Abstract
A parallel-processing four-station polymerase chain reaction (PCR) device has been developed, which performs continuous-flow PCR without optimization of the annealing temperature. Since the annealing temperature of each station can be controlled independently, the device covers an annealing temperature range of 50–68 °C, which is wide enough to perform PCR for any DNA fragment regardless of its optimum annealing condition. This arrangement lets us continuously obtain an amplified amount of a DNA fragment at least from one of the stations. The device consists of four identical cylindrical stations (diameter 20 mm, height 55 mm). A polytetrafluoroethylene capillary reactor (length 2 m, I.D. 100 μm, O.D. 400 μm) is wound helically up around each station. The whole assembly is designed to minimize the number of heating blocks (for providing temperatures of denaturation, annealing, and extension) to be seven and to shape a compact cube (height 55 mm, base 60 mm × 60 mm). The reproducibility for continuous-flow PCR is reasonably high (run-to-run and station-to-station relative standard deviation of their amplification is lower than 6 % and about 4 %, respectively). Performance on the optimization-free DNA amplification has been evaluated with four DNA samples with different annealing conditions and product sizes (323, 608, 828, and 1101 bp), which has demonstrated that in all cases, PCR is successful at least on one station. In addition, three DNA fragments with different lengths (323, 1101, and 2836 bp) have been successfully amplified in a segmented-flow mode without the carry-over contamination between segments. This result suggests that this device could serve as the PCR module of a continuous-flow high-throughput on-line total DNA analysis system integrating all necessary modules from cell lysis/DNA extraction to PCR product analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the introduction of polymerase chain reaction (PCR) in mid-1980s, it has become one of the most useful and versatile processes in biological science, and revolutionized a variety of fields including diagnostics of bacterial/viral infections and hereditary diseases, forensic science, biodefense, etc [1–5]. As the application area of PCR technology has expanded beyond laboratorial territory, there have been great interests and efforts in developing miniaturized PCR devices with merits for on-site analysis, such as small dimensions, minimal reagent consumption, rapid amplification, and low fabrication cost [6, 7]. Miniaturized PCR devices fall into two categories: stationary and continuous-flow types. In the stationary type, the mode of thermal cycling is similar to that generally employed in conventional benchtop PCR machines. The PCR solution is kept stationary in a microwell or microchamber, and then the whole device is heated and cooled to make the repetitive cycle of the three temperatures for denaturation, annealing, and extension [8–12]. This thermal cycling mode, however, causes delay in the temperature response of the solution because of the large thermal mass of the device itself. In the continuous-flow type, the PCR solution flows through a microchannel or capillary reactor designed for the solution to contact with the three fixed-temperature heating zones in sequence and to be subject to repetitive thermal cycling [13–19]. Since such a reactor of minimal cross-sectional area secures rapid thermal equilibrium of the solution with external heating zones, the time per cycle can be greatly shortened to perform fast DNA amplification. This continuous-flow microfluidic platform, in addition, has great potential to be the basis of a micro total DNA analysis system where other functionalities, for example, sample pretreatment (cell lysis and DNA preparation) and PCR product analysis are integrated along with PCR amplification [20–22].
PCR is a very efficient process because it can synthesize microgram levels of DNA even from a single DNA template under optimum conditions [23]. However, a number of variables should be considered in the optimization of PCR. The annealing temperature is one of primary factors. If the annealing temperature is too low, non-specific DNA fragments can be amplified, but when the annealing temperature is too high, the yield of the PCR product is decreased due to poor annealing of the primer pair [23]. Therefore, prior to obtaining an amplified amount of a DNA fragment, gradient PCR at the annealing temperature range of 50–65 °C is usually performed to find the optimum annealing temperature. Although commercial gradient PCR machines can generate a temperature gradient up to 12 different annealing temperatures spanning a range of 20 °C and show good performance, the PCR process usually takes about 2 h for 30 cycles; most of the reaction time is spent on heating and cooling during the reaction because of large thermal mass. In addition, these machines are too expensive, bulky, and energy intensive, making them hard to bring PCR to on-site analysis. Unfortunately, little attention has been paid to the development of miniaturized PCR device capable of carrying out parallel DNA amplifications at different annealing temperatures for expeditious optimization of the PCR condition. Recently, Da Xing group has demonstrated gradient PCR on a microfluidic device [24]. A nonlinear temperature gradient is made on a single copper plate, and three circular polytetrafluoroethylene (PTFE) tubes are partly embedded at two zones on the grooved plate so that a PCR solution in each tube can pass the two zones (hot (90–97 °C) and cool (60–70 °C) zones) alternatively. Although they have demonstrated three parallel PCR amplifications using the same DNA fragment of a 112 bp at different annealing temperatures ranging from 60 to 68 °C, still important challenges remain such as dealing with a large number of samples for high-performance DNA analysis system.
In this study, we present a cube-shaped compact four-station PCR device that provides independent temperature settings for annealing at each station. On this multi-station device, continuous-flow PCR of DNA samples is carried out without optimizing the annealing temperature because four different settings of annealing temperatures cover the common annealing-temperature range (50–65 °C), and therefore the amplified amount of the predesignated DNA fragment is continuously obtained at least from one station without non-specific PCR products. This optimization-free merit can be also applied to the segmented-flow mode for high-throughput DNA analysis. We have demonstrated the segmented-flow PCR of different DNA samples on the same station without carry-over contamination. To the best of our knowledge, this is the first report about miniaturized continuous-flow PCR device capable of carrying out parallel DNA amplifications at different annealing temperatures that are operated independently of each other.
Materials and methods
Reagents
The PCR reagents including Taq DNA polymerase 10× reaction buffer (500 mM KCl, 100 mM Tris-HCl (pH 9.0 at 25 °C), and 1 % Triton® X-100), MgCl2 (25 mM), dNTPs mixture (10 mM each of dATP, dGTP, dCTP, and dTTP in water), and Taq polymerase (5 units/μL) were purchased from Promega (Madison, WI) as a part of the PCR Core System II kit. Table 1 shows a list of primer sets for amplifying five DNA fragments of different sizes. The sets were synthesized by Bioneer (Daejeon, Korea). The template DNA for 323-bp PCR was obtained from Promega (positive control plasmid DNA, 1 ng/μL in TE buffer (pH 7.4)), and those for PCR of the rest four from Takara (ΦX174 RF I DNA (0.5 μg/μL), Otsu, Japan). A DNA size marker was purchased from Takara (ΦX174-Hae III digest), and Gel Red™ dye was obtained from Biotium (Hayward, CA). Ultrapure DI water produced from a Milli-Q System (Millipore, Billerica, MA) was autoclaved (SP510, Yamato, Tokyo, Japan) and then used in all experiments.
Construction of the continuous-flow multi-station PCR device
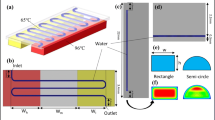
The multi-station PCR device has four cylindrical stations that can have different annealing temperatures, respectively. The stations are assembled into a compact cube-shaped device (60 mm × 60 mm × 55 mm in height). Figure 1a shows the arrangement of heating blocks for four-station thermal cycling. The blocks are made of oxygen-free copper to assure good thermal conduction. Each station has an independent annealing block, two stations share one extension block, and there is one denaturation block common to all stations. As a total, the device has seven heating blocks. All blocks except the one for denaturation are thermostated by using a pair of a heater cartridge (3 mm in diameter and 50 mm in length; Watlow, Richmond, IL) and a temperature sensor (1 mm in diameter and 50 mm in length; Watlow, Richmond, IL) that are connected to a temperature controller (DX4-KCSNR, Hanyoung Nux, Incheon, South Korea). Since the denaturation block has thin and long shape with large surface area, two heater/sensor pairs are used to achieve a homogeneous temperature distribution (Fig. 1c). A 2-m-long PTFE capillary (100 μm I.D. and 400 μm O.D.; EJ-06417-72, Cole-Parmer, Vernon Hills, IL) coils helically, with 30 turns at a fixed pitch per turn of helix, up around each 20-mm-diameter cylindrical structure assembled from an annealing copper block and a plastic thermal insulator block. The PTFE capillary reactor, whose net length without the inlet and outlet parts is 1 m, sits into grooves (400-μm width and 400-μm depth) formed on the surface of the cylindrical assembly. With arranging heating blocks and plastic thermal insulator, and winding capillaries as shown in Fig. 1a, thermal cycling for DNA amplification can be run repetitively in the sequence of denaturation, annealing, and extension. The contact lengths of a wound capillary with heating blocks in each cycle are 10 mm, 10 mm, and 20 mm for denaturation, annealing, and extension, respectively. Thermal isolation of each heating block is secured. Each annealing block, in particular, should be well isolated from the higher-temperature denaturation and extension blocks. It is accomplished by making a convective air space in between (Fig. 1c). The heat loss from the outer surface of the annealing block is minimized by covering the surface with a thick copper plate (Fig. 1b).
a Scheme of a continuous-flow multi-station PCR device (A annealing block, E extension block, D denaturation block, I insulator, S station). The arrows indicate flow directions of PCR mixture solution. b, c Photographic images of a continuous-flow multi-station PCR device. The annealing blocks are covered with copper plates to protect PCR mixture solution in the PTFE capillary from air cooling. Each heating block has holes for heat cartridges and temperature sensors
Multi-station PCR
The PTFE capillaries are washed with ethanol and DI water before and after use. PCR mixtures are prepared just before running PCR as follows: the PCR mixture for the 323-bp DNA amplification contained master mix (1× MgCl2-free polymerase buffer, 1.5 mM MgCl2, 0.2 mM of dNTPs mix, 1 unit Taq polymerase), 1 μM of forward and reverse primers, and 20 pg/µL of positive control plasmid DNA as a template. In the cases of the other DNA samples (1101, 828, and 608 bp), 0.5 μM of forward and reverse primers and 20 pg/µL of ΦX174 RFI DNA as a template were used with the same master mix. When running a multi-station PCR, 20-μL of the same PCR mixture is introduced into the upper end of each capillary mounted on the device. Denaturation and extension temperatures are common to all stations: 92 and 72 °C, respectively. Four different annealing temperatures are set on four stations, respectively, so as to span 15° with 5 °C differences between two adjacent stations: for example, 50, 55, 60, and 65 °C. The flow rate of PCR mixture solution through a capillary reactor, which varies with the length of a DNA fragment to be amplified, ranges from 0.2 to 3.0 μL/min. Four PCR product solutions coming out of the lower ends of capillaries are collected and analyzed by agarose gel electrophoresis. The performance of our multi-station PCR system has been compared with that of a commercial PCR machine (T Professional, Biometra, Göttingen, Germany) under the same experimental conditions.
Segmented-flow PCR
To establish segmented-flow PCR of different DNA samples on our continuous PCR device, checking of carry-over contamination in a capillary reactor should be preceded. Two 5-μL segments of the 323-bp PCR mixture (positive control) and negative controls mixture (containing all reagents in the PCR mixture but Taq polymerase (negative control A), the template DNA (negative control B), or the primer pair (negative control C)) are sequentially injected onto a hydrophobic carrier fluid of perfluorodecalin (Acros Organics, Geel, Belgium) using an HPLC injection valve (7725i, Rheodyne, Rohnert Park, CA) equipped with a 5-μL sample loop. This mode of injection lets a 2-μL perfluorodecalin segment intervene between two 5-μL aqueous segments without forming any air bubbles in the flow. Checking of carry-over contamination is made by analyzing all of the negative control segments by agarose gel electrophoresis after PCR. For negative controls contaminated by the preceding positive control, a 5-μL segment of washing solution (1× gel loading buffer, Takara) is interposed between the two segments in order to see that the washing solution can effectively rinse out the DNA or protein molecules adsorbed on the inner surface of the capillary reactor. Temperatures for thermal cycling are 92, 55, and 72 °C for denaturation, annealing, and extension, respectively, and the flow rate is 3.0 μL/min.
The PCR mixtures containing different templates and primers for amplifying 2836-, 1101-, and 323-bp fragments are sequentially injected with the same volume (5 μL) at regular intervals. The PCR segments are separated by a group of three segments in the sequence of 2-μL perfluorodecalin, 5-μL washing solution, and 2-μL perfluorodecalin. PCR amplifications in this segmented-flow mode are performed at a flow rate of 0.2 μL/min and 92-55-72 °C for denaturation-annealing-extension.
Results and discussion
Continuous-flow multi-station PCR device
We have developed a continuous-flow PCR device with four cylindrical stations, where PCR can be run in parallel at four different annealing temperatures. The design of this capillary-based device is based on the cylindrical single-station PCR device reported previously [25]. The single-station device is comprised of three thermostating metal blocks and a fused-silica capillary reactor coiled helically on the cylindrical heating block assembly. Since the single-station device is set at an annealing temperature during PCR thermal cycling, the PCR amplification using this device will be successful only for samples with known optimal annealing temperatures. Thus, the process for finding an optimal annealing temperature, such as gradient PCR, should precede PCR amplification on this device. With our multi-station device, on the other hand, parallel-processing PCR can be carried out at four different annealing temperatures so that the range could cover optimal annealing temperatures of most DNA samples. If we make a 5 °C gap between adjacent temperatures, the four temperatures can span a 15 °C range, for example, 50–65 °C. It is, therefore, expected that this arrangement will allow successful PCR at least on one station.
Our continuous-flow multi-station PCR device has three characteristic features in its design. Firstly, although the device has four, independent PCR thermal cyclers, the assembly is so compact that the device could be developed to be a portable system. The compact cube-shaped assembly (60 mm × 60 mm × 55 mm in width, length, and height) is realized by minimizing the number of heating blocks that needs to carry out parallel PCR amplification with different annealing temperatures. The assembly has only seven heating blocks: four for annealing, two for extension, and one for denaturation step. Each cylindrical station whose diameter is only 20 mm is positioned at each corner of the square plane (Fig. 1a). Secondly, the annealing blocks are thermally well isolated from nearby high-temperature extension and denaturation blocks by allowing convective air cooling all around each annealing block (Fig. 1c). Lastly, efficient thermal equilibrium between the capillary reactor and heating blocks is secured by sitting the capillary into fitted grooves on the surface of each cylindrical station, and by letting the capillary regions contacting with heating blocks not be exposed to the ambient air.
The surface temperature distribution of the PCR device has been measured by an infrared (IR) camera (P620, FLIR Systems, Inc., Wilsonville, OR) (see Electronic Supplementary Material (ESM), Fig. S1). Fifteen points from each heating block are selected, and temperatures are evaluated using and image analyzer (Tools, FLIR Systems, Inc., Wilsonville, OR). The standard deviations of measured temperatures from each heating zone are within ±0.2 °C (relative standard deviation (RSD) <0.21 %, n = 15). To check the temperature uniformity of heating blocks, we specified three areas (boxes i, ii, and iii in ESM Fig. S1a) from each heating block and analyzed all spots’ temperatures in the boxes. The surface temperatures of the three areas representing for denaturation, annealing, and extension block, respectively, are 91.6 ± 0.2 °C (0.23 % RSD, n = 4255), 55.2 ± 0.2 °C (0.30 % RSD, n = 3565), and 72.0 ± 0.2 °C (0.26 % RSD, n = 4255), indicating that a homogeneous temperature distribution is nearly achieved throughout the copper blocks. The surface temperatures of each heating block are also measured by using a thermocouple having small flat probe, and the result corresponds with that of IR camera (ESM Fig. S1c).
Multi-station PCR
Performance of our multi-station PCR device has been checked by running PCR amplifications for four DNA samples with different annealing conditions and PCR product sizes (Table 1 (a–d)), and then comparing its amplification yield with that of a commercial, batch-mode PCR machine under the same experimental conditions. The annealing temperatures of four stations are set with a 5 °C regular interval between two adjacent stations, so that the device can span an annealing temperature range of 15 °C (50 °C/55 °C/60 °C/65 °C or 53 °C/58 °C/63 °C/68 °C). The range is adjusted to cover optimal annealing temperatures used in most PCRs in biological or clinical fields. Figure 2 shows agarose gel electrophoresis results of four DNA fragments (323, 1101, 828, and 608 bp) amplified in the commercial batch system and the continuous-flow multi-station one at four different annealing temperatures. Fluorescence intensity of each DNA band is evaluated with image analysis software (ImageJ, NIH), where relative differences between two PCR systems are below 8 % except for 608-bp DNA fragment in Fig. 2d (see ESM Table S1). It is noted that results from our device are almost equal to those from the commercial one, which demonstrates that the PCR yield of our device is comparable to that of commercial one in a wide range of sample and annealing conditions.
Agarose gel electropherograms of amplified DNA fragments from a commercialized PCR machine and a continuous-flow multi-station PCR device at different annealing temperatures (M commercialized PCR machine, S stations of the continuous-flow multi-station PCR device). Panels (a–c) represent the amplifying results of DNA fragments with length 323, 1101, and 828 bp at 53, 58, 63, 68 °C for annealing, and panel (d) is for 608 bp of DNA fragment at 50, 55, 60, and 65 °C for annealing
In Fig. 2, different DNA samples show very different PCR behaviors at the set temperatures for annealing. The 323-bp DNA fragment is amplified without any non-specific DNA band in the whole range of annealing temperatures (Fig. 2a) while the optimal annealing temperatures of samples having longer PCR products fall in specific ranges: 53–63 and 53–58 °C for the 1101 bp and 828 bp DNA fragments, respectively (Fig. 2b, c). In the case of the DNA sample for amplifying 608-bp fragment, the optimal annealing temperature range is so narrow that amplification of 608-bp product is dominant only on a single station (Fig. 2d). Undesired DNA fragments are amplified below this temperature due to non-specific bindings of primers to the template DNA. We assume that the intensity difference of the DNA bands between M1 and S1 in Fig. 2d is caused by the slight temperature difference in the solution. The temperature profile of the reaction solution in our continuous-flow type device would be different from that of benchtop device which the reaction solution does not move. Even though temperatures of heating blocks are set at the same value, the difference in temperature profiles between the flowing and non-flowing liquid arises [26]. In addition, exposed cooling region of the capillary can also affect the temperature profile, because the whole device is not completely insulated from the ambient environment. For the delicate sample having very narrow optimal annealing temperature range, close attention to maintain constant environmental condition may be necessary.
The results in Fig. 2 indicate that when this four-station PCR device is employed, most of PCR would be successful at least on one station. Since our device is based on the use of a capillary reactor, it would be well coupled with microchip-based DNA analyzers [27, 28] that use only minimal amounts of DNA samples and is featured by rapid analysis. Therefore, it would be not difficult, by using a proper microchip analyzer, to check which station is the most successful in amplifying the desired DNA fragment for a DNA sample. On checking, all but the best station are turned off, and then we can continuously obtain the desired DNA fragment from our device as much as we want. In this preparation mode of PCR, we do not need a separate step for determining the optimal annealing temperature for a DNA sample, like gradient PCR, prior to amplification of the desired DNA fragment.
The flow rate of the PCR mixture solution through the capillary reactor varies with the size of the DNA fragment to be amplified: 3 μL/min for 323 bp DNA, 0.3 μL/min for 608 bp DNA, and 0.2 μL/min for 828, 1101, and 2836 bp DNA. The residence times of a PCR mixture on the three heating blocks of denaturation, annealing, and extension are 2, 2, and 4 s, respectively, at the rate of 3 μL/min. It is known that an extension rate of Taq polymerase is up to 100 nucleotides/s [29]. The 4-s residence on the extension block for amplifying 323-bp fragment, therefore, is matched with the polymerization rate. However, we need, in general, 1 min to extend a DNA strand by 1 kb with Taq polymerase in a batch system. The rapid extension of our system might be due to the use of a capillary reactor that has high surface-to-volume ratio and thus allows rapid heat transfer [30].
To check the station-to-station amplification efficiency variance of the multi-station PCR device, triplicate amplifications of the 323-bp fragment are done on each station. The amount of the PCR product is determined by measuring the fluorescence intensity of the 323-bp band on an electrophoresis gel image with ImageJ software. Figure 3 shows the relative amplification efficiency and the amplification reproducibility of each station. RSDs in amplification efficiency are 2.5, 3.8, 5.7, and 4.6 % for stations S1, S2, S3, and S4, respectively, and the station-to-station RSD is 4.2 %, which would be acceptable for practical use. Furthermore, PCR for 323 bp DNA fragment with serial diluted template DNA in a range of 4.4–4.4 × 106 copy/μL reaction solution are carried out through our multi-station PCR device in order to find out the minimum amount of initial DNA required for the reaction. As a result, 44 copy of template DNA/μL reaction solution is the minimum limit amount of initial DNA for visible band of PCR product in agarose gel electrophoresis.
Triplicate amplifications of 323 bp DNA fragments from four stations (S1–S4) of a continuous-flow multi-station PCR device are done to test the reproducibility. PCR products are analyzed by an agarose gel electrophoresis, and fluorescence intensities of the DNA bands are calculated by ImageJ software. RSDs of S1–S4 are 2.5, 3.8, 5.7, and 4.6 %, respectively, and RSD between the four stations in the three consecutive runs is 4.2 %. DNA amplification is done at 92-55-72 °C for denaturation-annealing-extension with a flow rate of 3 μL/min
Segmented-flow PCR
Our capillary-based PCR device can run PCR amplifications of different DNA samples in a sequential injection mode forming a segmented flow. However, in this case, negligible cross-contamination between different PCR segments should be secured. To cleanse the capillary surface contaminated by preceding segments, washing solution segments of 1× gel loading buffer are interposed between sample segments so that sample and washing solution segments are arranged alternately. Since the color of the washing solution is blue, this arrangement help to locate each sample segment in the segmented-flow mode. To check the possibility of carry-over contamination, we let a PCR mixture segment without the primers, the template DNA, or Taq polymerase for the 323 bp DNA (negative controls) follows the complete PCR mixture segment for 323-bp amplification without a washing solution segment between the complete and a negative control segments. We have found that carry-over contamination is not significant in the segment without Taq polymerase or the primers (Fig. 4, lanes 2 and 4, respectively), but serious in one without the template DNA (Fig. 4, lane 3). The DNA band in lane 3 is an amplified product of 323 bp, and a few template DNA molecules adsorbed onto the reactor surface may be served as template for the following PCR. Such carry-over is almost discouraged with 5 μL of the cleansing buffer (Fig. 4, lane 5). These results from the carry-over tests demonstrate the high resistance of Teflon to adsorption of biomolecules. When glass, silicon, and other polymers such as poly(dimethylsiloxane) are used for the material of PCR reactors, the inner surface of the reactor should be almost always chemically modified to gain resistance to adsorption [22, 31–36]. It is also noted that the Teflon capillary reactor is essentially the only expendable in our PCR system.
Agarose gel electropherogram of amplified DNA fragments from a continuous-flow multi-station PCR device. Lane 1, 323 bp amplified DNA fragment as a positive control; lanes 2–4, PCR products in the absence of Taq polymerase (negative control 1), template DNA (negative control 2), and primers (negative control 3) without washing after amplifying 323 bp DNA fragment; lane 5, PCR product of negative control 2 with washing (5 μL of 1× gel loading buffer) the capillary after amplifying 323 bp DNA fragment. DNA amplification is done at 92-55-72 °C for denaturation-annealing-extension with a flow rate of 3 μL/min
To demonstrate the segmented-flow PCR capability of our device, four PCR mixtures for amplifying different DNA fragments with a wide size range (2836, 1101, and 323 bp) are injected in series into the capillary PCR reactor. Figure 5 shows that different DNA fragments can be amplified sequentially (shown in the D lanes) with efficiency similar to that of the commercial, batch-mode PCR machine (shown in the M lanes). Fluorescence intensity of each DNA band is estimated with imageJ, where relative differences between two PCR systems are below 12 % (see ESM Table S2). The PCR product of the preceding segment does not appear in the following segment, as well as in the washing solution segment in between (shown in the B lanes), which confirms that carry-over contamination is effectively discouraged in this segmented-flow PCR with the intervening washing solution segments. The volume of each sample segment has been made constant by using an HPLC injection valve equipped with a fixed-volume sample loop. The volume is 5 μL, which is nearly the lowest limit that can be provided by using such an HPLC injection system. In the sequence of samples at our present segmented-flow pattern, one period corresponding to the interval of sample injection consists of four segments: a sample, a perfluorodecalin, a washing solution, and another perfluorodecalin segment. Since the total volume of one period is about 14 μL and the maximum flow rate used in this work is 3 μL/min, the maximum throughput of PCR amplification with the present setup would be about 5 min per sample.
Agarose gel electropherogram of amplified DNA fragments in a segmented-flow mode (M commercialized PCR machine, D continuous-flow multi-station PCR device, B washing buffer). 1–3 indicate amplified DNA fragments with length of 2836, 1101, and 323 bp, and each DNA fragment comes from distinct regions of the same template DNA or from different template DNA. DNA amplification is done at 92-55-72 °C for denaturation-annealing-extension with a flow rate of 0.2 μL/min
Conclusions
Our multi-station device that covers the general annealing-temperature range simplifies the conventional two-step DNA amplification process (optimum annealing temperature determination and then PCR amplification) into a single-step one, where the solution of an amplified DNA fragment free from non-specific PCR products flows continuously from one station at least. The device has been designed to minimize the number of heating blocks in the miniaturized format and to ensure the reproducibility in DNA amplification at every station. Although the sequential PCR of different DNA samples under a segmented-flow mode has been demonstrated to show potential applicability to high-throughput DNA analysis without optimum annealing temperatures provided, the present sample volume of each segment, 5 μL, is not small enough to claim a high-throughput. The sample volume should be reduced to the level of nL or pL by a combination with an adequate segmentation technique [37, 38] to attain the full applicability. Since the device is based on the use of capillary reactors, it has great potential for expanding into a micro total DNA analysis system including modules of cell lysis/DNA extraction, optimization-free PCR, PCR product analysis, etc. Our group has reported a continuous DNA analysis microchip that can be interfaced with a segmented-flow PCR system [39], and successfully demonstrated on-line analysis of different DNA samples amplified under a segmented-flow mode from the multi-station PCR device [40].
References
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, et al. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–4.
Jung JH, Choi SJ, Park BH, Choi YK, Seo TS. Ultrafast rotary PCR system for multiple influenza viral RNA detection. Lab Chip. 2012;12:1598–600.
Butler JM. Forensic STR typing: biology, technology and genetics of STR markers. 2nd ed. New York: Elsevier; 2005.
Liu P, Seo TS, Beyor N, Shin K-J, Scherer JR, Mathies RA. Integrated portable polymerase chain reaction-capillary electrophoresis microsystem for rapid forensic short tandem repeat typing. Anal Chem. 2007;79:1881–9.
Regan JF, Makarewicz AJ, Hindson BJ, Metz TR, Gutierrez DM, Corzett TH, et al. Environmental monitoring for biological threat agents using the autonomous pathogen detection system with multiplexed polymerase chain reaction. Anal Chem. 2008;80:7422–9.
Zhang Y, Ozdemir P. Microfluidic DNA amplification–a review. Anal Chim Acta. 2009;638:115–25.
Zhang Y, Jiang H-R. A review on continuous-flow microfluidic PCR in droplets: advances, challenges and future. Anal Chim Acta. 2016;914:7–16.
Phaneuf CR, Pak N, Forest CR. Modeling radiative heating of liquids in microchip reaction chambers. Sensors Actuators A. 2011;167:531–6.
Amasia M, Cozzens M, Madou MJ. Centrifugal microfluidic platform for rapid PCR amplification using integrated thermoelectric heating and ice-valving. Sensors Actuators B. 2012;161:1191–7.
Sugumar D, Kong LX, Ismail A, Ravichandran M, Yin LS. Rapid multi sample DNA amplification using rotary-linear polymerase chain reaction device (PCRDisc). Biomicrofluidics. 2012;6:014119.
Zhang R, Gong H-Q, Zeng X, Lou CP, Sze CC. A microfluidic liquid phase nucleic acid purification chip to selectively isolate DNA or RNA from low copy/single bacterial cells in minute sample volume followed by direct on-chip quantitative PCR assay. Anal Chem. 2013;85:1484–91.
Houssin T, Cramer J, Grojsman R, Bellahsene L, Colas G, Moulet H, et al. Ultrafast, sensitive and large-volume on-chip real-time PCR for the molecular diagnosis of bacterial and viral infections. Lab Chip. 2016;16:1401–11.
Hartung R, Brösing A, Sczcepankiewicz G, Liebert U, Häfner N, Dürst M, et al. Application of an asymmetric helical tube reactor for fast identification of gene transcripts of pathogenic viruses by micro flow-through PCR. Biomed Microdevices. 2009;11:685–92.
Peham JR, Grienauer W, Steiner H, Heer R, Vellekoop MJ, Nöhammer C, et al. Long target droplet polymerase chain reaction with a microfluidic device for high-throughput detection of pathogenic bacteria at clinical sensitivity. Biomed Microdevices. 2011;13:463–73.
Wu W, Lee NY. Three-dimensional on-chip continuous-flow polymerase chain reaction employing a single heater. Anal Bioanal Chem. 2011;400:2053–60.
Chung KH, Choi YH, Park S. Development of a continuous-flow polymerase chain reaction device utilizing a polymer disk with a spiral microchannel of gradually varying width. Sensors Actuators B. 2014;191:75–85.
Moschou D, Vourdas N, Kokkoris G, Papadakis G, Parthenios J, Chatzandroulis S, et al. All-plastic, low-power, disposable, continuous-flow PCR chip with integrated microheaters for rapid DNA amplification. Sensors Actuators B. 2014;199:470–8.
Shu B, Zhang C, Xing D. Segmented continuous-flow multiplex polymerase chain reaction microfluidics for high-throughput and rapid foodborne pathogen detection. Anal Chim Acta. 2014;826:51–60.
Tachibana H, Saito M, Tsuji K, Yamanaka K, Hoa LQ, Tamiya E. Self-propelled continuous-flow PCR in capillary-driven microfluidic device: microfluidic behavior and DNA amplification. Sensors Actuators B. 2015;206:303–10.
Liu P, Li X, Greenspoon SA, Scherer JR, Mathies RA. Integrated DNA purification, PCR, sample cleanup, and capillary electrophoresis microchip for forensic human identification. Lab Chip. 2011;11:1041–8.
Jha SK, Chand R, Han D, Jang Y-C, Ra G-S, Kim JS, et al. An integrated PCR microfluidic chip incorporating aseptic electrochemical cell lysis and capillary electrophoresis amperometric DNA detection for rapid and quantitative genetic analysis. Lab Chip. 2012;12:4455–64.
Jiang X, Jing W, Zheng L, Liu S, Wu W, Sui G. A continuous-flow high-throughput microfluidic device for airborne bacteria PCR detection. Lab Chip. 2014;14:671–6.
Rychlik W, Spencer WJ, Rhoads RE. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990;18:6409–12.
Zhang C, Xing D. Microfluidic gradient PCR (MG-PCR): a new method for microfluidic DNA amplification. Biomed Microdevices. 2010;12:1–12.
Park N, Kim S, Hahn JH. Cylindrical compact thermal-cycling device for continuous-flow polymerase chain reaction. Anal Chem. 2003;75:6029–33.
Fuchiwaki Y, Saito M, Wakida S-I, Tamiya E, Nagai H. A practical liquid plug flow-through polymerase chain-reaction system based on a heat-resistant resin chip. Anal Sci. 2011;27:225–30.
Njoroge SK, Witek MA, Battle KN, Immethun VE, Hupert ML, Soper SA. Integrated continuous flow polymerase chain reaction and micro-capillary electrophoresis system with bioaffinity preconcentration. Electrophoresis. 2011;32:3221–32.
Zhang P, Nan H, Lee M-J, Kang SH. Ultra-fast separation of infectious disease-related small DNA molecules by single- and multi-channel microchip electrophoresis. Talanta. 2013;106:388–93.
Wittwer CT, Garling DJ. Rapid cycle DNA amplification: time and temperature optimization. BioTechniques. 1991;10:76–83.
Li Y, Zhang C, Xing D. Fast identification of foodborne pathogenic viruses using continuous-flow reverse transcription-PCR with fluorescence detection. Microfluid Nanofluid. 2011;10:367–80.
Sugumar D, Ismail A, Ravichandran M, Aziah I, Kong LX. Amplification of SPPS150 and Salmonella typhi DNA with a high throughput oscillating flow polymerase chain reaction device. Biomicrofluidics. 2010;4:024103.
Shaw KJ, Joyce DA, Docker PT, Dyer CE, Greenway GM, Greenman J, et al. Development of a real-world direct interface for integrated DNA extraction and amplification in a microfluidic device. Lab Chip. 2011;11:443–8.
Wu W, Kang K-T, Lee NY. Bubble-free on-chip continuous-flow polymerase chain reaction: concept and application. Analyst. 2011;136:2287–93.
Chen JJ, Shen CM, Ko YW. Analytical study of a microfludic DNA amplification chip using water cooling effect. Biomed Microdevices. 2013;15:261–78.
Trinh KTL, Wu W, Lee NY. Planar poly(dimethylsiloxane) (PDMS)–glass hybrid microdevice for a flow-through polymerase chain reaction (PCR) employing a single heater assisted by an intermediate metal alloy layer for temperature gradient formation. Sensors Actuators B. 2014;190:177–84.
Chen JJ, Liao MH, Li KT, Shen CM. One-heater flow-through polymerase chain reaction device by heat pipes cooling. Biomicrofluidics. 2015;9:014107.
Lim K, Kim S, Hahn JH. Roller-type squeezing pump with picoliter handling capability. Sensors Actuators B. 2003;92:208–14.
Ha JW, Hahn JH. Acupuncture sample injection for microchip capillary electrophoresis and electrokinetic chromatography. Anal Chem. 2016;88:4629–34.
Hahn JH, Kwak BJ, Kim H-O, Microchip for analyzing nucleic acid and method of nucleic acid analysis using the same. Patent 9,017,941. 2015.
Kim H-O, Kwak B-J, Hahn JH. Compact continuous-flow PCR system and on-line DNA analysis. Orlando: Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy 2012: Proceedings of the PITTCON 2012 Symposium; 2012. 11–15 March 2012 (PITTCON 2012).
Acknowledgments
This research was supported by the R&D Center for Green Patrol Technologies through the R&D for Global Top Environmental Technologies (RE201604086) funded by the Ministry of Environment, Republic of Korea (MOE). This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2015R1A2A2A04005445).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 152 kb)
Rights and permissions
About this article
Cite this article
Kim, H., Park, N. & Hahn, J.H. Parallel-processing continuous-flow device for optimization-free polymerase chain reaction. Anal Bioanal Chem 408, 6751–6758 (2016). https://doi.org/10.1007/s00216-016-9798-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9798-z