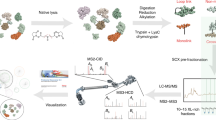
Abstract
Chemical cross-linking combined with mass spectrometry (MS) and computational modeling has evolved as an alternative method to address fundamental questions in structural biology. The constraints revealed by the cross-links yield valuable distance information and allow one to deduce three-dimensional structural information on very large and transient protein complexes. During the past few years, technical advances in the cross-linking/MS approach have been enormous, mainly owing to the fantastic advances in MS technology, and it is easily overlooked that significant progress has been made in the design of novel cross-linking reagents. In this review, the advent of cleavable cross-linking reagents will be highlighted. In particular, gas-phase (MS-) cleavable cross-linkers offer unique properties for an automated, data-dependent assignment of cross-linked products based on the generation of characteristic fragment ion signatures in MS/MS and MS3 spectra. Therefore, MS-cleavable cross-linkers are envisioned to hold the key for proteome-wide applications of the chemical cross-linking/MS approach, not only to delineate the conformation of single proteins but also to decipher protein interaction networks.

Outline of the analytical strategy using cleavable cross-linkers in combination with MS to conduct protein conformational and protein interaction studies
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chemical cross-linking combined with a mass spectrometric analysis of the generated products has emerged as an alternative approach that is able to provide structural insights into proteins [1–5]. The chemical cross-linking/mass spectrometry (MS) approach complements MS-based techniques for studying structure and dynamics of proteins, such as native MS [6, 7] and footprinting methods [8, 9]. In chemical cross-linking, a chemical cross-linker is covalently inserted between two functional groups within a protein or a protein complex to derive three-dimensional structure information. As the cross-linker possesses a defined length, it acts as a kind of “molecular ruler” by connecting functional groups of amino acid side chains, which are then identified by MS and MS/MS analysis. Usually, chemical cross-linking studies are performed in a “bottom-up” fashion, involving an enzymatic digestion of the covalently connected proteins followed by LC/ESI-MS/MS (liquid chromatography–electrospray ionization tandem mass spectrometry) analysis. The positions of the cross-linked amino acids, together with the cross-linker length, yield distance constraints that allow one to deduce 3D structural information about the proteins under investigation, i.e., by computational modeling with Rosetta [10, 11]. The most commonly used cross-linking principles are N-hydroxysuccinimide (NHS) esters that react preferentially with lysine residues. However, there are also some disadvantages associated with targeting lysine residues: The lysine side chains are highly flexible; therefore the structural information gained from these experiments is “soft”, i.e., cross-linkers possessing spacer lengths of ca. 8 Å might bridge Cα–Cα distances of up to ca. 27 Å. Also, hydrophobic protein regions lacking lysines are not susceptible to the cross-linking reaction.
In general, proteolytic digests of proteins cross-linked with NHS esters will contain, besides many unmodified peptides, three types of cross-linker-modified peptides: Cross-links within the same peptide (intrapeptide, type 1 or “loop-link”) or between different peptides (interpeptide, type 2), and peptides modified by a partially hydrolyzed cross-linker (type 0 or “dead-end” cross-link or “mono-link”). Although a systematic nomenclature has been proposed for the different cross-linked products [12], it has not received a widespread acceptance yet.
The increasing importance of the cross-linking/MS approach for 3D structural studies of protein conformation and protein–protein interactions is mainly attributed to the inherent strengths of MS as a sensitive and rapid method for protein analysis. Also, the protein’s conformation and flexibility are accurately reflected as the cross-linking reaction can be executed at physiological pH. The reaction times of cross-linkers range from micro- or milliseconds (for photo-reactive diazirines or benzophenones) to seconds (for amine-reactive NHS esters). Moreover, membrane proteins, post-translationally modified proteins, and splice variants can be studied and transient interactions can be captured. The large number of cross-linking reagents with different specificities covering a range of spacer lengths (“zero-length” up to 20 Å) allows one to adapt the reaction conditions to the respective protein system under investigation.
In the past 5 years, the number of reviews on the cross-linking/MS approach has massively increased, reflecting the rising popularity of the technique for in vitro protein conformational studies as well as for in vivo protein network analyses. As these reports, e.g., [13–19], highlight the latest advances in chemical cross-linking/MS quite comprehensively, I will focus in my contribution on the design and application of selected cleavable—particularly MS-cleavable cross-linkers. Usually, the covalent connection of tryptic peptides via cross-linking results in large precursor ions (m/z > 1500), which resist an efficient fragment ion formation in tandem MS experiments. To overcome this limitation, an MS-cleavable cross-linker should preferably exhibit a labile covalent bond, guaranteeing a favored fragmentation over the peptide backbone during collisional activation. This makes MS-cleavable cross-linkers especially useful for conducting data-dependent analyses of cross-linked products based on the fragment ion signatures in collision-induced dissociation (CID) experiments. MS-cleavable cross-linkers are just about to reveal their full potential for designing fully automated workflows and their availability will most likely advance the cross-linking/MS approach to become more and more a routine technique for structural proteomics and protein interaction analyses.
Cross-linkers cleavable by reduction
One of the first examples that employed chemical cross-linking in conjunction with MS was presented in 2000 where the authors used the cross-linker 3,3′-dithio-bis(sulfosuccinimidyl propionate) (DTSSP; Table 1) that is cleavable by reduction for mapping protein interfaces [20]. When employing such a cross-linker, the protein binding partners are covalently connected, separated by non-reducing SDS-PAGE, and the protein complex is enzymatically digested. Aftwards, comparative MS peptide mapping is performed under reducing and non-reducing conditions (Fig. 1). Peptide maps obtained prior to and after reduction, i.e., cleavage of the cross-linker, are compared and signals that disappear after reduction of the linker are assigned as putative cross-linked peptides. The presence of a specific cross-link is confirmed if one or both halves of the cross-linked peptides are observed in the peptide map after reduction of the cross-linker. This approach impresses by its simplicity as it can also be conducted on low-resolution mass spectrometers.
Analytical strategy for chemical cross-linking with reductive cleavable reagents. The strategy is exemplified for the thiol-cleavable cross-linker DTSSP and differential peptide mapping. Figure adapted from [20]
A heterobifunctional amine-/photo-reactive cross-linker is succinimidyl 2-([4,4′-azipentanamido]ethyl)-1,3′-dithiopropionate (SDAD). While for most structural proteomics applications homobifunctional amine-reactive cross-linkers are applied, heterobifunctional amine-/photo-reactive linkers exhibit distinct advantages. As such, the bait protein can be activated by reaction with the amine-reactive site of the cross-linker (the NHS ester) before potential interaction partners are added, and the formation of a covalent bond is induced by UV irradiation via the photo-reactive site of the linker (the diazirine). SDAD has been successfully applied to study protein–protein interactions of E. coli formate dehydrogenases [21].
A peculiar cross-linker is bis(succinimidyl)-3-azidomethyl glutarate (BAMG) that, upon reduction at its azide group, exhibits CID cleavability (Table 1) [22]. After reaction of proteins with BAMG and proteolytic digestion, two competing pathways will proceed in the presence of a reducing agent: (i) The azide in the spacer of BAMG is reduced to an amine without cleavage of the cross-link, and (ii) the amide bonds between amino acid and linker that have been formed during the cross-linking reaction are cleaved by CID. One of the features of the BAMG linker is that cross-linked peptides can be efficiently separated from the bulk of unmodified peptides by reversed-phase diagonal chromatography as their retention times are quite different. This analytical strategy has been coined “identification of cleavable cross-links by diagonal chromatography”. BAMG has been described for identifying cross-links in HeLa cell lysates [23]; however, as a result of the existence of two competing pathways, a more general application of BAMG for mapping protein interaction networks in complex systems seems to be a daunting task.
MS-cleavable cross-linkers creating characteristic fragment ion signatures
More promising than cross-linkers cleavable by reduction are gas-phase cleavable cross-linkers, regarding their potential for fully automated analyses. These cross-linkers contain one or more labile bonds in the spacer, and, as such, are cleaved in the gas phase in tandem MS (MS/MS) experiments. Most of these linkers are cleaved using CID conditions, while only a few examples exist for reagents with IRMPD (infrared multiphoton dissociation) [24], ETD (electron transfer dissociation) [25], or radical-driven [26] cleavability. The examples presented in this contribution will be restricted to CID-cleavable cross-linkers.
One of the first reports on a cross-linker releasing a specific reporter ion upon CID to facilitate the identification of cross-linked products was published as early as 2001 [27]. Here, the authors described the design and characterization of the homobifunctional amine-reactive cross-linker N-benzyliminodiacetoyloxy succinimide (BID) that yields a benzyl cation at m/z 91 as characteristic marker ion in CID-MS/MS experiments (Table 2).
Several years later, two cross-linkers were designed containing labile aspartyl–prolyl bonds, such as disuccinimidylsuccinamyl aspartyl proline (SuDP; Table 2). The novel feature of incorporating an Asp–Pro bond near the center of the linker region provides a specific gas-phase cleavage site [28, 29]. The Asp–Pro bond is known to preferentially fragment during CID and requires less energy to effectively achieve fragmentation compared to other peptide bonds [30]. This decreased peptide bond stability is mediated by the transfer of a labile proton from the aspartic acid side chain to the basic backbone amine of the neigboring proline residue. Therefore, an Asp–Pro moiety incorporated into the spacer region of a cross-linking reagent allows a single-site fragmentation to produce two unique peptides, each modified with a portion of the fragmented cross-linker. In the initial report, the dissociation of cross-linked complexes was performed in the source region of the mass spectrometer (in-source CID), followed by MS/MS experiments to identify the connected peptides on the basis of the created b- and y-type ions [28]. To improve the method, the cross-linker’s selective Asp–Pro bond dissociation was employed for an effective cleavage of cross-linked complexes by CID-MS/MS in an ion trap mass spectrometer with subsequent fragmentation of the individual peptides by MS3 [29].
Inspired by these Asp–Pro-containing cross-linkers, a cross-linker was designed that is based on Edman degradation chemistry (Table 2). This so-called Edman linker harbors a thiourea moiety with a highly nucleophilic sulfur atom allowing attack at the adjacent Gly–Pro amide carbonyl, initiating the cleavage of the linker molecule [31]. The linker is efficiently cleaved in the low (5–100 eV) as well as in the high energy regime (keV) of CID experiments. On the basis of the Edman linker’s structure, the thiourea-based concept of a dissociative cross-linker was extended to create a simplified analogue [32]. Finally, the synthesis efforts resulted in a urea-based symmetric linker (BuUrBu or disuccinimidyl dibutyric urea (alternatively DSBU); Table 2) that exhibits exquisite properties for automated data analysis as different cross-link types, i.e., intra- and interpeptide and “dead-end” products, can be discriminated [33]. The BuUrBu cross-linker creates characteristic fragment ion patterns attributed to the mass increases of 85 u (Bu) or 111 u (BuUr) at the cross-linked peptides in CID-MS/MS experiments (Fig. 2). As such, an interpeptide cross-link will show two 26-u doublets as cleavage occurs at either of the two NH − C = O bonds in the central urea moiety. Therefore, both peptides will be modified with Bu and BuUr fragments of the linker in MS/MS spectra, resulting in two doublet signals in MS/MS. The characteristic fragment ion patterns of the BuUrBu linker simplify the identification of cross-linked species from complex mixtures and greatly reduce the potential of identifying false-positive cross-links. BuUrBu has been successfully employed to study the structure of the intrinsically disordered tetrameric tumor suppressor p53 that has so far not been amenable to a structural characterization in its full-length form [34].
Proposed fragmentation scheme of BuUrBu-cross-linked peptides. a Chemical structure of BuUrBu and theoretical fragmentation pathway after cleavage under collision-induced dissociation conditions (CID- and HCD-MS/MS). Bu and BuUr specific fragments of the cross-linker, P 1 peptide 1, P 2 peptide 2. b Doublet patterns (Δm ~ 26 u) of reporter ions in the fragment ion mass spectra indicate the presence of a cross-link; a “dead-end” cross-link will give one doublet, while two doublets are indicative of an interpeptide cross-link
Another class of MS-cleavable cross-linkers is based on the presence of a fixed charge sulfonium ion in the spacer chain of the linker [35]. Initial studies demonstrated that peptide ions containing fixed-charge sulfonium ions in the side chains of certain amino acid residues, e.g., methionine and cysteine, show an exclusive loss of a dialkylsulfide moiety via selective cleavage at the site of the fixed charge. On the basis of these studies, an amine-reactive cross-linker (S-methyl 5,5′-thiodipentanoylhydroxysuccinimide; Table 2) was designed, in which a sulfonium ion is incorporated into the spacer arm.
A structurally related cross-linker is disuccinimidyl sulfoxide (DSSO) that contains a central sulfoxide moiety [36, Table 2]. Similar to BuUrBu, DSSO-cross-linked peptides can be discrimininated on the basis of their distinct fragmentation patterns in MS/MS spectra that are unique for different cross-linking types and allow one to conduct automated data analyses. During CID analysis of an interpeptide cross-link, cleavage of one C–S bond next to the sulfoxide will give a pair of peptide fragments, in which one peptide fragment is an alkene (+54 u) and one peptide fragment is modified with a sulfenic acid (+104 u) losing a water molecule (+86 u; Fig. 3). In the case of an intermolecular, i.e. type 2 cross-link, two doublets will be observed owing to the cleavage of either of the two symmetric C–S bonds, resulting in four individual peaks in the MS/MS spectrum. In that respect, the DSSO linker creates similar fragmentation patterns to the BuUrBu linker. The efficacy of DSSO was impressively demonstrated for the structural characterization of the yeast 20S proteasome complex [37], the characterization of a CRISPR-CAS complex [36], and for identifying cross-links from HeLa cell lysates, including the ribosome, the proteasome, the TCP-1 ring complex, and the eukaryotic elongation factor 1 complex [38].
Proposed fragmentation scheme of DSSO-cross-linked peptides. a DSSO synthesis and structure. MS/MS fragmentation patterns of the three types of DSSO-cross-linked peptides: b interpeptide, c “dead-end”, and d intrapeptide cross-link. e Conversion of a sulfenic acid-modified fragment to an unsaturated thiol-modified fragment after water loss. f Mass relationships between MS/MS fragment ions shown in b–d and their precursor ions. DCC N,N′-dicyclohexylcarbodiimide, MCPBA m-chloroperbenzoic acid. Figure reprinted from [36]
Trifunctional cross-linkers creating reporter ions
The bifunctional cross-linking strategy can be extended towards trifunctional linkers that in addition to two NHS esters contain a biotin label, such as in CBDPS [39] or PIR [40] (Table 3). The third function can then be used for a selective enrichment of cross-linked products by affinity purification [41]. Trifunctional cross-linkers are mainly used to decipher protein interaction networks and they have been successfully employed for studying protein assemblies in vivo. To avoid disturbing the protein interfaces by a bulky biotin moiety, an azide group can be incorporated into the cross-linker, allowing the coupling of a biotin group by click chemistry for subsequent affinity enrichment after the cross-linking reaction. Azide-A-DSBSO incorporates this principle of a clickable third function and is analoguous in its fragmentation behavior to DSSO where cleavage occurs at the sulfoxide group (Table 3) [42]. Conclusively, this subclass of trifunctional cross-linkers combines two distinct advantages: (i) selective MS/MS cleavage creating characteristic marker ions; (ii) enrichment of cross-linked peptides by affinity purification.
One class of cleavable trifunctional cross-linkers that have been introduced in 2005 are protein interaction reporters (PIR; Table 3) [40]. Since then, the development of several different PIR molecules has been reported using a variety of activation methods to enable peptide release, design of bioinformatics strategies, and in vivo applications [43, 44]. The size of the PIR and its spacer length (43 Å) are quite large, but apparently PIR cross-linkers can bridge much shorter distances owing to their high flexibility [45].
Just recently, a remarkable strategy has been presented that relies on the use of so-called Leiker linkers (Fig. 4) [46]. As with other trifunctional reagents, this cross-linker class contains a biotin tag for affinity purification of cross-linked peptides. It also harbors a cleavage site to release cross-linked peptides after enrichment on streptavidin beads. Chemical cleavage of the Leiker linker at the N = N bond releases the biotin group so it does not interfere with subsequent LC/MS/MS analysis. The innovative aspect is that chemical cleavage yields at the same time an aromatic amine, giving rise to specific reporter ions during tandem MS measurements. As such, intra- and interpeptide as well as “dead-end” cross-links yield a reporter ion of m/z 122.06 in higher-energy collision-induced dissociation (HCD) spectra (Fig. 4d). For quantitative cross-link analysis, an isotope-labeled version of the Leiker linker was synthesized, in which six hydrogen atoms in the spacer arm are replaced with deuterium.
Scheme of cross-linking/MS workflow using the trifunctional cleavable Leiker reagent. a The Leiker linker contains a biotin moiety (magenta), a cleavage site (arrows), and six hydrogen atoms that are accessible to isotope labeling (asterisks). b The workflow for purification of Leiker-linked peptides. c Three types of Leiker-linked peptides. d Cross-linked peptides generate a reporter ion of m/z 122.06 in HCD, as shown in the spectrum of an inter-linked peptide NYQEAKDAFLGSFLYEYSR-LAKEYEATLEECCAK (+4 charged, M + H+ 4433.0553), in which C denotes carbamidomethylated cysteine. Figure reprinted from [46]
Strategies for automated data analysis
As outlined above, MS-cleavable cross-linkers allow an automated assignment of cross-links based on characteristic reporter ions. In general, one of the greatest challenges for an MS analysis of cross-linked products consists in their often poor fragmentation. When MS-cleavable cross-linkers are employed, the characteristic reporter ion patterns created from interpeptide cross-links serve as a basis for unambiguous assignment. Also, an exhaustive backbone fragmentation of the connected peptides is crucial for an unambiguous assignment of cross-linked products. To achieve this goal, two-step collisional activation is usually performed meaning that the cleavable cross-linker is first cleaved at its labile bond(s) resulting in the characteristic fragment ion patterns indicative of a cross-link. Then, MS3 experiments can be performed on the selected cross-linker reporter ions yielding backbone fragmentation of the connected peptides. Alternatively, consecutive CID and ETD experiments can be performed with the benefit of combining information from two complementary fragmentation principles. As such, the resulting fragment ion spectra will include high-intensity signals of the characteristic cross-linker fragments as well as backbone coverage of the cross-linked peptides (c- and z-type ions produced by ETD, supported by b- and y-type ions produced by CID). That workflow was recently proposed for the identification of cross-links from protein complexes in human cellular lysates [38].
Two software solutions, MeroX [47] and XLinkX [38], are currently available for an automated data analysis based on the characteristic fragment ion signatures created by MS-cleavable linkers. MeroX scans MS/MS spectra for two matching doublet signals indicative of an interpeptide cross-link if, for example, the cleavable linkers DSSO or BuUrBu are used (Figs. 2, 3, and 5). In the newly developed RISE (reporter ion scan event) mode, MeroX calculates the masses of the two cross-linked peptides from the matching doublets and compares the peptide masses to in silico digested proteins to identify a cross-link candidate. The quality of MS/MS signal assignment (presence of two doublets and signals originating from fragmentation of the peptide backbone) determines the score [34]. XlinkX is quite similar as it retrieves the precursor mass of each linked peptide based on the characteristic fragmentation pattern created by the MS-cleavable cross-linker. Then, spectra that contain at least one doublet signature are considered as potential cross-link, and every cross-link pair is subjected to further peptide sequence analysis. XlinkX matches fragment ions for each MS/MS spectrum on the basis of the determined masses of the two cross-linked peptides and all fragment ions generated from the precursor ion [38].
Workflow for automated data analysis using the BuUrBu linker. a Structure of an interpeptide cross-link with BuUrBu. b Cross-links are screened at the MS level, precursor ions are selected and c in MS/MS spectra, the two characteristic 26-u doublets indicate an interpeptide cross-link. On the basis of the masses of the doublet signals, the mass of the cross-linked product is calculated with MeroX (RISE mode) [34]. Backbone fragments in the MS/MS spectrum allow one to sequence the connected peptides and locate the exact cross-linking sites
BuUrBu is different from DSSO as the energy required for cleaving the cross-linker is higher for the central urea moiety in BuUrBu compared to the sulfoxide group in DSSO (compare Figs. 2 and 3). Therefore, DSSO shows predominately cross-linker fragments and almost no backbone fragments in MS/MS spectra, requiring MS3 or sequential CID/ETD experiments to delineate the amino acid sequences of the connected peptides. If the BuUrBu-linker is employed, the characteristic doublets of the cross-linker as well as exhaustive backbone fragments of the connected peptides are already observed in one single collisional activation step, i.e., at the MS/MS level (Fig. 5). This eliminates the need for conducting MS3 experiments, which are often of poor quality when low-intensity MS/MS precursors are fragmented. An in-depth comparison of both software packages revealed that XlinkX is optimized for the fragmentation characteristics of DSSO, while MeroX performs better for BuUrBu [34]. XLinkX is reported to handle proteome-wide datasets, while MeroX reliably calculates cross-links from samples containing up to ca. 5000 proteins [34, 38]. On the other hand, MeroX is more versatile than XLinkX regarding the choice of cross-linkers, proteases, and reaction sites.
Summary and outlook
Currently, MS-cleavable cross-linkers seem to be the most promising strategy for an automated analysis to derive three-dimensional structural information of proteins and to map protein–protein interactions in complex systems. The latter application will become more and more important as powerful methods are needed to study protein networks within the cell, yielding insights into a protein’s function in the natural cellular environment. Just recently, software solutions have become available that make use of the full potential of MS-cleavable cross-linkers based on the characteristic signatures created by a selective fragmentation of the linker in the gas phase. The incorporation of reactive groups other than NHS esters can be envisioned, such as photo-, sulfhydryl-, or carboxyl-reactive groups that will allow amino acids other than lysines to be targeted. One very recent example of an alternative cross-linking chemistry is the DSSO-based, carboxyl-reactive sulfoxide-containing MS-cleavable homobifunctional cross-linker dihydrazide sulfoxide [48]. Also, isotope-labeled, i.e., deuterated, MS-cleavable cross-linkers will by beneficial for a targeted selection of cross-linked products at the MS level. We envision that the number of available MS-cleavable cross-linkers will increase within the next few years and extend the arsenal of available reagents for conducting cross-linking experiments at the proteome level.
Abbreviations
- Azide-A-DSBSO:
-
Azide-tagged, acid-cleavable disuccinimidyl bissulfoxide
- BAMG:
-
Bis(succinimidyl)-3-azidomethyl glutarate
- BID:
-
N-benzyliminodiacetoyloxy succinimide
- BuUrBu:
-
4-{3-[3-(2,5-dioxo-pyrrolidine-1-yloxycarbonyl) propyl]ureido}butyric acid 2,5-dioxo-pyrrolidine-1-yl ester
- CBDPS:
-
Cyanurbiotindipropionyl succinimide
- CID:
-
Collision-induced dissociation
- DSBU:
-
Disuccinimidyl dibutyric urea
- DSSO:
-
Disuccinimidyl sulfoxide
- DTSSP:
-
3,3′-dithiobis(sulfosuccinimidyl propionate)
- ESI:
-
Electrospray ionization
- ETD:
-
Electron transfer dissociation
- HCD:
-
Higher-energy collision-induced dissociation
- IRMPD:
-
Infrared multiphoton dissociation
- LC:
-
Liquid chromatography
- MALDI:
-
Matrix-assisted laser desorption/ionization
- MS:
-
Mass spectrometry
- MS/MS:
-
Tandem mass spectrometry
- NHS:
-
N-hydroxysuccinimide
- PIR:
-
Protein interaction reporter
- RISE:
-
Reporter ion scan event
- SDAD:
-
Succinimidyl 2-([4,4′-azipentanamido]ethyl)-1,3′-dithiopropionate
- SDS-PAGE:
-
Sodium dodecyl polyacrylamide gel electrophoresis
- SuDP:
-
Disuccinimidylsuccinamyl aspartyl proline
References
Young MM, Tang N, Hempel JC, et al. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci U S A. 2000;97(11):5802–6.
Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25(4):663–82.
Leitner A, Walzthoeni T, Kahraman A, et al. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9(8):1634–49.
Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom Rev. 2010;29(6):862–7.
Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173(3):530–40.
Sharon M, Robinson CV. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu Rev Biochem. 2007;76:167–93.
Heck AJ. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods. 2008;5(11):927–33.
Konermann L, Pan Y. Exploring membrane protein structural features by oxidative labeling and mass spectrometry. Expert Rev Proteomics. 2012;9(5):497–504.
Yan Y, Chen G, Wei H, et al. Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. J Am Soc Mass Spectrom. 2014;25(12):2084–92.
Kahraman A, Herzog F, Leitner A, Rosenberger G, Aebersold R, Malmström L. Cross-link guided molecular modeling with ROSETTA. PLoS One. 2013;8(9), e73411. doi:10.1371/journal.pone.0073411.
Kalkhof S, Haehn S, Paulsson M, Smyth N, Meiler J, Sinz A. Computational modeling of laminin N-terminal domains using sparse distance constraints from disulfide bonds and chemical cross-linking. Proteins. 2010;78(16):3409–27.
Schilling B, Row RH, Gibson BW, Guo X, Young MM. MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J Am Soc Mass Spectrom. 2003;14(8):834–50.
Bruce JE. In vivo protein complex topologies: sights through a cross-linking lens. Proteomics. 2012;12(10):1565–75.
Paramelle D, Miralles G, Subra G, Martinez J. Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics. 2013;13(3-4):438–56.
Sinz A. The advancement of chemical cross-linking and mass spectrometry for structural proteomics: from single proteins to protein interaction networks. Expert Rev Proteomics. 2014;11(6):733–43.
Holding AN. XL-MS: protein cross-linking coupled with mass spectrometry. Methods. 2015;89:54–63.
Liu F, Heck AJ. Interrogating the architecture of protein assemblies and protein interaction networks by cross-linking mass spectrometry. Curr Opin Struct Biol. 2015;35:100–8.
Leitner A, Faini M, Stengel F, Aebersold R. Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem Sci. 2016;41(1):20–32.
Tran BQ, Goodlett DR, Goo YA. Advances in protein complex analysis by chemical cross-linking coupled with mass spectrometry (CXMS) and bioinformatics. Biochim Biophys Acta. 2016;1864(1):123–9.
Bennett KL, Kussmann M, Björk P, et al. Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping–a novel approach to assess intermolecular protein contacts. Protein Sci. 2000;9(8):1503–18.
Zorn M, Ihling CH, Golbik R, Sawers RG, Sinz A. Mapping cell envelope and periplasm protein interactions of Escherichia coli respiratory formate dehydrogenases by chemical cross-linking and mass spectrometry. J Proteome Res. 2014;13(12):5524–35.
Kasper PT, Back JW, Vitale M, et al. An aptly positioned azido group in the spacer of a protein cross-linker for facile mapping of lysines in close proximity. ChemBioChem. 2007;8(11):1281–92.
Buncherd H, Roseboom W, Ghavim B, et al. Isolation of cross-linked peptides by diagonal strong cation exchange chromatography for protein complex topology studies by peptide fragment fingerprinting from large sequence databases. J Chromatogr A. 2014;1348:34–46.
Gardner MW, Vasicek LA, Shabbir S, Anslyn EV, Brodbelt JS. Chromogenic cross-linker for the characterization of protein structure by infrared multiphoton dissociation mass spectrometry. Anal Chem. 2008;80(13):4807–19.
Gardner MW, Brodbelt JS. Preferential cleavage of N-N hydrazone bonds for sequencing bis-arylhydrazone conjugated peptides by electron transfer dissociation. Anal Chem. 2010;82(13):5751–9.
Hage C, Ihling CH, Götze M, Schäfer M, Sinz A. Dissociation behavior of a TEMPO-active ester cross-linker for peptide structure analysis by free radical initiated peptide sequencing (FRIPS) in negative ESI-MS. J Am Soc Mass Spectrom. 2016. doi:10.1007/s13361-016-1426-9.
Back JW, Hartog AF, Dekker HL, Muijsers AO, de Koning LJ, de Jong L. A new crosslinker for mass spectrometric analysis of the quaternary structure of protein complexes. J Am Soc Mass Spectrom. 2001;12(2):222–7.
Soderblom EJ, Goshe MB. Collision-induced dissociative chemical cross-linking reagents and methodology: applications to protein structural characterization using tandem mass spectrometry analysis. Anal Chem. 2006;78(23):8059–68.
Soderblom EJ, Bobay BG, Cavanagh J, Goshe MB. Tandem mass spectrometry acquisition approaches to enhance identification of protein-protein interactions using low-energy collision-induced dissociative chemical crosslinking reagents. Rapid Commun Mass Spectrom. 2007;21(21):3395–408.
Mák M, Mezö G, Skribanek Z, Hudecz F. Stability of Asp-Pro bond under high and low energy collision induced dissociation conditions in the immunodominant epitope region of herpes simplex virion glycoprotein D. Rapid Commun Mass Spectrom. 1998;12(13):837–42.
Dreiocker F, Müller MQ, Sinz A, Schäfer M. Collision-induced dissociative chemical cross-linking reagent for protein structure characterization: applied Edman chemistry in the gas phase. J Mass Spectrom. 2010;45(2):178–89.
Müller MQ, Dreiocker F, Ihling CH, Schäfer M, Sinz A. Fragmentation behavior of a thiourea-based reagent for protein structure analysis by collision-induced dissociative chemical cross-linking. J Mass Spectrom. 2010;45(8):880–91.
Müller MQ, Dreiocker F, Ihling CH, Schäfer M, Sinz A. Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal Chem. 2010;82(16):6958–68.
Arlt C, Götze M, Ihling CH, Hage C, Schäfer M, Sinz A. An integrated workflow for structural proteomics studies based on cross-linking/mass spectrometry with an MS/MS cleavable cross-linker. Anal Chem. 2016;88(16):7930–7.
Lu Y, Tanasova M, Borhan B, Reid GE. Ionic reagent for controlling the gas-phase fragmentation reactions of cross-linked peptides. Anal Chem. 2008;80(23):9279–87.
Kao A, Randall A, Yang Y, et al. Mapping the structural topology of the yeast 19S proteasomal regulatory particle using chemical cross-linking and probabilistic modeling. Mol Cell Proteomics. 2012;11(12):1566–77.
Benda C, Ebert J, Scheltema RA, et al. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell. 2014;56(1):43–54.
Liu F, Rijkers DT, Post H, Heck AJ. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods. 2015;12(12):1179–84.
Petrotchenko EV, Serpa JJ, Borchers CH. An isotopically coded CID-cleavable biotinylated cross-linker for structural proteomics. Mol Cell Proteomics. 2011;10(2):M110.001420. doi:10.1074/mcp.M110.001420.
Tang X, Munske GR, Siems WF, Bruce JE. Mass spectrometry identifiable cross-linking strategy for studying protein-protein interactions. Anal Chem. 2005;77(1):311–8.
Trester-Zedlitz M, Kamada K, Burley SK, Fenyö D, Chait BT, Muir TW. A modular cross-linking approach for exploring protein interactions. J Am Chem Soc. 2003;125(9):2416–25.
Kaake RM, Wang X, Burke A, et al. A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol Cell Proteomics. 2014;13(12):3533–43. doi:10.1074/mcp.M114.042630.
Chowdhury SM, Munske GR, Tang X, Bruce JE. Collisionally activated dissociation and electron capture dissociation of several mass spectrometry-identifiable chemical cross-linkers. Anal Chem. 2006;78(24):8183–93.
Weisbrod CR, Chavez JD, Eng JK, Yang L, Zheng C, Bruce JE. In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J Proteome Res. 2013;12(4):1569–79.
Zhang H, Tang X, Munske GR, Tolic N, Anderson GA, Bruce JE. Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol Cell Proteomics. 2009;8(3):409–20.
Tan D, Li Q, Zhang MJ, Liu C, Ma C, Zhang P, et al. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. Elife. 2016;5. doi:10.7554/eLife.12509.
Götze M, Pettelkau J, Fritzsche R, Ihling CH, Schäfer M, Sinz A. Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J Am Soc Mass Spectrom. 2015;26(1):83–97.
Gutierrez CB, Yu C, Novitsky EJ, Huszagh AS, Rychnovsky SD, Huang L. Developing a novel acidic residue reactive and sulfoxide-containing MS-cleavable homobifunctional cross-linker for probing protein-protein interactions. Anal Chem. 2016;88(16):8315–22.
Acknowledgments
AS gratefully acknowledges financial support from the Deutsche Forschungsgemeinschaft (DFG project Si 867/15-2), the region of Saxony-Anhalt, and the EU (COST Action BM1403). AS thanks Dr. C. Ihling for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that she has no conflict of interest.
Rights and permissions
About this article
Cite this article
Sinz, A. Divide and conquer: cleavable cross-linkers to study protein conformation and protein–protein interactions. Anal Bioanal Chem 409, 33–44 (2017). https://doi.org/10.1007/s00216-016-9941-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9941-x