Abstract
In the present study, the application of a liquid chromatography high-resolution mass spectrometry (LC-HRMS) analytical assay for the quantitative analysis of a recombinant human immunoglobulin G1 (hIgG1) in rat serum is reported using three generic peptides GPSVFPLAPSSK (GPS), TTPPVLDSDGSFFLYSK (TTP), and VVSVLTVLHQDWLNGK (VVS). Moreover, the deamidation site of a fourth peptide FNWYVDGVEVHNAK (FNW) was identified and further excluded from the assay evaluation due to the inaccuracy of the quantitative results. The rat serum samples were spiked with a fully labeled hIgG1 as internal standard (ISTD). The digestion with trypsin was performed onto the pellet prior to peptide analysis by LC-HRMS using a quadrupole time of flight (QTOF) mass analyzer operating in selected reaction monitoring (SRM) mode with enhanced duty cycles (EDC). The assay linearity for the three investigated peptides was established for a hIgG1 (hIgG1A) from 1.00 to 1000 μg mL−1 with a mean coefficient of determination (R 2) higher than 0.9868. The inter-day accuracy and precision obtained in rat serum over 3 days were ≤11.4 and ≤10.5 %, respectively. Short-term stability on the auto-sampler at 6 °C for 30 h, at RT for 48 h, and a 100-fold dilution factor were demonstrated. In addition, QC samples prepared in cynomolgus monkey serum and measured with the present method met the acceptance criteria of ±20.0 and ≤20.0 % for all three peptides regarding accuracy and precision, respectively. The LC-HRMS method was applied to the analysis of samples from five individual cynomolgus monkeys dosed with a second hIgG1 (hIgG1B) and consistent data were obtained compared to the LC-MS/MS method (conventional triple quadrupole (QqQ) mass analyzer operating in SRM). The present data demonstrate that LC-HRMS can be used for the quantitative analysis of hIgG1 in both species and that quantification is not only limited to classical QqQ instruments.

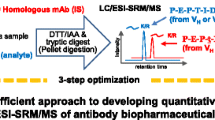
Method workflow
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The analysis of therapeutic proteins in complex biological matrices by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) has emerged during the last years in the field of bioanalysis as an alternative to conventional ligand binding assays [1–5]. In this respect, several bioanalytical groups have reported the use of mass spectrometry-based methods for the quantitative analysis of different type of proteins including high molecular weight proteins such as monoclonal antibodies (mAb) [6–8], chimeric mAbs [9], antibody-drug conjugates [10], or PEGylated proteins [11–13] as well as smaller proteins/peptides such as the parathyroid hormone, insulin analogues or prion proteins [14–16]. More recently, universal analytical assays for the quantification of mAbs in pre-clinical species using generic peptides originating from the human fragment crystallizable (Fc) region were extensively developed [17–19]. Nowadays, the majority of quantitative assays for protein analysis are still routinely performed (similar to small molecules) with LC systems hyphenated to triple quadrupole (QqQ) mass analyzers operating in selected reaction monitoring (SRM) acquisition mode due to their high specificity, dynamic range and ease of operation [20–22]. The selection of one or more surrogate peptides after trypsin digestion is the most common approach in proteomics for quantification of a whole protein in various matrices. Although LC-MS/MS provides great selectivity and sensitivity, interferences from other generated tryptic peptides in complex matrices (serum or plasma) at the retention time of the surrogate peptide(s) of interest cannot be excluded, even though appropriate sample cleanup strategies such as solid phase extraction (SPE) can reduce the sample complexity [23]. As a result, an increased lower limit of quantification (LLOQ) is generally observed due to high background noise during the quantitative analysis of a specific protein.
The use of high resolution mass spectrometry (HRMS) performed on quadrupole time of flight (QTOF), orbitrap or Fourier transform ion cyclotron resonance (FTICR) mass analyzers is a promising way to improve the method selectivity for the quantitative analysis [24–26]. The high mass accuracy between 0.1 and 5.0 ppm as well as the highly resolved isotopic pattern with a resolution ranging from 25,000 to 1,000,000 (depending on the type of HRMS instrument) are the major advantages compared to conventional LC-MS/MS.
General limitations associated in the past with HRMS instruments can nowadays be overcome to comply with a reliable quantitation since the new generation of HRMS instruments exhibit enhanced acquisition speed, increased sensitivity, and linear dynamic range [27].
Recently, the use of HRMS for the quantification of small molecules and larger peptides in human plasma was successfully demonstrated [28, 29]. Moreover, the selectivity of HRMS methods has been improved by decreasing the background in complex human plasma resulting in a better signal to noise (S/N) ratio [30]. Mekhssian et al. [31] analyzed a mAb quantitatively in human plasma over a calibration range from 1.00 to 200 μg mL−1 using a LC system coupled to a TripleTOF™ 5600 mass spectrometer.
The present study aims at exploring the capabilities of HRMS for the quantitative analysis of human immunoglobulin G1 (hIgG1) in pre-clinical species. The development of a generic LC-HRMS method for the quantification of a hIgG1 in rat serum is described using pellet digestion as sample preparation combined with HRMS detection of generic peptides. In the quantitative data obtained on three different days, the dynamic ranges and LLOQs for the selected peptides were determined. Furthermore, the method developed in rat serum was applied to the quantitative analysis of another hIgG1 in cynomolgus monkey serum.
Material and methods
Chemicals and reagents
The recombinant hIgG1A used for the preparation of calibration standards (Cs) and quality control (QC) samples, hIgG1B (pre-clinical study samples), and the stable isotope-labeled protein internal standard (ISTD, hIgG1C) were produced at Novartis Pharma AG (Basel, Switzerland). The latter was labeled with [13C]-lysine/arginine moieties using the stable isotope labeling with amino acids in cell culture (SILAC) approach. The reference peptides GPSVFPLAPSSK (GPS), TTPPVLDSDGSFFLYSK (TTP), and VVSVLTVLHQDWLNGK (VVS) used for MS tuning and SPE optimization were synthesized by Thermo Fisher Scientific (Ulm, Germany). Phosphate buffered saline (PBS; 12.0 mM phosphate, 137 mM sodium chloride, 2.70 mM potassium chloride, pH 7.4) and formic acid (FA) were purchased from Amresco (Solon, OH, USA) and Merck (Darmstadt, Germany), respectively. Sodium iodide and leucine enkephalin solutions were obtained from Waters (Milford, MA, USA). dl-Dithiothreitol (DTT), iodoacetamide (IAA), ammonium bicarbonate (ABC), bovine pancreas trypsin, trifluoroacetic acid (TFA), acetic acid, ammonium hydroxide (NH4OH, 28–30 %), acetonitrile (ACN), methanol (MeOH), and MS grade water were provided by Sigma-Aldrich (Buchs, Switzerland). All solvents (LC-MS grade) as well as reagents were of high analytical grade (≥99 %) and were used without further purification. The drug-free batches of rat and cynomolgus monkey sera used for the preparation of Cs and QC samples were delivered from Fisher Clinical Services (Allschwil, Switzerland).
Preparation of Cs and QC samples with hIgG1A
Working solutions were prepared by serial dilution of hIgG1A stock solution (20.0 mg mL−1 in PBS, storage at 2–8 °C) into PBS. Subsequently, the resultant solutions were spiked into blank rat serum (1:20, v/v) yielding in nine Cs concentrations of 1.00, 5.00, 10.0, 20.0, 50.0, 200, 500, 800, and 1,000 μg mL−1 as well as four QC levels at 3.00, 15.0, 450, and 750 μg mL-1. Additionally, one set of hIgG1A QC samples was prepared with the same concentration levels for the method cross-check in cynomolgus monkey serum.
Pre-clinical study samples from cynomolgus monkey with hIgG1B
A single dose of hIgG1B (5.00 mg kg−1) was administrated intravenously (i.v.) to five different female cynomolgus monkeys. Blood samples were taken at designated time points (pre-dose, 1, 4, 24, 72, 120, and 168 h post-dose). At each sampling time point, approximately 2.0 mL of blood was drawn into tubes containing no anticoagulant and was allowed to clot at room temperature for at least 30 min. Subsequently, the samples were centrifuged for 10 min at 1,500×g and 4 °C. The resulting serum was aliquoted (200 μL) and stored ≤ −70 °C pending analysis. The pre-clinical study was conducted in compliance with the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, the Office of Laboratory Animal Welfare, and in accordance with the Novartis Animal Care and Use Committee (NACUC).
Sample preparation
The sample preparation protocol was slightly modified from the one published by Ouyang et al. [32]. Briefly, 50 μL of serum either from study samples, Cs, QC, or blank samples was pipetted in a 2.0-mL Protein LoBind 96-well plate from Eppendorf (Hamburg, Germany). Then, 50 μL of ISTD solution at 20.0 μg mL−1 in 100 mM ABC in water was added, whereas for blank samples, 50 μL of 100 mM ABC in water was added instead. Zero samples referred to a later stage in this paper are blank samples spiked with ISTD. Afterwards, 20 μL of 100 mM DTT prepared in water was added to each well in order to reduce intra- and inter-disulfide bonds by incubating the samples on a ThermoMixer for 60 min at 60 °C. Subsequently, resulting free thiol groups were alkylated by pipetting 10 μL of 100 mM IAA in water into the plate being incubated at room temperature for 30 min in darkness. For the generation of the pellet, 400 μL of MeOH was added to the samples. The resulting samples were mixed using a ThermoMixer and centrifuged at 4 °C for 5 min at 900×g. The supernatant was removed by inverting the plate on a blotter, and the pellet was re-suspended in 200 μL of 200 mM ABC buffer in 10 % MeOH. A volume of 50 μL of trypsin at 8.00 mg mL-1 in 100 mM ABC in water was added to each sample, and the digestion was performed for 1 h at 60 °C. The digestion process was terminated by the addition of 50 μL 15 % TFA. The samples were centrifuged at 900×g for 5 min at 4 °C. An off-line SPE was performed by passing 250 μL of the digested sample through an Oasis MCX 96-well plate (30 mg, 60 μm) from Waters (Milford, MA, USA) being pre-washed two times with 1.0 mL of ACN followed by 2 × 1.0 mL of 1 % acetic acid. After loading, the cartridges were washed once with 1.0 mL ACN/1 % acetic acid (50:50, v/v). Finally, the peptides were eluted with 900 μL NH4OH/ACN/water (10:70:20, v/v/v) into a clean 1.0 mL Protein LoBind 96-well plate. The eluent was evaporated to dryness under a stream of nitrogen at 60 °C. The samples were reconstituted in 100 μL of 0.1 % FA in ACN/water (10:90, v/v) followed by centrifugation for 5 min at 4 °C and 900×g prior to LC-MS analysis.
LC-HRMS
Chromatographic separation of the tryptic peptides was achieved with an ACQUITY UPLC I-Class system from Waters (Milford, MA, USA). Twenty microliters of sample was loaded on an ACE C18, 150 × 4.6 mm, 3 μm column from Hichrom (Berkshire, UK) which was maintained at 60 °C. The flow rate was set to 1.0 mL min−1. The mobile phases consisted of 0.1 % FA in water (A) and 0.1 % FA in ACN (B) with an optimized elution gradient program set as follows: 0.0–1.0 min, 5 % B; 1.0–1.5 min, 5–20 % B; 1.5–7.0 min; 20–35 % B; 7.0–8.0 min, 35–90 % B; 8.0–10.0 min, 90 % B; 10.0–10.5 min, 90–5 % B; 10.5–13.0 min, 5 % B.
The UPLC system was hyphenated to a SYNAPT G2-Si HD high-resolution mass spectrometer from Waters (Milford, MA, USA) whereas the effluent was split (MS/Waste, 1:8, v/v) prior to MS detection. The peptides were ionized with a Zspray™ ion source using electrospray ionization (ESI) in positive mode. The MS was operating in TOF-SRM (sensitivity mode) with enhanced duty cycles (EDC) resulting in a resolution of 20,000 at full width at half maximum (FWHM). The parameters were as follows: mass range m/z 400–1500, capillary voltage 2.2 kV, source temperature 120 °C, sampling cone voltage 50 V, cone gas flow 25 L h−1, desolvation temperature 300 °C, the flow rate of desolvation gas (N2) 1000 L h−1. The optimized TOF-SRM parameters for each peptide including the m/z values for the selected precursor and the most abundant fragment ions as well as their identification and charge states are summarized in Table 1. The asterisk indicates the enhanced fragment for each peptide. The quadrupole mass isolation window for the precursor selection was 1 Da (low and high mass resolution set to 15 arbitrary units). An ion current extraction window (XIC) of 50 mDa was used for the fragment ions to reconstruct the chromatograms. The difference between exact (theoretical) and accurate (experimental) m/z values for the selected product ions was usually on the third decimal even though the m/z ratios were reported with four decimals. This was in agreement with the observed mass accuracy of the QTOF mass analyzer since the mass accuracy achieved upon calibration with sodium iodide was below 5 ppm. Leucine enkephalin was used as lock mass during data acquisition.
Data acquisition and processing
The LC-HRMS system was controlled by MassLynx 4.1 whereas peak area integration, construction of the calibration curve, and the back-calculation of the concentrations were performed with TargetLynx XS from Waters (Milford, MA, USA). Graphical illustration was conducted with OriginPro (version 9.1.0) from OriginLab Coorperation (Northhampton, MA, USA).
Evaluation of analytical assay performance
Selectivity
The selectivity of the method was determined by comparing the mean apparent analytical response for each peptide at the expected retention time and measured SRM transition(s) in blank samples (three different batches per species) relative to the peptide signal at the LLOQ. The comparison between the mean peak area at the SRM transition(s) of the surrogate peptide in a zero sample relative to the mean analytical response obtained for the surrogate peptide at the LLOQ was used to assess potential contribution of [13C6]-labeled peptide to the light version of the peptide. The contribution of surrogate peptide to heavy labeled version of the peptide was assessed by comparing the mean analytical response for [13C6] peptide in a blank sample spiked with the protein at the upper limit of quantification (ULOQ) to the mean analytical response obtained for zero samples. The acceptance criterion for each peptide was set to ≤20.0 % of the analytical response at the LLOQ and for the heavy labeled peptides ≤5.0 % of the analytical response at the working concentration of the ISTD.
Linearity and sensitivity
Calibration curves (in duplicate on each day) were constructed using a quadratic mathematical model (y = ax 2 + bx + c) with a weighting factor of 1/x 2 to calculate the concentrations where y represented the peak area ratio of the response for the peptide to the response of [13C6]-lysine labeled peptide version and x was the nominal concentration of the protein in the Cs samples. The acceptance criteria were ±20.0 % (±25.0 % at the LLOQ and ULOQ) for 75.0 % of the Cs from nominal values (with a minimum of six different levels). Additionally, at least 50.0 % of the Cs tested at each concentration level should meet the abovementioned acceptance criteria and the derived coefficient of determination (R 2) should be at least 0.95. The lowest concentration meeting the acceptance criteria of ±25.0 and ≤25.0 % regarding accuracy and precision was set as the LLOQ.
Carry-over
A series of three blank samples were injected in one run directly after the ULOQ sample at 1000 μg mL−1 to evaluate any carry-over. The extent of carry-over should be ≤20.0 % of the response of the tryptic peptide observed at the LLOQ and ≤5.0 % of the response observed for the corresponding isotopically labeled peptide at the working concentration of the ISTD.
Accuracy and precision
The accuracy of the analytical assay was evaluated by the deviation (% bias) from the nominal value of at least three QC concentration levels (3× LLOQ, mid and high) whereas the percentage of the coefficient of variation (CV) determined the precision of the method. Each QC level was analyzed in triplicate on each day to evaluate the intra-day accuracy and precision whereas in total nine replicates from three different days were utilized to calculate the inter-day accuracy and precision. A mean bias within ±20.0 % of the nominal values and a precision of ≤20.0 % were set as acceptance criteria.
Dilution
In case pre-clinical samples exhibit a higher concentration than the ULOQ, an appropriate dilution factor has to be assessed. Thus, an additional QC sample at five times the ULOQ was prepared and diluted 100-fold with blank rat serum in replicates of five resulting in a nominal concentration of 50.0 μg mL−1. The back-calculated mean concentration (with the dilution factor incorporated) should be ±20.0 % of the initial concentration with a precision ≤20.0 %. Additionally, three out of five individual concentrations should meet the acceptance criteria.
Stability
The protein stability during this method evaluation was assessed with two QC levels (15.0 and 750 μg mL−1) at room temperature for a period of 48 h. Stability on the auto-sampler of the generated peptides was investigated at 6 °C for two storage periods (16 and 30 h). After storage, the samples were measured against a freshly prepared calibration curve including an independent set of QC samples. The stability was deemed acceptable if the deviation from the initial concentration was ±20.0 %. Stability data of the mAb bulk material and stock solution were evaluated previously and were not part of this investigation.
Method comparison with LC-MS/MS
Analytical method comparison between LC-HRMS and an existing LC-MS/MS assay was performed on in vivo samples from a toxicokinetic study. The LC-MS/MS system consisted of a Symbiosis Pro from Spark Holland B.V (Emmen, Netherlands) equipped with a Reliance unit (conditioned stack and auto-sampler) and a Mistral column oven coupled to an API 6500 QTRAP mass spectrometer controlled by Analyst 1.6 from Applied Biosystems (Foster City, CA, USA). The sample preparation was similar to the one described in section Sample preparation. A detailed description of the LC-MS/MS parameters is summarized in the Electronic Supplementary Material (ESM, Table S1).
Results and discussion
Surrogate peptide selection
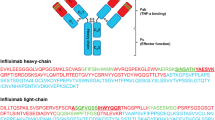
Besides the light and the heavy chain, the structure of antibodies can be further subdivided into variable and constant regions. The latter is only suitable for a universal MS-based assay in pre-clinical species as the majority of the amino acid sequence is conserved over all human antibodies exhibiting common peptides after tryptic digestion, whereas the former contains antibody specific peptides located in the complementarity determining region (CDR) being responsible for specific target binding. For the identification of conserved peptides, the amino acid sequences of the Fc region from several mAbs were aligned with each other and an in-silico digestion was performed. Fifteen common peptides were identified and were further selected by the following criteria: (1) oxidation sites such as methionine (M), cysteine (C), or tryptophan (W) should be avoided if possible; (2) peptides with glycosylation sites on nitrogen (NxS and NxT, whereas x can be any amino acid except proline (P), serines (S), or threonines (T)) [33], on oxygen (S and T rich domains) [34], or carbon-linked motifs (WxxW, WS/TC) [35, 36] were excluded; (3) if P was located downstream from lysine (K) or arginine (R) as well as two basic amino acids were located next to each other (RR, KK, or RK), the peptides were not taken into account to prevent miscleavage of peptides; and (4) the peptide should have at least 8 amino acids and its isoelectric point should not be too high or too low to guarantee adequate retention under reversed-phase chromatographic conditions. Out of the initial 15 peptides GPS (from the CH1 region), TTP (from the CH3 region) and two peptides from the CH2 region (FNW and VVS) were identified as candidates for the generic assay development.
Deamidation site in both CH2 peptides
During kinetic studies of the digestion, a decrease in signal intensity for both CH2 peptides was observed, whereas two additional peaks were generated during overnight digestion at 37 °C. Chelius et al. [37] identified several amino acid motifs causing deamidation on asparagine (N) via a cyclic intermediate state (succinimide) to isoaspartic (isoD) and aspartic acid (D) under certain temperature and pH conditions. As a result, two additional peaks with a ratio of approximately 1:3 appear over time [38]. It has been shown that the LNG motif in the VVS peptide is subjected for deamidation [39]. Concerning the FNW peptide, the motif FNW and HNA are likely to be deamidated, but occurrence is less prone. In order to confirm the potential deamidation of the FNW peptide, the ion chromatogram of the single deamidated form was extracted from the full scan resulting in the appearance of two additional peaks with a retention time shift of 0.15 and 0.35 min from the unmodified FNW.
In addition, the MS/MS data obtained on the precursor ion of the FNW peptide (m/z 559.9373) showed that all y-fragment ions (beyond the y11 + ion) from the isoaspartic acid (Fig. 1a) and aspartic acid forms (Fig. 1b) differ by approximately 1 Da in mass in comparison with the non-deamidated form (Fig. 1c). The same experiments were performed on the doubly deamidated versions, but no additional peaks were identified. Thus, the FNW peptide underwent a single deamidation on the C-terminal asparagine in the HNA motif. The three individual chromatographic peaks of the FNW peptide that originated from the deamidation could not be combined in a single peak under the selected conditions. As a result of inadequate peak integration, inaccurate quantitative data with the FNW peptide was observed. Consequently, this peptide was not included in the final method.
Selection of QTOF quantification mode
Three different acquisition modes were assessed for quantitative purposes with the SYNAPT G2-Si HD HRMS QTOF instrument (i) TOF-MS, (ii) TOF-MS/MS, and (iii) TOF-SRM. The general advantages and working principles for each mode were previously reported by Morin et al. [29], whereas the modes used with their TripleTOF™ 5600 instrument were entitled TOF-MS, SRMHR/HS, and SRMHS enhance. In the first acquisition mode, quantification was performed by extracting the accurate mass of the analyte of interest from the full scan. This resulted in high signal intensities for each surrogate peptide as no signal was lost due to fragmentation (Table 2). However, sensitivity was affected as well since high additional background noise was obtained in full scan mode resulting in low S/N ratios (peak to peak model) ranging from 7–42. In TOF-MS/MS, a precursor was selected by the quadrupole and fragmented in the collision cell. Subsequently, a single or multiple product ion(s) was/were used for quantitative purposes. Although the signal intensities in TOF-MS/MS were not as high as in TOF-MS due to possible distribution of the signal over several product ions, the resultant S/N ratios were significantly improved (up to 24-fold increase with the TTP peptide) as the chemical noise decreased. The working principle of TOF-SRM is in general based on TOF-MS/MS. However, the pusher region of a TOF analyzer was synchronized with the release of one specific product ion for each analyte from the TWaveTM collision cell allowing maximum transmission. As a result, TOF-SRM with maximized duty cycles for a specific m/z range gave similar signal intensities compared to TOF-MS, whereas the S/N ratios were comparable to the TOF-MS/MS mode. This trend was consistent for each surrogate peptide and was also in agreement with the results for other peptides reported by Morin et al. [29]. This demonstrated that TOF-SRM increased not only selectivity but also sensitivity of the analytical method.
Selectivity improvement using the monoisotopic [M + 2H]2+ ion of the VVSd y14 2+ fragment ion
Since the deamidation of the VVS peptide was expected during the tryptic digestion process, six fragment ions covering the non-deamidated (VVS) and deamidated isoform (VVSd) were included for the quantitative analysis (Table 1). As the mass difference of the triply charged precursor for both isoforms was approximately 0.3 Da, the quadrupole could also not distinguish between the corresponding precursor for each isoform due to the mass isolation window of 1 Da. During selectivity investigations, an interfering peak was found in the extracted ion chromatogram using the exact (theoretical) m/z ratio of six fragments (three from each isoform) in blank rat serum (Fig. 2a). The MS/MS spectrum of the interfering peak revealed a doubly charged fragment originating from an unknown endogenous compound with a monoisotopic [M + 2H]2+ ion at m/z 804.4498, a [(M + 1) + 2H]2+ ion at m/z 804.9492 and a [(M + 2) + 2H]2+ ion at m/z 805.4466 (Fig. 2b). The interference was caused by the [(M + 2) + 2H]2+ ion being close to the exact monoisotopic mass of the VVS y14 2+ fragment at m/z 805.4385. The advantage of HRMS in the SRM acquisition mode compared to classical QqQ instruments is that the isotopic pattern of a given compound is highly resolved offering the possibility to select the most appropriate m/z value for quantification. Hence, the interference could significantly be reduced by excluding the y14 2+ fragment of the VVS peptide at m/z 805.4385 during chromatogram extraction (Fig. 2c). As the [(M + 1) + 2H]2+ ion of the VVS fragment at m/z 805.9353 represented 91.6 % of the signal intensity of its monoisotopic y14 2+ product ion at m/z 805.4349 (Fig. 2d), no significant loss of sensitivity was observed by excluding the [M + 2H]2+ ion of the VVS peptide. The reason was that both isoforms were still extracted with a XIC window of 50 mDa as the accurate mass of the [(M + 1) + 2H]2+ ion of the VVS y14 2+ fragment at m/z 805.9353 (Fig. 2d) differed by approximately 9 mDa compared to the accurate mass of the selected [M + 2H]2+ ion of the VVSd y14 2+ product ion at m/z 805.9268 (Fig. 2f). Both isoforms were not fully baseline separated using in total five fragments and the retention time of VVS and VVSd was at 7.1 and 7.4 min, respectively (Fig 2e). This example demonstrated how HRMS can be used to solve selectivity issues in complex matrices.
Selectivity improvement for VVS peptide in rat serum by using the [M + 2H]2+ ion of the deamidated VVS y14 2+ fragment instead of the [M + 2H]2+ ion of the non-deamidated VVS y14 2+ fragment. a extracted chromatogram of blank rat serum using three fragments from each isoform, b MS/MS spectrum from interfering peak showing interference from the [(M + 2) + 2H]2+ ion at m/z 805.4466 of an unknown endogenous compound in rat serum, c extracted chromatogram of blank rat serum after exclusion of VVS y14 2+ fragment , d MS/MS spectrum (zoom into m/z 803–810) of the triply charged non-deamidated VVS precursor ion at m/z 603.3, e extracted chromatogram of both VVS isoforms (in total five fragments) generated from hIgG1A spiked in rat serum at 10.0 μg mL−1, and f MS/MS spectrum (zoom into m/z 803–810) of the triply charged deamidated VVS precursor ion at m/z 603.7
Evaluation of analytical assay performance
Selectivity
The mean percentage of the endogenous interference for TTP was 3.4 ± 1.4 %, VVS resulted in an interference below 7.3 % by using the [M + 2H]2+ ion of the VVSd y14 2+ fragment as described in the previous section, whereas no endogenous interference was detected for GPS for rat serum using three different batches (ESM, Table S2). Moreover, the interferences detected on the SRM transitions for the isotopically labeled peptides were for TTP and VVS below 0.2 and 0.4 %, respectively. Only for [13C6]-GPS, the interference was slightly higher with maximum 1.8 %. The working concentration of the stable isotope-labeled protein internal standard was set to 20.0 μg mL−1 to fulfill the acceptance criterion regarding the ISTD contribution to the surrogate peptide signal. No significant contribution of the analyte to the signal of the heavy peptides was observed. As the acceptance criteria were fulfilled, it was demonstrated that the analytical method was not only highly selective for the generic tryptic peptides but also for their [13C6]-lysine labeled versions in rat serum. The only interferences at the LLOQ of 1.00 μg mL−1 in cynomolgus monkey serum was caused by the SRM transitions of the GPS peptide confirming the results by Zhang et al. [23]. Consequently, the GPS peptide is not suitable for the quantitative analysis in cynomolgus monkey serum with a LLOQ of 1.00 μg mL−1.
Linearity and sensitivity
The slope of the calibration curve reached a plateau with higher concentrations (>800 μg mL-1) especially with the TTP surrogate peptide. The calibration curves were described more adequately by a quadratic instead of a linear regression model with a weighting factor of 1/x2 resulting in a better curve fit [better coefficient of determination (R 2) values for each surrogate peptide with good accuracy and precision data over the anticipated calibration range]. The resultant R 2 values obtained over 3 days ranged from 0.9868 to 0.9911 for the three surrogate peptides (Table 3). For two out of three generic peptides, the highest inter-day precision with a maximum of 14.8 % was observed at 1.00 μg mL−1 whereas the accuracy at this concentration ranged from 0.8 to 1.7 %. As this concentration level met the set acceptance criteria of ±25.0 % and ≤25.0 % regarding accuracy and precision, it was set as the LLOQ. The accuracy of the eight remaining concentration levels ranged from −8.2 to 5.2 % for GPS, for TTP from −7.3 to 5.9 % and for VVS from −9.8 to 3.4 %. The precision was below 14.2, 9.5, and 12.4 % for GPS, TTP, and VVS, respectively. Thus, the constructed calibration curves of the surrogate peptides could be used to reliably determine the protein concentration in rat serum in a consistent fashion over the anticipated concentration range suitable for the routine analysis.
Carry-over
No carry-over for all three peptides was found in the second and third blank rat serum sample directly after the injection of a sample at the ULOQ (ESM, Table S3). However, the extent of carry-over for TTP and VVS exceeded in the first blank the acceptance criterion of ≤20.0 % compared to the signal at the LLOQ with 64.8 and 71.2 %, respectively. This result demonstrated that an injection of at least one rinse or blank sample after highly concentrated samples is required prior to low concentration samples to avoid overestimation caused by carry-over.
Accuracy and precision in rat serum
The accuracy and precision for the four QC levels at 3.00, 15.0, 450, and 750 μg mL−1 in three different rat serum batches fulfilled the acceptance criteria. For the investigated peptides, the inter-day accuracy ranged from -5.4 to 11.4 % whereas the precision was between 4.4 and 10.5 % (Table 4). The intra-run bias and precision across three different batches ranged from −13.3 to 16.8 % and 1.3 to 14.5 %, respectively (ESM, Table S4). The results indicated that the developed LC-HRMS method was accurate and precise not only between different serum batches but also within individual batches. As the same inter-peptide concentration values with a variation ≤3.0 % at each QC level (Table 4) were obtained, it was demonstrated that the peptides were generated in a similar fashion during tryptic digestion and that the same concentration values were observed regardless of the tryptic surrogate peptide selected.
Dilution factor
The QC sample at 5.00 mg mL−1 was diluted by a factor of 100-fold using blank rat serum resulting in a nominal concentration of 50.0 μg mL−1. The mean back calculated concentration was 5.34 ± 0.31 mg mL−1 with a bias and precision ranging from 3.1 to 14.0 % and 1.9 to 6.0 %, respectively (ESM, Table S5). These results indicated that pre-clinical samples exhibiting a higher concentration than the qualified calibration range can be diluted with blank matrix prior to analysis.
Stability
The short-term stability data determined with two QC levels at 750 and 15.0 μg mL−1 (ESM, Table S6) revealed that the recombinant hIgG1A, was stable at room temperature up to 48 h, since the % bias compared to the expected concentration at t0 was between −8.6 and 4.8 % depending which peptide was considered. It was further demonstrated that the generated peptides were stable on the auto-sampler after tryptic digestion for at least 30 h, as the calculated mean bias was within ±20.0 % acceptance criterion (GPS ≤9.7 %, TTP ≤4.8 %, and VVS ≤1.7 %).
Accuracy and precision in cynomolgus monkey serum
The question if mAbs can be quantified in a consistent manner when spiked in a different matrix than rat serum was also addressed in our investigations to further expand the method. In this respect, hIgG1A was spiked at four QC concentration levels in cynomolgus monkey serum. Subsequently, the QC concentrations were back-calculated against a calibration curve and an additional set of QC samples prepared in rat serum (Table 5). The resultant accuracies for GPS, TTP, and VVS ranged from −6.9 to 13.0 % fulfilling the acceptance criterion of ±20.0 %. High matrix interference on the SRM transitions of GPS caused inaccurate results at low concentrations in cynomolgus monkey serum with the QqQ instruments reported by Zhang et al. [23]. In contrast, a low bias with −6.9 % was observed with TOF-SRM for the GPS peptide at the low QC level (3× LLOQ) demonstrating its advantage over QqQ instruments. Moreover, the precision with the HRMS method was also within the acceptance criterion of ≤20.0 % ranging from 1.0 to 13.3 %. Only the precision in the GPS sample for the low QC level at 3.00 μg mL−1 exceeded the criterion by 7.1 % and was therefore excluded for the analysis of hIgG1B in cynomolgus monkey samples.
Application to pre-clinical study
The present method was applied to one pre-clinical study including five female cynomolgus monkeys which were dosed i.v. once with the hIgG1B at 5.00 mg kg−1. In total, 35 serum samples were either analyzed by LC-HRMS or with the conventional LC-MS/MS approach. The mean concentration-time profile of five individual monkeys taking the results from both peptides (TTP and VVS) into account was typical for i.v. administration of a therapeutic protein (Fig. 3a). The data found with the GPS peptide were not included in the present comparison due to the high variability in the cynomolgus monkey QC samples. The reason for this high variability remained unknown. No hIgG1B was detected in the pre-dose samples for each individual cynomolgus monkey. The obtained hIgG1B serum concentration-time profiles were identical at each time point regardless the analytical assay. The variation in the mean concentrations of five individual cynomolgus monkeys obtained either with the TTP or the VVS peptide was 2.7 ± 1.7 % CV for the LC-HRMS analysis. In contrast, the variation between both surrogate peptides was slightly increased for the LC-MS/MS assay ranging from 3.7 to 8.5 % CV.
When the individual concentrations for each cynomolgus monkey instead of the mean concentration were considered, no significant deviation was revealed by plotting the mean concentration for each surrogate peptide from the LC-HRMS and LC-MS/MS assay against the bias between both analytical assays (Fig. 3b). The mean bias between both analytical instruments observed with TTP and VVS was 6.5 ± 6.7 and −0.1 ± 6.6 %, respectively. All samples with exception of one sample for TTP were within the acceptance criterion of ±20.0 % (dashed lines). These results indicated that equivalent data was generated on both platforms regardless of the peptide selected. Thus, the developed LC-HRMS method was quantifying the hIgG1B in a reliable fashion as already demonstrated with QC samples in cynomolgus monkey serum and could be used as an alternative to conventional analytical assays using QqQ mass analyzers.
Conclusion
The application of HRMS instruments in the field of qualitative proteomics provides valuable information regarding drug-antibody ratios or the determination of various glycan forms of a mAb. However, therapeutic proteins can also be quantified in a reliable fashion with HRMS instruments using the bottom-up approach as demonstrated. Consequently, the combination of qualitative/quantitative approaches in targeted and non-targeted analysis by one single instrument has a significant impact and opens new opportunities to the pharmaceutical industry to support pre-clinical and clinical studies during their drug development process of biotherapeutics in a regulated environment. The major advantage associated with HRMS instruments is the highly resolved isotopic pattern which can increase the method selectivity, whereas the same linear range, accuracy, and precision were obtained compared to classical low resolution QqQ mass analyzers widely used for quantification. Thus, more quantitative HRMS methods will be established in the future as quantitative bioanalytical LC-MS based assays are generally no longer limited to QqQ instruments only due to the evolution of recent HRMS instruments. As four different peptides were incorporated in the described method, the approach can easily be adapted to other modalities such as hIgG4, antibody-drug conjugates, or even chimeric/bispecific antibodies based on human immunoglobulin G. Since the samples can be prepared within one working day and the analysis can run overnight by LC-HRMS, a certain throughput is also associated being an important aspect for pharmaceutical industry.
References
An B, Zhang M, Qu J. Toward sensitive and accurate analysis of antibody biotherapeutics by liquid chromatography coupled with mass spectrometry. Drug Metab Dispos. 2014;42(11):1858–66. doi:10.1124/dmd.114.058917.
Bults P, van de Merbel NC, Bischoff R. Quantification of biopharmaceuticals and biomarkers in complex biological matrices: a comparison of liquid chromatography coupled to tandem mass spectrometry and ligand binding assays. Expert Rev Proteomics. 2015;12(4):355–74. doi:10.1586/14789450.2015.1050384.
Hopfgartner G, Lesur A, Varesio E. Analysis of biopharmaceutical proteins in biological matrices by LC-MS/MS II. LC-MS/MS analysis. Trends Anal Chem. 2013;48:52–61.
van den Broek I, Niessen WM, van Dongen WD. Bioanalytical LC-MS/MS of protein-based biopharmaceuticals. J Chromatogr B. 2013;929:161–79. doi:10.1016/j.jchromb.2013.04.030.
Zheng J, Mehl J, Zhu Y, Xin B, Olah T. Application and challenges in using LC–MS assays for absolute quantitative analysis of therapeutic proteins in drug discovery. Bioanalysis. 2014;6(6):859–79. doi:10.4155/bio.14.36.
Hagman C, Ricke D, Ewert S, Bek S, Falchetto R, Bitsch F. Absolute quantification of monoclonal antibodies in biofluids by liquid chromatography − tandem mass spectrometry. Anal Chem. 2008;80(4):1290–6. doi:10.1021/ac702115b.
Heudi O, Barteau S, Zimmer D, Schmidt J, Bill K, Lehmann N, et al. Towards absolute quantification of therapeutic monoclonal antibody in serum by LC-MS/MS using isotope-labeled antibody standard and protein cleavage isotope dilution mass spectrometry. Anal Chem. 2008;80(11):4200–7. doi:10.1021/ac800205s.
Ladwig PM, Barnidge DR, Snyder MR, Katzmann JA, Murray DL. Quantification of serum IgG subclasses by use of subclass-specific tryptic peptides and liquid chromatography–tandem mass spectrometry. Clin Chem. 2014;60(8):1080–8. doi:10.1373/clinchem.2014.222208.
Willrich MAV, Murray DL, Barnidge DR, Ladwig PM, Snyder MR. Quantitation of infliximab using clonotypic peptides and selective reaction monitoring by LC–MS/MS. Int Immunopharmacol. 2015;28(1):513–20. doi:10.1016/j.intimp.2015.07.007.
Myler H, Rangan VS, Wang J, Kozhich A, Cummings JA, Neely R, et al. An integrated multiplatform bioanalytical strategy for antibody-drug conjugates: a novel case study. Bioanalysis. 2015;7(13):1569–82. doi:10.4155/bio.15.80.
Dawes ML, Gu H, Wang J, Schuster AE, Haulenbeek J. Development of a validated liquid chromatography tandem mass spectrometry assay for a PEGylated adnectin in cynomolgus monkey plasma using protein precipitation and trypsin digestion. J Chromatogr B. 2013;934:1–7. doi:10.1016/j.jchromb.2013.06.027.
Gong J, Gu X, Achanzar WE, Chadwick KD, Gan J, Brock BJ, et al. Quantitative analysis of polyethylene glycol (PEG) and PEGylated proteins in animal tissues by LC-MS/MS coupled with in-source CID. Anal Chem. 2014;86(15):7642–9. doi:10.1021/ac501507g.
Liu Q, De Felippis MR, Huang L. Method for characterization of PEGylated bioproducts in biological matrixes. Anal Chem. 2013;85(20):9630–7. doi:10.1021/ac401921z.
Kumar V, Barnidge DR, Chen LS, Twentyman JM, Cradic KW, Grebe SK, et al. Quantification of serum 1–84 parathyroid hormone in patients with hyperparathyroidism by immunocapture in situ digestion liquid chromatography-tandem mass spectrometry. Clin Chem. 2010;56(2):306–13. doi:10.1373/clinchem.2009.134643.
Peterman S, Niederkofler EE, Phillips DA, Krastins B, Kiernan UA, Tubbs KA, et al. An automated, high-throughput method for targeted quantification of intact insulin and its therapeutic analogs in human serum or plasma coupling mass spectrometric immunoassay with high resolution and accurate mass detection (MSIA-HR/AM). Proteomics. 2014;14(12):1445–56. doi:10.1002/pmic.201300300.
Van Dorsselaer A, Carapito C, Delalande F, Schaeffer-Reiss C, Thierse D, Diemer H, et al. Detection of prion protein in urine-derived injectable fertility products by a targeted proteomic approach. PLoS One. 2011;6(3):e17815. doi:10.1371/journal.pone.0017815.
Furlong MT, Ouyang Z, Wu S, Tamura J, Olah T, Tymiak A, et al. A universal surrogate peptide to enable LC-MS/MS bioanalysis of a diversity of human monoclonal antibody and human Fc-fusion protein drug candidates in pre-clinical animal studies. Biomed Chromatogr. 2012;26(8):1024–32. doi:10.1002/bmc.2759.
Law WS, Genin J-C, Miess C, Treton G, Warren AP, Lloyd P, et al. Use of generic LC–MS/MS assays to characterize atypical PK profile of a biotherapeutic monoclonal antibody. Bioanalysis. 2014;6(23):3225–35. doi:10.4155/bio.14.167.
Li H, Ortiz R, Tran L, Hall M, Spahr C, Walker K, et al. General LC-MS/MS method approach to quantify therapeutic monoclonal antibodies using a common whole antibody internal standard with application to preclinical studies. Anal Chem. 2012;84(3):1267–73. doi:10.1021/ac202792n.
Gallien S, Duriez E, Domon B. Selected reaction monitoring applied to proteomics. J Mass Spectrom. 2011;46(3):298–312. doi:10.1002/jms.1895.
Lebert D, Picard G, Beau-Larvor C, Troncy L, Lacheny C, Maynadier B, et al. Absolute and multiplex quantification of antibodies in serum using PSAQ™ standards and LC-MS/MS. Bioanalysis. 2015;7(10):1237–51. doi:10.4155/bio.15.56.
Yost RA, Enke CG. Triple quadrupole mass spectrometry for direct mixture analysis and structure elucidation. Anal Chem. 1979;51(12):1251–64. doi:10.1021/ac50048a002.
Zhang Q, Spellman DS, Song Y, Choi B, Hatcher NG, Tomazela D, et al. Generic automated method for liquid chromatography–multiple reaction monitoring mass spectrometry based monoclonal antibody quantitation for preclinical pharmacokinetic studies. Anal Chem. 2014;86(17):8776–84. doi:10.1021/ac5019827.
Gallien S, Domon B (2015) Advances in high-resolution quantitative proteomics: implications for clinical applications. Expert Rev Proteomics; 1–10. doi:10.1586/14789450.2015.1069188
Gallien S, Domon B. Detection and quantification of proteins in clinical samples using high resolution mass spectrometry. Methods. 2015;81:15–23. doi:10.1016/j.ymeth.2015.03.015.
Gallien S, Duriez E, Demeure K, Domon B. Selectivity of LC-MS/MS analysis: implication for proteomics experiments. J Proteomics. 2013;81:148–58. doi:10.1016/j.jprot.2012.11.005.
Huang MQ, Lin ZJ, Weng N. Applications of high-resolution MS in bioanalysis. Bioanalysis. 2013;5(10):1269–76. doi:10.4155/bio.13.100.
Dillen L, Cools W, Vereyken L, Lorreyne W, Huybrechts T, de Vries R, et al. Comparison of triple quadrupole and high-resolution TOF-MS for quantification of peptides. Bioanalysis. 2012;4(5):565–79. doi:10.4155/bio.12.3.
Morin LP, Mess JN, Garofolo F. Large-molecule quantification: sensitivity and selectivity head-to-head comparison of triple quadrupole with Q-TOF. Bioanalysis. 2013;5(10):1181–93. doi:10.4155/bio.13.87.
Ramagiri S, Garofolo F. Large molecule bioanalysis using Q-TOF without predigestion and its data processing challenges. Bioanalysis. 2012;4(5):529–40. doi:10.4155/bio.12.10.
Mekhssian K, Mess JN, Garofolo F. Application of high-resolution MS in the quantification of a therapeutic monoclonal antibody in human plasma. Bioanalysis. 2014;6(13):1767–79. doi:10.4155/bio.14.111.
Ouyang Z, Furlong MT, Wu S, Sleczka B, Tamura J, Wang H, et al. Pellet digestion: a simple and efficient sample preparation technique for LC–MS/MS quantification of large therapeutic proteins in plasma. Bioanalysis. 2011;4(1):17–28. doi:10.4155/bio.11.286.
Mellquist JL, Kasturi L, Spitalnik SL, Shakin-Eshleman SH. The amino acid following an Asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry. 1998;37(19):6833–7. doi:10.1021/bi972217k.
Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33(3):151–208. doi:10.1080/10409239891204198.
Krieg J, Hartmann S, Vicentini A, Glasner W, Hess D, Hofsteenge J. Recognition signal for C-mannosylation of Trp-7 in RNase 2 consists of sequence Trp-x-x-Trp. Mol Biol Cell. 1998;9(2):301–9.
Pan Y, Karagiannis K, Zhang H, Dingerdissen H, Shamsaddini A, Wan Q, et al. Human germline and pan-cancer variomes and their distinct functional profiles. Nucleic Acids Res. 2014;42(18):11570–88. doi:10.1093/nar/gku772.
Chelius D, Rehder DS, Bondarenko PV. Identification and characterization of deamidation sites in the conserved regions of human immunoglobulin gamma antibodies. Anal Chem. 2005;77(18):6004–11. doi:10.1021/ac050672d.
Zhang YT, Hu J, Pace AL, Wong R, Wang YJ, Kao YH. Characterization of asparagine 330 deamidation in an Fc-fragment of IgG1 using cation exchange chromatography and peptide mapping. J Chromatogr B. 2014;965:65–71. doi:10.1016/j.jchromb.2014.06.018.
Ren D, Pipes GD, Liu D, Shih LY, Nichols AC, Treuheit MJ, et al. An improved trypsin digestion method minimizes digestion-induced modifications on proteins. Anal Biochem. 2009;392(1):12–21. doi:10.1016/j.ab.2009.05.018.
Acknowledgments
The authors would like to thank Charlotte Hagman, Laurence Masson and Fanny Deglave for providing the pre-clinical study samples and for the preparation of QC samples during cross-check. Furthermore, we would like to express our deep gratitude to Christine Carapito (CNRS-University of Strasbourg) for critical reviewing this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The pre-clinical study was conducted in compliance with the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, the Office of Laboratory Animal Welfare and in accordance with the Novartis Animal Care and Use Committee (NACUC).
Conflict of interest
In case of Christian Lanshoeft, the work was mainly conducted in fulfillment for the degree of Ph.D. from the University of Strasbourg under the academic supervision of Dr. Sarah Cianférani. Novartis Pharma AG completely funded this project financially. The work was performed at the facilities of Novartis only for research purposes and was not related to any Novartis project currently in development.
No relevant affiliations, financial involvement with any organization, entity with a financial interest in or financial conflict with the subject matter was associated with the remaining authors. This includes employment, consultancies, honoraria, stock ownership, expert testimony, grants, patents or royalties.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 41 kb)
Rights and permissions
About this article
Cite this article
Lanshoeft, C., Wolf, T., Heudi, O. et al. The use of generic surrogate peptides for the quantitative analysis of human immunoglobulin G1 in pre-clinical species with high-resolution mass spectrometry. Anal Bioanal Chem 408, 1687–1699 (2016). https://doi.org/10.1007/s00216-015-9286-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-9286-x