Abstract
Massive production of nanomaterials poses a high risk to environmental ecology and human health. However, comprehensive understanding of nanotoxicity is still a major challenge due to the limitations of assessment methods, especially at the molecular level. We developed a new, sensitive, and robust fingerprinting surface-enhanced Raman spectroscopy (SERS) approach to interrogate both dose- and time-dependent phenotypic bacterial responses to zinc oxide nanoparticles (ZnO NPs). SERS enhancement was provided by biocompatible Au NPs. Additionally, a novel vacuum filtration-based strategy was adopted to fabricate bacterial samples with highly uniform SERS signals, ensuring the acquisition of robust and independent spectral changes from ZnO NPs-impacted bacteria without undesirable spectral variations. Combined with multivariate analysis, clear and informative spectral alteration profiles were obtained. Much greater alterations were found in low-dose ranges than high-dose ranges, indicating a reduction in the bioavailability of ZnO NPs with doses. Time-resolved bacterial responses provided important information on toxic dynamics, i.e., rapid action of ZnO NPs within 0.5 h was identified, and ZnO NPs at low doses and long exposure time exerted similar effects to high doses, indicating the concerns associated with low-dose exposure. Further analysis of biochemical changes revealed metabolic activity decrease over both incubation time and doses. Meanwhile, a short-term protection strategy of bacteria by producing lipid-containing outer membrane vesicles to mitigate the cell of toxic NPs was suggested. Finally, Zn2+ ions released from NPs were demonstrated to be irrelevant to bacterial responses on both dose and time scales. The new SERS methodology can potentially profile a large variety of toxic NPs and advance our understanding of nanotoxicity.

A highly uniform SERS signal of bacteria negating undesired spectral variation via a novel vacuum filtration-based strategy, combined with multivariate PCA-LDA analysis, was utilized to interrogate both dose- and time-dependent antibacterial effect of zinc oxide nanoparticles, and can be extended to a variety of other toxic nanoparticles.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Massive industrial production of engineered nanoparticles (NPs) raises great concerns on the health of living organisms. Inorganic nanoparticles are among the most widely used nanomaterials (http://www.nanotechproject.org) in both consumer and industrial products, such as cosmetics (ZnO, TiO2), antimicrobial reagents (Ag), environmental remediation (Fe x O y , TiO2), and semiconductors (CeO2, Al2O3, SiO2) [1]. They can easily become environmental nano-contaminants during production, utilization, or disposal processes. A great variety of inorganic nanomaterials have been demonstrated to be toxic to microorganisms [1–6]. As engineered nanoparticles are not formed naturally, environmental microorganisms that play important roles in biodegradation, nutrient cycling, and ecological balance may have not developed appropriate coping strategies. To achieve sustainable use of nanomaterials, comprehensive understanding of antibacterial effects and their mechanism is crucial for protecting ecological and human health.
Traditional culture-based analysis such as growth curves [2, 7], enumeration of colony forming units (CFUs) [8, 9], and determination of minimum inhibitory concentration (MIC) [10] can clearly indicate toxic dosages, but provides no information on action mechanism. Morphology analysis including scanning electron microscopy (SEM) and transmission electron microscopy (TEM) is limited to visualization of severe damage to cell membrane and release of intracellular content [8, 9]. Correlations of reactive oxygen species (ROS) production and/or dissolved metal ions of NPs with antibacterial activity of NPs provide a way to seek the origin of nanotoxicity [11, 12]. However, these conventional toxicology methods do not address the action mechanisms and dynamics of NPs at molecular level. For this purpose, reverse transcription-quantitative PCR (RT-qPCR) [8], genomics [7], and proteomics are applied to reveal nanotoxicity at molecular level [13]. For example, RT-qPCR analysis showed that the expression level of two oxidative stress genes and a general stress response gene increased, indicating that ZnO NPs induced oxidative stress in Campylobacter [8]. In another study, a total of 46 proteins using a proteomics-based approach displayed significant changes after bronchoalveolar lavage fluid in rats was exposed to ZnO NPs, indicating that ZnO NPs mainly impacted the immune and inflammatory processes [14]. However, these methodologies are complex, costly, and time-consuming. Clearly, new tools are required in order to strengthen the arsenal of molecular level-based techniques to study antibacterial NPs.
Recent progress in Raman spectroscopy has provided new research opportunities in in situ and non-destructively studying bacterial metabolic and physiological responses to toxic NPs [9]. Raman spectroscopy is based on the measurement of the vibrational energy of chemical bonds, and has been employed to investigate antibacterial mechanism of various NPs including ZnO [9], Ag, Fe3O4 [15], and carbon nanomaterials [5, 16, 17]. Alterations in the specific Raman bands of bacteria-related biomolecules, such as nucleic acids, proteins, lipids, carbohydrates, and metabolites, are indicative of phenotypic cellular responses to NPs [18]. For example, Liu et al. proposed the destruction of the cell membrane and the leakage of intracellular contents after exposure to ZnO NPs due to increase of lipid and protein Raman bands [9]. However, normal Raman spectroscopy suffers from two major challenges for nanotoxicity studies: (1) disadvantages of weak signal and relatively long acquisition time due to the very low Raman scattering efficiency (one Raman photon out of 108 incident photons) [19], which is even more prominent when a large number of spectra and samples are required for statistical analysis in toxicology studies; (2) fluorescence emitted from some NPs, such as carbon-based nanomaterials, may interfere and even overwhelm the Raman signal of bacteria [17].
Surface-enhanced Raman scattering (SERS) addresses these problems by dramatically enhancing the Raman signal, reducing acquisition time and quenching fluorescence interference. Relying on the significant electromagnetic enhancement provided by silver or gold NPs, molecules adsorbed or in close proximity to NPs can be enhanced by 106–1014 fold over normal Raman scattering [20–24]. This gives SERS an ultra-high detection sensitivity even down to the single molecule level. Additionally, its ability to quench fluorescence from nanomaterials or impurities broadens its application to more diverse NPs versus normal Raman. For these reasons, SERS has recently demonstrated its first application in studying the nanotoxicity of Ag NPs against bacteria [6]. Taking advantage of the self-provided electromagnetic enhancement by Ag NPs, SERS achieved in situ, non-destructive, and rapid interrogation of size and medium-dependent antibacterial activity of Ag NPs [25]. Furthermore, the impact of Ag NPs on the metabolic processes of purine and protein synthesis was revealed based on dramatic spectral changes in protein, hypoxanthine, adenosine, and guanosine [6]. However, strong SERS enhancements were mainly from several specific metallic nanoparticles such as Ag, Au, Cu, while a large majority of engineered NPs are not SERS-active, such as ZnO, TiO2, carbon nanomaterials, quantum dots, etc. Instead, they exhibit Raman and/or fluorescence signatures which might interfere with the Raman signatures of bacteria.
To apply SERS to NPs more universally, biocompatible and SERS-active Au NPs were utilized to enhance the SERS signal of bacteria impacted by toxic NPs. The biocompatibility of Au NPs has been demonstrated in our previous works by observing that Au NPs exerted no effect on bacterial viability, growth curve and SERS spectral signatures [6, 26]. In addition, to acquire robust and independent ZnO-induced biochemical changes, undesired intensity variations caused by uneven sample geometry or SERS enhancement have to be eliminated. To achieve this, a new sample fabrication strategy featuring highly uniform bacterial sample thickness and SERS signal across multiple samples was developed via vacuum suction filtration. By employing this method and SERS enhancement provided by Au NPs, SERS combined with principal component analysis and linear discriminant analysis (PCA-LDA) was applied to interrogate both dose- and time-dependent antibacterial effect of ZnO NPs, one of the most widely used nanomaterials in both consumer and industrial products [27, 28]. Bacterial responses to sublethal and lethal doses of ZnO NPs over time were monitored. This not only allowed study of dosage effect based on spectral changes at identical exposure time, but also revealed toxic dynamics by monitoring time-resolved spectral changes. Furthermore, the relationship between dose- and time-dependent toxic effects and action mechanism of ZnO NPs was also examined. These studies will advance and supplement our understanding of nanotoxicity by providing new insights with a new technique.
Materials and methods
Synthesis of Au NPs
Au NPs were prepared by using trisodium citrate as the reductant and capping reagent according to Frens’ method [29]. Detailed procedure was described in Electronic Supplementary Material (ESM). The prepared Au NPs exhibit an oval shape with the dominant size at long axis being around 120 nm (Fig. 2b), the same as that in our previous work, where more detailed information on SEM image, size distribution, and biocompatibility were displayed [6, 26]. Before being used for SERS enhancement, Au NPs were washed once by ultrapure water (Millipore, USA) and concentrated through centrifugation at 2900 rpm for 6 min (Eppendorf centrifuge 5430R, Germany). The supernatant was discarded, and the precipitate was collected for SERS sample preparation.
Bacterial viability assay
Escherichia coli were chosen as the model microorganism and grown in Luria-Bertani broth (LB, containing 5 g/L NaCl, 10 g/L tryptone, and 5 g/L yeast extract) at 37 °C with constant shaking for 12–13 h. The cultures were then diluted with fresh LB to reach an initial OD at 600 nm (OD600) of 0.2, washed once with sterile ultrapure water and resuspended in the same volume of ultrapure water. A proper volume of ZnO NPs (Beijing DK nano technology Co., Ltd, China) with size of 278 ± 133 nm at the long axis (TEM observation in Fig. 1a) was suspended in water and added to bacterial solution to reach a final concentration of 0.6, 3, 15, 60, 240, 480 mg/L. The mixtures were then incubated for different time intervals of 0.5, 1.5, 3, 6, 12, 24 h at 37 °C with constant shaking in a dark environment. Bacterial cultures in ZnO NPs-free ultrapure water under the same conditions were used as control. At each predetermined time, treated cultures were serially diluted, and then spread onto LB agar plates in duplicate and incubated overnight at 37 °C. Each viable cell formed a visible colony, allowing enumeration of viable cells by our naked eyes.
Preparation of uniform SERS sample
After each predetermined treatment time as described above in the viability assay, treated bacteria were centrifuged at 8000 rpm for 5 min to discard supernatant with bacterial pellet left in 40–70 μL water depending on the pellet size. Fourteen microliters of concentrated Au NPs (∼8000 mg/L) was added to cell pellet, and thoroughly mixed cell and Au NPs solution was then dropped on a 6-mm-diameter mixed cellulose ester membrane (MCE, Catno.VCWP02500, Millipore) with a pore size of 0.1 μm placed on a glass vacuum filter. After a rapid vacuum suction filtration with only water passing through, bacteria and Au NPs mixture dried rapidly and homogenously on membrane surface, and used for further SERS and SEM tests. All treatments were performed in triplicate.
SERS measurement
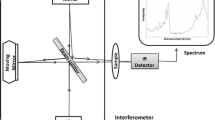
SERS spectra were acquired from a LabRAM Aramis (HORIBA Jobin-Yvon) confocal micro-Raman system equipped with a 600 g/mm grating. Excitation was provided by a He-Ne 632.8 nm laser with a power of 70 μW on the sample. A ×50 objective (Olympus) with a numerical aperture of 0.55 and a working distance of 8 mm was used to focus laser beam and collect Raman signal. DuoScan in the micro-mapping mode with a scanning area of 30 μm × 30 μm based on the combination of two mirrors scanning rapidly the laser beam across the chosen area, was used in order to reduce the possible damage of laser to bacteria. Spectral acquisition time was 5 s. Five SERS spectra from five different areas of one sample and 15 spectra from triplicate samples following one treatment were acquired.
Spectra data pre-processing and PCA-LDA analysis
SERS spectra were pre-processed first via baseline correction by fitting a five-order polynomial to the baseline and subtracting from spectra using LabSpec5 software (HORIBA Jobin-Yvon), and then wavelet de-noising via IrootLab toolbox (https://code.google.com/p/irootlab/) running on MATLAB 2012a [30]. Following pre-processing, class means spectra with standard deviation and PCA-LDA were preformed on all spectra dataset using IrootLab toolbox. PCA-LDA is a statistical analysis method where PCA is applied to reduce the dimensionality of spectral data to 10–20 factors accounting for more than 99 % of the total variance, and LDA is subsequently employed to derive orthogonal variables from which the between-class variance is maximized over the within-class variance. PCA-LDA output data were visualized as 1D scores plots. Nearness or distance of plot indicates similarity or dissimilarity of spectra between treatment class and control class. This allows dose- or time-dependent bacterial response to be visualized based on distance of each treatment in the PCA-LDA scores plot derived from SERS spectra.
SEM test
For SEM characterization, the above prepared bacterial samples were fixed with 2.5 % glutaraldehyde in 0.1 M phosphate buffer (PBS, pH 7.2) for 2 h, and then dehydrated with sequential treatments by 30, 50, 70, 90, and 100 % ethanol for 15 min, followed by critical point drying overnight (Auto-Samdri 815 Automatic Critical Point Dryer; Tousimis, Rockville, MD, USA). The dried samples were sputtered and coated with gold for SEM imaging by using a scanning electron microscopy (SEM, Hitachi S-4800).
Results and discussion
Dose- and time-dependent viability assay of bacteria exposed to ZnO NPs
To assess the effect of ZnO NPs on bacterial viability, E. coli at an initial colony forming unit (CFU)/mL of 3 × 107 were incubated with different concentrations of ZnO NPs (0, 0.6, 3, 15, 60, 240, 480 mg/L) for different time in water at 37 °C. Change of bacterial viability was shown in Fig. 1b. Compared with the control that displaying the same bacterial viability after even 24 h, ZnO NPs induced an obvious decline in bacterial viability with both increased concentration and prolonged treating time, demonstrating the antibacterial activity of ZnO NPs. When the concentration of ZnO NPs was no more than 0.6 mg/L, no significant change in bacterial viability was observed before 12 h, and only 1-log10 reduction was induced during 12 to 24 h. When the concentration of ZnO NPs increased to 3 to 60 mg/L, viable cells were reduced by 4.5-log10 after 12 h and almost eliminated after 24 h. A further increase of ZnO NPs to 240 and 480 mg/L induced a rapider bacteria elimination after 12 h treatment. Additionally, bacterial viability in the range of 3–60 mg/L or 240–480 mg/L of ZnO NPs was very close, respectively, possibly owing to the reduced bioavailability of ZnO NPs, because obvious aggregation and precipitation of NPs were clearly observed during incubation, especially at high concentrations.
Highly uniform and robust SERS preparation method proposed for antibacterial study of ZnO NPs
To apply rapid and sensitive SERS technique to interrogate biochemical changes of bacteria impacted by toxic ZnO NPs, not only fingerprinting SERS spectra of bacteria are required, the more important thing is to achieve robust and independent spectral alterations consistent with actual cellular biochemical changes. However, conventional drop-air dry method normally generates bacterial samples with highly uneven thickness due to coffee ring effect [21, 31–33], resulting in some spectral variations unrelated to biochemical changes of bacteria. It is therefore hard to use these spectral information to analyse action mechanism, even with spectral normalization which will also distort spectral information to some extent.
For this purpose, vacuum filtration method by homogeneously and rapidly retaining mixture of ZnO NPs-treated bacteria and SERS-active Au NPs, was employed here to achieve uniform sample thickness and also SERS enhancement. Performance of three types of microfiltration membrane in retaining Au NPs, i.e., 0.1 μm MCE, 0.1 μm polyvinylidene fluoride membrane (PVDF), and 0.22 μm MCE, were compared and the results were shown in Fig. 2a. Three membranes after Au NPs filtration were moved to a new position at the top row in order to observe the leakage at the bottom row (Fig. 2a). It was clear that 0.1 μm MCE rejected all Au NPs, with nothing left on the underneath 8 μm MCE membrane, while both 0.1 μm PVDF and 0.22 μm MCE just partly retained Au NPs with other part penetrating through the membrane and leaking onto the underneath membrane. In addition, the wetting of 8 μm MCE membrane clearly illustrated the penetration of water from mixture during filtration. Hence, MCE with a pore size of 0.1 μm was chosen as the carrier for all bacterial sample preparation. SEM images illuminated the morphology and distribution of Au NPs (Fig. 2b), as well as the mixture of bacteria treated by different dosages of ZnO NPs and Au NPs (Fig. 2c–f) on membrane surface. It can be clearly observed that Au NPs were distributed closely around bacteria cells to provide SERS enhancement with an obvious increase of ZnO NPs at higher ZnO NPs dosage.
SERS spectra were obtained directly on these samples. To evaluate the advantage of SERS over normal Raman in offering high-quality signal with minimized interference from NPs or substrates, normal Raman and SERS spectra of E. coli cells exposed to 0, 3, 60, 480 mg/L of ZnO NPs for 0.5 h were compared (Fig. 3a, b). Normal Raman samples were prepared using the same vacuum filtration without adding Au NPs. ZnO NPs exhibited a distinctive Raman band at 438 cm−1, which also appeared at cells treated with the highest 480 mg/L of ZnO NPs (ZnO480) (Fig. 3a). In addition, MCE membrane displayed two obvious Raman bands at 856 and 1290 cm−1, which showed up in all cell samples but decreased with increasing concentrations of ZnO NPs, due to the coverage of ZnO NPs on MCE. Unfortunately, Raman peaks associated with bacteria were too weak to be observed by normal Raman, even with two-fold higher laser power (140 μW) and six-fold longer acquisition time (30 s) than SERS. In contrast, SERS spectra of bacteria exhibited rich Raman bands representing typical biochemical components of bacteria, such as 733, 1320, and 1580 cm−1 (adenosine-related), 1000, 1028, 1241, and 1634 cm−1 (protein-related), and 1453 and 1469 cm−1 (CH2 of lipid or protein), consistent with previous publications on SERS spectra of bacteria [34]. In addition, no Raman bands of ZnO or MCE membrane were observed in SERS spectra. This is due to the short-distance enhancement effect of SERS which only allows Raman signal of substance adsorbed or in close proximity to Au NPs to be enhanced. Compared with rigid ZnO NPs or MCE membrane underneath, bacteria-related compositions can adsorb on or contact with Au NPs much better, enabling a more efficient enhancement to bacteria by Au NPs but minimizing interference of ZnO NPs or substrates. More importantly, SERS spectra experienced prominent changes with the increasing concentrations of ZnO NPs (Fig. 3b), demonstrating the sensitivity of SERS in monitoring biochemical changes of bacteria in response to toxic ZnO NPs.
a Normal Raman and b SERS spectra of E. coli exposed to 0, 3, 60, 480 mg/L of ZnO NPs for 0.5 h. c Histogram of integrated SERS intensity at 733 cm−1 from bacteria treated with different concentrations of ZnO NPs in triplicate. The calculated RSD values from either one sample or triplicate samples were listed in the inset table. ZnO0 was the control without treatment; ZnOX represents treatment with X mg/L of ZnO NPs
The other most important thing is to evaluate the uniformity of SERS signal. Bacterial samples with and without ZnO NPs treatment were prepared in triplicate via vacuum filtration. Twenty-one SERS spectra were acquired from different regions of sample indicated as red squares in the inset of Fig. 3c. Integrated SERS intensity of 733 cm−1 band was used to assess SERS uniformity by calculating relative standard deviation (RSD) from one sample (21 spectra) and also triplicate samples (21 × 3 = 63 spectra), and displayed as a histogram in Fig. 3c with related RSD values listed in the inset table. SERS signal with RSD <10–20 % was generally regarded to be uniform in many previous publications [35, 36]. In our case, RSD of either single sample or triplicate samples varied in a small range between 6.3 and 12.4 %, in addition, RSD of triplicate samples (63 spectra) was very close to that of single sample (21 spectra), demonstrating excellent SERS uniformity and reproducibility across multiple samples. Similar intensity across triplicate samples also indicated a very close enhancement provided by the same amount of Au NPs rejected on MCE membrane. Meanwhile, with concentration of ZnO NPs increasing, uniformity dropped by a small degree with total RSD increasing from 9.2 to 11.2 %, indicating that ZnO NPs negatively impacted the uniform enhancement of Au NPs. This was also confirmed by observing a higher SERS signal uniformity via vacuum filtration in our previous work studying bacterial responses to antibiotic [37]. Even so, RSD value less than 12 % still ensured a high uniformity of SERS signal. SERS spectra of cells prepared by conventional drop-evaporation method on a glass slide (ESM Fig. S1a) were obviously far less uniform than that via vacuum filtration (ESM Fig. S1b) with the calculated RSD of around 20 % being double of that the latter. Apparently, homogenous vacuum filtration avoided large signal variations inevitably appearing in samples fabricated by conventional drop-evaporation method. Therefore, independent spectral changes without interfering variations can represent the actual phenotypic biochemical alterations of bacteria in response to toxic ZnO NPs. The outcome will contribute to understand both dose- and time-dependent antibacterial properties of ZnO NPs.
Dose-dependent antibacterial properties of ZnO NPs
Mean SERS spectra with standard deviation of E. coli cells treated with different concentrations of ZnO NPs for 0.5 h to 24 h were shown in Fig. 4 and ESM Fig. S2. The narrow shadow representing the standard deviation around each mean spectrum indicated the high uniformity of SERS signal. Clear spectral alterations representing the phenotype bacterial response to toxic ZnO NPs at each treatment time were observed, such as bands at 733, 1028, 1320, and 1469 cm−1. To illustrate the dose-induced bacterial response in a more visible and efficient way, each SERS spectrum was plotted as a point in 1D PCA-LDA scores plot (Fig. 4d), where the segregation distance of each point was proportional to spectral dissimilarity. A lot of information was revealed from PCA-LDA scores plot. For instance, at the same treatment time, spectral changes in the low-dose range (0–3 mg/L) was much greater than that in the broad high-dose range (3–480 mg/L). This was consistent with cell viability test (Fig. 1b) that also varied more significantly within 3 mg/L than above. To further illuminate the correlation, PCA-LDA scores representing toxic dose-dependent SERS responses were plotted against the viability assay results (Fig. 5). Pearson’s correlation coefficient (r) derived from the linear fitting via Origin 9.1 was used to evaluate the degree of correlation. Under a confidence level of 95 %, the r values obtained here (Fig. 5) were between −0.99 and −0.72, indicating high and negative correlations between toxicity specific SERS responses and the viability assay [38]. Additionally, dose-induced bacterial responses in shorter treatment time (0.5–3 h) were more variable than longer time (6–24 h). The shift of spectral feature at 0 and 0.6 mg/L to those of high doses after 6 h was contributed to the reduced difference, indicating that bacteria without or with sublethal ZnO NPs (0.6 mg/L) treatment were also physiologically deteriorated with time. After a careful inspection of spectra (Fig. 4a–c and ESM Fig. S2), dose-induced spectral change pattern was summarized in Table 1.
As the concentration of ZnO NPs increased, bands at 733, 965, 1241, and 1580 cm−1 decreased, while 1028 and 1469 cm−1 bands increased, and 1000, 1634 cm−1 were almost unchanged. Moreover, spectral changes could be observed even when cell viability was not affected, such as bands at 733 cm−1, which decreased after treatment with 0.6 mg/L of ZnO NPs for 0.5 h to 6 h without CFU decline (Fig. 4a, b). This indicated that SERS could sensitively monitor the impact of ZnO NPs on cell changes prior to cell death. Liu and Roselli also pointed out that ZnO NPs may alter the intracellular metabolic system in bacteria cells [9, 40].
Time-dependent antibacterial properties of ZnO NPs
Investigation on time-dependent antibacterial properties provides important information on dynamics of cellular response to toxic NPs. As far as we know, studies on dynamic effect were far less than dose effect. In addition, most of dynamic studies focused on temporal evolution of cellular viability and properties of NPs at nano-bio interface, such as dissolution or release of metallic ions, aggregation and sedimentation of NPs, and endocytosis rate [41–44]. However, very few work focused on temporal biochemical responses of bacteria exposed to NPs. Recently, a genome-wide methodology was used to track time-resolved response of genes and pathway to toxicity of zinc-containing nanomaterials [7]. Through sequential screening over 24 h, the results suggested two distinctively early and later bacterial response with toxicity mechanisms changing from pathways related to general metabolic function, transport, signaling, and metal ion homeostasis to that related to membrane synthesis over time [7]. SERS is also a potential way to monitor time-resolved phenotypic bacterial responses. Here, aiming at providing more insights into toxic dynamics, time-dependent SERS spectra of bacteria treated with ZnO NPs from 0.5 h to 24 h were obtained (Fig. 6a–c and ESM Fig. S3). Nonlinear spectral segregation following treatment time was observed in 1D PCA-LDA score plot (Fig. 6d). It was interesting to find that cells without ZnO NPs (ZnO0) treatment experienced a dramatic spectral change, despite no decline in cell viability until 24 h (Fig. 1b). Considering that no nutrient was supplied, the dramatic spectral change should be related to the adaptive response of cells to the unfavorable deionized water environment. With the addition of low-dose ZnO NPs (0.6 mg/L), time-dependent spectral variation amplitude declined with all points moving up, indicating that ZnO NPs aggravated the unhealthy state of cells. When ZnO NPs was imposed at 3 mg/L and above, all spectral points shifted to a similar position a bit higher than that of control at 24 h (ZnO0), resulting in a greatly reduced time-dependent spectral variations. This indicated that toxic action of ZnO NPs at as low as 3 mg/L could rapidly perform within 0.5 h, and deteriorate cells to a state that cells required 24 h to reach in DI water without ZnO NPs. Because 0.5 h was the shortest time to get bacterial sample ready, the action time of toxic ZnO NPs could be even shorter. For the lower dose of ZnO NPs at 0.6 mg/L (ZnO0.6), no spectral difference from the control could be observed at 0.5 h, instead, the obvious action started from 1.5 h. The reason could be related to the diffusion time required for the contact between cells and ZnO0.6. Even though, the spectral point from ZnO0.6 at 24 h was closer to that of higher dosage than control, indicating that low dosage and long exposure time of ZnO NPs imposed a similar effect to high dosage. Hence, low-dosage effect of NPs could also be a major concern in health assessment especially after long-term exposure.
Comparison of time- and dose-induced biochemical changes in bacteria
Phenotypic spectral changes of bacteria over time with and without toxic ZnO NPs treatment were also summarized in Table 1 to facilitate comparison with dosage-induced changes. Variations over time at ZnO0 represented bacterial response to nutrient deprivation. Although cell viability at ZnO0 was not affected with incubation time, obvious spectral changes were observed throughout the period of 24 h of starvation. Interestingly, most of the bands displayed an increase during the initial 3 h and then decreased with the prolonged time (Fig. 6a–c). Among them, 733 and 1320 cm−1 bands were attributed to adenosine-related substances including adenosine triphosphate (ATP), because of their almost identical spectral features (ESM Fig. S4). ATP is the energy source for bacterial activity and bacterial cells have been demonstrated to secrete extracellular ATP [45, 46], explaining their appearance in bacterial SERS spectra where only Raman signal of biomolecules in contact with Au NPs outside of bacteria can be enhanced. Other bands with such parabola-like change were mainly protein-related, i.e., 1000, 1241, 1453, 1580, and 1634 cm−1. Increased bacterial activities in respiratory and protein-synthesizing systems were found during the first hours of nutrient and energy depletion and then decreased in the long term [47]. In addition, bacteria adopted a survival strategy by maintaining a relatively low energy consumption to cope with long-term nutrient deficit. These findings were consistent with our observation of bands decreasing subsequent to an initial increase of extracellular ATP and protein bands, indicative of an active cellular response to the deteriorating conditions in the initial phase before a decrease in bacterial metabolic activity.
Compared with the above single nutrient deprivation-induced bacterial response, the addition of toxic ZnO NPs (0.6–480 mg/L) induced a direct decrease in adenosine- and protein-related bands with a prolonged incubation time (Table 1). These findings may indicate that toxic ZnO NPs accelerated and aggravated bacterial response, resulting in the absence of initially raised bacterial activity observed at ZnO0. Combined with the declined bacterial viability with time at the presence of ZnO NPs (Fig. 1b), the decreased extracellular ATP and protein bands were related to the decreased metabolic activity of cells. The application of numerical simulation in the future may facilitate the visualization of the intimate link between dose- and time-dependent toxic response [48].
For spectral variation over dosage of toxic ZnO NPs (Table 1), the same decline in ATP- and protein-related bands was observed, indicating a similar decrease in the metabolic activity of bacteria to that over time. However, differences between spectral alteration profiles over dosage and over time were also found. The most prominent differences were the two lipid-related bands at 1469 and 1028 cm−1, which kept increasing with dosage, contrary to the parabolic-like change with time. The increased intensity of lipid bands was previously observed by Liu et al. when using normal Raman spectroscopy to study antibacterial effect of ZnO NPs against pathogen E. coli [9]. Combined with the observation of damaged cell membrane and no viable cell, lipid secretion due to the destruction of cell membrane was suggested to account for increased intensity of lipid band [9]. In our work, increased lipid band intensity was observed as early as 0.5 h without significant decline in bacterial viability (Fig. 4a), it therefore should not be related to cell membrane damage and cell lysis, instead, it could be more likely from active bacterial response to toxic ZnO NPs by secreting more lipids. Production of outer membrane vesicles (OMV) mainly composed of lipids was reported as a bacterial stress response to antibacterial treatments, and protected bacterial survival by quickly relieving cell of toxic materials [49, 50]. This may explain the increase of lipid bands over increasing dosage of antibacterial ZnO NPs. In addition, production of OMV is an energy-consuming process, suggesting that OMV-mediated lipid production is an immediate and short-term solution to antibacterial reagents [49]. This is also consistent with our finding of time-dependent change of lipid bands which increased at the first 3 h but decreased subsequently with a prolonged time (Fig. 6b, c).
Presently, there are still discrepancies on whether the nanotoxicity origins primarily from the released Zn2+ ions or ZnO NPs [51–53]. To understand this problem, we compared dissolved Zn2+ ions and SERS-revealed phenotypic bacterial responses at both dosage and time scales. The concentration of Zn2+ ions released from 3, 60, and 480 mg/L of ZnO NPs increased rapidly from 0.5 to 6 h and then became stable until 24 h (Fig. 7), with that from 60 and 480 mg/L of ZnO NPs being much closer than 3 mg/L. This trend was not consistent with changes in cellular biochemical responses, which were rapidly triggered within 0.5 h and then became relatively stable (Fig. 6d), and were much closer for 3 and 60 mg/L than 480 mg/L (Fig. 4d). Therefore, toxicity of ZnO NPs was not directly correlated with Zn2+ ions released from NPs.
Conclusions
SERS was successfully employed to study the antibacterial properties of inorganic and non-SERS-active ZnO NPs, by adding biocompatible and SERS-active Au NPs to enhance the SERS signatures of bacteria impacted by toxic NPs. More importantly, a novel vacuum filtration-based sample fabrication strategy was developed to acquire robust and independent biochemical changes in bacteria without undesired spectral variations. Combined with PCA-LDA multivariate analysis, clear dose- and time-dependent spectral alterations representing phenotypic bacterial responses to ZnO NPs were observed. Spectral changes in the low-dose range (0–3 mg/L) was found to be much greater than the broad high-dose range (3–480 mg/L), indicating a reduced bioavailability of ZnO NPs at higher doses. Time-dependent antibacterial properties provided important information on toxic dynamics. Toxic action of ZnO NPs as low as 3 mg/L executed rapidly within 0.5 h, and deteriorated cells to a state in which cells required 24 h to reach in DI water without ZnO NPs. In addition, ZnO NPs at low dose and long exposure time gave a similar effect to high-dosage samples, indicating low-dose effect of NPs could also be a major concern to health after long-term exposure.
Time- and dose-induced biochemical changes in bacteria in response to ZnO NPs were also compared. A decrease in metabolic activity with the incubation time and doses of ZnO NPs was suggested by inspecting the common decline in ATP- and protein-related bands. In addition, lipid-related bands increased over doses but displayed a parabolic-like change over time. This indicated that bacteria possibly adopted a short-term protection strategy by producing outer membrane vesicles containing lipids to expel toxic NPs. Finally, a comparison between spectral changes and Zn2+ ions released from NPs on both dose and time scales showed that the toxicity of ZnO NPs was not caused by the released ions.
The new SERS method presented here is also applicable for diverse inorganic nanomaterials. Considering the rapid growth of nanotechnology and the emergence of new nanomaterials, SERS can be used to compare and categorize the nanotoxicity of presently existing and/or newly emerged toxic NPs.
References
Maurer-Jones MA, Gunsolus IL, Murphy CJ, Haynes CL. Toxicity of engineered nanoparticles in the environment. Anal Chem. 2013;85:3036–49.
Jones N, Ray B, Ranjit KT, Manna AC. Antibacterial activity of ZnO nanoparticle suspensions on a broad spectrum of microorganisms. FEMS Microbiol Lett. 2008;279:71–6.
Adams LK, Lyon DY, Alvarez PJ. Comparative eco-toxicity of nanoscale TiO2, SiO2, and ZnO water suspensions. Water Res. 2006;40:3527–32.
Thuptimdang P, Limpiyakorn T, McEvoy J, Prüß BM, Khan E. Effect of silver nanoparticles on Pseudomonas putida biofilms at different stages of maturity. J Hazard Mater. 2015;290:127–33.
Li J, Strong R, Trevisan J, Fogarty SW, Fullwood NJ, Jones KC, et al. Dose-related alterations of carbon nanoparticles in mammalian cells detected using biospectroscopy: potential for real-world effects. Environ Sci Technol. 2013;47:10005–11.
Cui L, Chen P, Chen S, Yuan Z, Yu C, Ren B, et al. In situ study of the antibacterial activity and mechanism of action of silver nanoparticles by surface-enhanced Raman spectroscopy. Anal Chem. 2013;85:5436–43.
Reyes VC, Li M, Hoek EM, Mahendra S, Damoiseaux R. Genome-wide assessment in Escherichia coli reveals time-dependent nanotoxicity paradigms. ACS Nano. 2012;6:9402–15.
Xie Y, He Y, Irwin PL, Jin T, Shi X. Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Appl Environ Microbiol. 2011;77:2325–31.
Liu Y, He L, Mustapha A, Li H, Hu ZQ, Lin M. Antibacterial activities of zinc oxide nanoparticles against Escherichia coli O157:H7. J Appl Microbiol. 2009;107:1193–201.
Siddique S, Shah ZH, Shahid S, Yasmin F. Preparation, characterization and antibacterial activity of ZnO nanoparticles on broad spectrum of microorganisms. Acta Chim Slov. 2013;60:660–5.
Xia T, Kovochich M, Liong M, Mädler L, Gilbert B, Shi H, et al. Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties. ACS Nano. 2008;2:2121–34.
Feris K, Otto C, Tinker J, Wingett D, Punnoose A, Thurber A, et al. Electrostatic interactions affect nanoparticle-mediated toxicity to gram-negative bacterium Pseudomonas aeruginosa PAO1. Langmuir. 2009;26:4429–36.
Ivask A, Suarez E, Patel T, Boren D, Ji Z, Holden P, et al. Genome-wide bacterial toxicity screening uncovers the mechanisms of toxicity of a cationic polystyrene nanomaterial. Environ Sci Technol. 2012;46:2398–405.
Juang YM, Lai BH, Chien HJ, Ho M, Cheng TJ, Lai CC. Changes in protein expression in rat bronchoalveolar lavage fluid after exposure to zinc oxide nanoparticles: an iTRAQ proteomic approach. Rapid Commun Mass Spectrom. 2014;28:974–80.
Candeloro P, Tirinato L, Malara N, Fregola A, Casals E, Puntes V, et al. Nanoparticle microinjection and Raman spectroscopy as tools for nanotoxicology studies. Analyst. 2011;136:4402–8.
Knief P, Clarke C, Herzog E, Davoren M, Lyng FM, Meade AD, et al. Raman spectroscopy—a potential platform for the rapid measurement of carbon nanotube-induced cytotoxicity. Analyst. 2009;134:1182–91.
Alimohammadi M, Xu Y, Wang D, Biris AS, Khodakovskaya MV. Physiological responses induced in tomato plants by a two-component nanostructural system composed of carbon nanotubes conjugated with quantum dots and its in vivo multimodal detection. Nanotechnology. 2011;22:295101.
Huang WE, Griffiths RI, Thompson IP, Bailey MJ, Whiteley AS. Raman microscopic analysis of single microbial cells. Anal Chem. 2004;76:4452–8.
Kneipp J, Kneipp H, Kneipp K. SERS—a single-molecule and nanoscale tool for bioanalytics. Chem Soc Rev. 2008;37:1052–60.
Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–6.
Athamneh AIM, Alajlouni RA, Wallace RS, Seleem MN, Senger RS. Phenotypic profiling of antibiotic response signatures in Escherichia coli using Raman spectroscopy. Antimicrob Agents Chemother. 2014;58:1302–14.
Huang JY, Zong C, Xu LJ, Cui Y, Ren B. Clean and modified substrates for direct detection of living cells by surface-enhanced Raman spectroscopy. Chem Commun. 2011;47:5738–40.
Huefner A, Kuan WL, Barker RA, Mahajan S. Intracellular SERS nanoprobes for distinction of different neuronal cell types. Nano Lett. 2013;13:2463–70.
Kubryk P, Kolschbach JS, Marozava S, Lueders T, Meckenstock RU, Niessner R, et al. Exploring the potential of stable isotope (resonance) Raman microspectroscopy and surface-enhanced Raman scattering for the analysis of microorganisms at single cell level. Anal Chem. 2015;87:6622–30.
El-Zahry MR, Mahmoud A, Refaat IH, Mohamed HA, Bohlmann H, Lendl B. Antibacterial effect of various shapes of silver nanoparticles monitored by SERS. Talanta. 2015;138:183–9.
Cui L, Chen SD, Zhang KS. Effect of toxicity of Ag nanoparticles on SERS spectral variance of bacteria. Spectrochim Acta A. 2015;137:1061–6.
Hansen SF, Michelson ES, Kamper A, Borling P, Stuer-Lauridsen F, Baun A. Categorization framework to aid exposure assessment of nanomaterials in consumer products. Ecotoxicology. 2008;17:438–47.
Piccinno F, Gottschalk F, Seeger S, Nowack B. Industrial production quantities and uses of ten engineered nanomaterials in Europe and the world. J Nanoparticle Res. 2012;14:1109.
Frens G. Controlled nucleation for regulation of particle-size in monodisperse gold suspensions. Nat Phys Sci. 1973;241:20–2.
Trevisan J, Angelov PP, Scott AD, Carmichael PL, Martin FL. IRootLab: a free and open-source MATLAB toolbox for vibrational biospectroscopy data analysis. Bioinformatics. 2013;29:1095–7.
Walter A, Reinicke M, Bocklitz T, Schumacher W, Rosch P, Kothe E, et al. Raman spectroscopic detection of physiology changes in plasmid-bearing Escherichia coli with and without antibiotic treatment. Anal Bioanal Chem. 2011;400:2763–73.
Liu TT, Lin YH, Hung CS, Liu TJ, Chen Y, Huang YC, et al. A high speed detection platform based on surface-enhanced Raman scattering for monitoring antibiotic-induced chemical changes in bacteria cell wall. Plos One. 2009;4, e5470.
Anyfantakis M, Geng Z, Morel M, Rudiuk S, Baigl D. Modulation of the coffee-ring effect in particle/surfactant mixtures: the importance of particle-interface interactions. Langmuir. 2015;31:4113–20.
Chen PY, Cui L, Zhang KS. Surface-enhanced Raman spectroscopy monitoring the development of dual-species biofouling on membrane surfaces. J Membr Sci. 2015;473:36–44.
Wang HY, Zhou YF, Jiang XX, Sun B, Zhu Y, Wang H, et al. Simultaneous capture, detection, and inactivation of bacteria as enabled by a surface-enhanced Raman scattering multifunctional chip. Angew Chem Int Ed. 2015;54:5132–6.
Sun B, Jiang XX, Wang HY, Song B, Zhu Y, Wang H, et al. Surface-enhancement Raman scattering sensing strategy for discriminating trace mercuric iron (II) from real water samples in sensitive, specific, recyclable, and reproducible manners. Anal Chem. 2015;87:1250–6.
Cui L, Zhang Y-J, Huang WE, Zhang B-FM, Francis L, Li J-Y, et al. Surface-enhanced Raman spectroscopy for identification of heavy metal arsenic(V)-mediated enhancing effect on antibiotic resistance. Anal Chem. 2016. doi:10.1021/acs.analchem.5b04490.
Salleh FHM, Arif SM, Zainudin S, Firdaus-Raih M. Reconstructing gene regulatory networks from knock-out data using gaussian noise model and Pearson correlation coefficient. Comput Biol Chem. 2015;59:3–14.
Talari ACS, Movasaghi Z, Rehman S, Rehman IU. Raman spectroscopy of biological tissues. Appl Spectrosc Rev. 2014;50:46–111.
Roselli M, Finamore A, Garaguso I, Britti MS, Mengheri E. Zinc oxide protects cultured enterocytes from the damage induced by Escherichia coli. J Nutr. 2003;133:4077–82.
Sharma D, Ashaduzzaman M, Golabi M, Shriwastav A, Bisetty K, Tiwari A. Studies on bacterial proteins corona interaction with saponin imprinted ZnO nanohoneycombs and their toxic responses. ACS Appl Mater Interfaces. 2015;7:23848–56.
Manshian BB, Pfeiffer C, Pelaz B, Heimerl T, Gallego M, Moller M, et al. High-content imaging and gene expression approaches to unravel the effect of surface functionality on cellular interactions of silver nanoparticles. ACS Nano. 2015;9:10431–44.
Liu S, Hu M, Zeng TH, Wu R, Jiang R, Wei J, et al. Lateral dimension-dependent antibacterial activity of graphene oxide sheets. Langmuir. 2012;28:12364–72.
Limbach LK, Li Y, Grass RN, Brunner TJ, Hintermann MA, Muller M, et al. Oxide nanoparticle uptake in human lung fibroblasts: effects of particle size, agglomeration, and diffusion at low concentrations. Environ Sci Technol. 2005;39:9370–6.
Mempin R, Tran H, Chen C, Gong H, Kim Ho K, Lu S. Release of extracellular ATP by bacteria during growth. BMC Microbiol. 2013;13(301).
Hironaka I, Iwase T, Sugimoto S, Okuda K, Tajima A, Yanaga K, et al. Glucose triggers ATP secretion from bacteria in a growth-phase-dependent manner. Appl Environ Microbiol. 2013;79:2328–35.
Nyström T, Mårdén P, Kjelleberg S. Relative changes in incorporation rates of leucine and methionine during starvation survival of two bacteria isolated from marine waters. FEMS Microbiol Lett. 1986;38:285–92.
Maher MA, Naha PC, Mukherjee SP, Byrne HJ. Numerical simulations of in vitro nanoparticle toxicity—the case of poly(amido amine) dendrimers. Toxicol in Vitro. 2014;28:1449–60.
Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–84.
Manning AJ, Kuehn MJ. Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 2011;11:258.
Li M, Zhu LZ, Lin DH. Toxicity of ZnO nanoparticles to Escherichia coil: mechanism and the influence of medium components. Environ Sci Technol. 2011;45:1977–83.
Sawai J. Quantitative evaluation of antibacterial activities of metallic oxide powders (ZnO, MgO and CaO) by conductimetric assay. J Microbiol Methods. 2003;54:177–82.
Zhang LL, Jiang YH, Ding YL, Daskalakis N, Jeuken L, Povey M, et al. Mechanistic investigation into antibacterial behaviour of suspensions of ZnO nanoparticles against E. coli. J Nanoparticle Res. 2010;12:1625–36.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (21173208), Natural Science Foundation of Ningbo (2014A610107), and Natural Science Foundation of Fujian Province (2015J01067). We are grateful to Miss Catherine Yang Fan at Oxford University for her help in polishing the language of this paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 515 kb)
Rights and permissions
About this article
Cite this article
Zhang, B., Cui, L. & Zhang, K. Dosage- and time-dependent antibacterial effect of zinc oxide nanoparticles determined by a highly uniform SERS negating undesired spectral variation. Anal Bioanal Chem 408, 3853–3865 (2016). https://doi.org/10.1007/s00216-016-9478-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9478-z