (e.g., using bi-functional reagents containing the protein selective antibody bound to metal nanoparticles [32]), extremely sensitive and selective ICP-MS imaging methods can be envisaged for MMP-11.
References
Weigelt B, Peterse JL, Van’t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–02.
Kang HS, Lee SC, Park YS, et al. Protein and lipid MALDI profiles classify breast cancers according to the intrinsic subtype. BMC Cancer. 2011;11:465–74.
Kinsella MD, Nassar A, Siddiqui MT, Cohen C. Estrogen receptor (ER), progesterone receptor (PR), and HER2 expression pre- and post- neoadjuvant chemotherapy in primary breast carcinoma: a single institutional experience. Int J Clin Exp Pathol. 2012;5:530–6.
ASY L, Zhuang Z. The changing role of pathology in breast cancer diagnosis and treatment. Pathobiology. 2011;78:99–114.
Yersal O, Barutca S. Biological subtypes of breast cancer: prognostic and therapeutic implications. World J Clin Oncol. 2014;5:412–24.
Silva MP, Soave DF, Ribeiro-Silva A, Poletti ME. Trace elements as tumor biomarkers and prognostic factors in breast cancer: a study through energy dispersive x-ray fluorescence. BMC Res Note. 2012;5:194–204.
Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–90.
Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–77.
Meyer BS, Rademann J. Extra- and intracellular imaging of human matrix metalloprotease 11 (hMMP-11) with a cell-penetrating FRET substrate. J Biol Chem. 2012;287:37857–67.
Eiró N, Fernandez-Garcia B, Vázquez J, Del Casar JM, González LO, Vizoso FJ. A phenotype from tumor stroma based on the expression of metalloproteases and their inhibitors, associated with prognosis in breast cancer. Oncoimmunology. 2015;4:e992222.
Vizoso FJ, González LO, Corte MD, Rodriguez JC, Vázquez J, Lamelas ML, Junquera S, Merino AM, García-Muñiz JL. Study of matrix metalloproteinases and their inhibitors in breast cancer. Br J Cancer. 2007;96:906–11.
Eiró N, Fernández-García B, González LO, Vizoso FJ. Cytokines related to MMP-11 expression by inflammatory cells and breast cancer metastasis. OncoImmunology. 2013;2:e24010.
Buache E, Thai R, Wendling C, Alpy F, Page A, Chenard MP, Dive V, Ruff M, Dejaegere A, Tomasetto C, Rio MC. Functional relationship between matrix metalloproteinase-11 and matrix metalloproteinase-14. Cancer Med. 2014;3:1197–210.
Römpp A, Schäfer KC, Guenther S, Wang Z, Köstler M, Leisner A, Paschke C, Schramm T, Spengler B. High-resolution atmospheric pressure infrared laser desorption/ionization mass spectrometry imaging of biological tissue. Anal Bioanal Chem. 2013;405:6959–68.
Wang HAO, Grolimund D, Giesen C, Borca CN, Shaw-Stewart JRH, Bodenmiller B, Günther D. Fast chemical imaging at high spatial resolution by laser ablation inductively coupled plasma mass spectrometry. Anal Chem. 2016;85:10107–16.
Giesen C, Mairinger T, Khoury L, Waentig L, Jakubowski N, Panne U. Multiplexed immunohistochemical detection of tumor markers in breast cancer tissue using laser ablation inductively coupled plasma mass spectrometry. Anal Chem. 2011;83:8177–83.
Niehoff AC, Kettling H, Pirkl A, Chiang YN, Dreisewerd K, Yew JY. Analysis of drosophila lipids by matrix-assisted laser desorption/ionization mass spectrometric imaging. Anal Chem. 2014;86:11086–92.
Becker JS, Matusch A, Wu B. Bioimaging mass spectrometry of trace elements—recent advance and applications of LA-ICP-MS: a review. Anal Chim Acta. 2014;835:1–18.
Konz I, Fernández B, Fernández ML, Pereiro R, González-Iglesias H, Coca-Prados M, Sanz-Medel A. Quantitative bioimaging of trace elements in the human lens by LA-ICP-MS. Anal Bioanal Chem. 2014;406:2343–8.
Bishop DP, Clases D, Fryer F, Williams E, Wilkins S, Hare DJ, Cole N, Karst U, Doble PA. Elemental bio-imaging using laser ablation-triple quadrupole-ICP-MS. J Anal At Spectrom. 2016;31:197–202.
Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom. 2007;42:254–62.
Blaze MTM, Aydin B, Carlson R, Hanley L. Identification and imaging of peptides and proteins on enterococcus faecalis biofilms by matrix assisted laser desorption ionization mass spectrometry. Analyst. 2012;137:5018–25.
Harris GA, Nicklay JJ, Caprioli RM. Localized in situ hydrogel-mediated protein digestion and extraction technique for on-tissue analysis. Anal Chem. 2013;85:2717–23.
Stauber J, MacAleese L, Franck J, Claude E, Snel M, Kaletas BK, Wiel IM, Wisztorski M, Fournier I, Heeren RM. On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2010;21:338–47.
González de Vega R, Fernández Sanchez ML, Pisonero J, Eiró N, Vizoso FJ, Sanz-Medel A. Quantitative bioimaging of Ca, Fe, Cu and Zn in breast cancer tissues by LA-ICP-MS. J Anal Atomic Spectrom. 2017;32:671–7.
Silva MP, Soave DF, Ribeiro-Silva A, Poletti ME. Trace elements as tumor biomarkers and prognostic factors in breast cancer: a study through energy dispersive x-ray fluorescence. BMC Res Notes. 2012;5:194–204.
Mulware SJ. Comparative trace elemental analysis in cancerous and noncancerous human tissues using PIXE. J. Biophysics 2013;2013:8. doi:10.1155/2013/192026.
Mills BJ, Broghamer WL, Higgins PJ, Lindeman RD. Inhibition of tumor growth by zinc depletion of rats. J Nutr. 1984;114:746–52.
Cid S, Eiro N, González LO, Beridze N, Vazquez J, Vizoso FJ. Expression and clinical significance of metalloproteases and their inhibitors by endothelial cells from invasive breast carcinomas. Clin Breast Cancer. 2016;16:83–91.
Eiró N, Pidal I, Fernandez-Garcia B, Junquera S, Lamelas ML, del Casar JM, González LO, López-Muñiz A, Vizoso FJ. Impact of CD68/(CD3+CD20) ratio at the invasive front of primary tumors on distant metastasis development in breast cancer. PLoS One. 2012;7:e52796.
Sanz-Medel A. “Heteroatom-tagged” quantification of proteins via ICP-MS. Anal Bioanal Chem. 2016;408:5393–5.
Garcia-Cortes M, Encinar JR, Costa-Fernandez JM, Sanz-Medel A. Highly sensitive nanoparticle-based immunoassays with elemental detection: application to prostate-specific antigen quantification. Biosens Bioelectron. 2016;85:128–34.
Acknowledgments
This work was supported by projects FC-15-GRUPIN14-092 (Principado de Asturias) and MINECO-13-CTQ2013-49032-C2-1-R (Ministerio de Educación y Ciencia). Parts of this study were supported by the Cells in Motion Cluster of Excellence (CiM-EXC 1003), Münster, Germany (project FF-2013-17).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The study was performed with approval of the Ethics Committee of Regional Clinical Research of the Principado de Asturias in accordance with the ethical standards. Written informed consents were obtained from all patients according to the institutional regulations of the Biobank of Principado de Asturias.
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Published in the topical collection celebrating ABCs 16th Anniversary.
Rights and permissions
About this article
Cite this article
de Vega, R.G., Sanchez, M.L.F., Eiro, N. et al. Multimodal laser ablation/desorption imaging analysis of Zn and MMP-11 in breast tissues. Anal Bioanal Chem 410, 913–922 (2018). https://doi.org/10.1007/s00216-017-0537-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0537-x