Abstract
Abnormal lipid metabolism is a common feature in most solid tumors, and occurs in early stages of the tumor progression. As benign breast tumor is different from malignant tumor of breast cancer, it is particularly important to take benign breast tumor into consideration when investigating cancer biomarkers. In this study, by using a normal-phase/reversed-phase two-dimensional liquid chromatography-mass spectrometry (NP/RP 2D LC-MS) method, we conducted comprehensive lipid profiling in human plasma obtained from six benign breast tumor patients and five breast cancer patients, as well as nine healthy controls. As a result, 512 lipid species were successfully identified. Principal component analysis allowed clear separation of the three groups. Quantitative analysis revealed that many lipid contents were similar in benign and malignant breast tumors compared with controls, and these were proposed as potential breast tumor biomarkers other than breast cancer biomarkers. Two phosphatidylinositol (PI) species, including PI (16:0/16:1) and PI (18:0/20:4), could differentiate between benign and malignant breast tumors, as well as breast cancer patients and healthy controls, indicating that they could be utilized as potential breast cancer biomarkers. In addition, PI (16:0/18:1), phosphatidylglycerol (36:3), and glucosylceramide (d18:1/15:1) were demonstrated to be potential biomarkers to evaluate the level of malignancy of breast tumor. Taken together, our results indicate the usefulness of lipid profiling in the discrimination between patients with breast cancer and non-carcinoma lesions, which might provide assistance in clinical diagnosis.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most prevalent form of cancer in women worldwide [1]. It is also one of the most aggressive malignant tumors associated with poor prognosis, rapid clinical progression, and a high rate of metastasis with high mortality [2]. Thus, finding new biomarkers, which are capable of detecting cancer in its early stage, is of vital importance. Similar to breast cancer, benign breast tumor represents a group of histologically heterogeneous lesions [3]. As it is definitely different from malignant tumor, it may be used as a good representative group when searching for cancer biomarkers.
Lipids are a kind of hydrophobic or amphipathic small molecules. They play crucial roles in cellular energy storage, structure, and signaling [4]. Till now, numerous studies have shown that lipid defects are central to the pathogenesis of many important and devastating diseases, including cancer. Thus, lipidomics was put forward, which aims at the full characterization of lipid molecular species and of their biological roles with respect to expression of proteins involved in lipid metabolism and function [5]. Investigating breast tumor in lipidomics level may not only elucidate the lipid molecular composition of interested samples but also deliver clues about the mechanisms behind the control of lipid homeostasis [6].
In recent years, several studies have investigated the application of lipidomics in breast cancer research. Phospholipids were revealed to be altered in breast cancer cells [7, 8], patients’ urine, and tissue [9–11]. Apart from these, sphingolipids and glycosphingolipids were demonstrated to be either pro- or antitumorigenic [12]. However, only a few of these studies included the samples from benign breast tumor patients. As a cancer biomarker should differentiate breast cancer from healthy control, as well as benign breast tumor, the proposed biomarkers for breast cancer in these studies need further confirmation. If significant differences only exist between breast cancer and healthy control, but do not exist between benign and malignant tumors, they are tumor biomarkers other than cancer biomarkers. Though earlier studies have demonstrated that both benign and malignant proliferation of breast tissue is associated with change in plasma lipids and lipoprotein levels [13, 14], details about the difference of individual lipid alternation with respect to benign and malignant breast tumors remain very limited. We believe that the study on the difference in lipid profiling between benign and malignant breast tumors would be helpful for finding more relevant cancer biomarkers.
In addition, most of the previous studies could only detect several classes of lipids, which limited people to gain a comprehensive understanding of the role of lipids played in cancer. In our previous work, an online normal-phase/reversed-phase two-dimensional lipid chromatography-mass spectrometry (NP/RP 2D LC-MS) method was developed [15–17], which could realize the simultaneous detection of lipid species from 12 lipid classes. This method has been used for lipid profiling of human plasma from peritoneal dialysis patients [18] and modified for comprehensive lipid profiling of human plasma from atherosclerosis patients, including free fatty acid (FFA), phosphatidylinositol (PI), phosphatidylglycerol (PG), lysophosphatidylglycerol (LPG), phosphatidylethanolamine (PE), lysophosphatidylethanolamine (LPE), phosphatidylserine (PS), phosphatidylcholine (PC), lysophosphatidylcholine (LPC), sphingomyelin (SM), ceramide (Cer), galactosylceramide (GalCer), glucosylceramide (GluCer), lactosylceramide (LacCer), monoacylglycerol (MG), diacylglycerol (DG), and triacylglycerol (TG), showing GalCers increasing in plasma of atherosclerotic stroke patients [19].
In this study, we employed the method to measure the lipid species in plasma taken from healthy people and patients with benign breast tumor or breast cancer before treatment. To the best of our knowledge, this is the first comprehensive evaluation of plasma lipid profiles with benign breast tumor patients. Unlike the work based on the comparison between healthy controls and breast cancer patients, the introduction of benign breast tumor group provides us a more detailed classification of lipid biomarkers, including potential biomarkers for breast tumor and breast cancer, and the biomarkers to evaluate the malignancy of breast tumor. As the diagnosis of breast tumors is based on visual inspection of stained tissue sections by an experienced pathologist now [20], our results indicated the potential diagnostic efficacy of lipids in breast tumors, which is much less harmful to people than traditional surgery.
Materials and methods
Materials
HPLC-grade chloroform was purchased from Mallinckrodt Baker (Phillipsburg, NJ, USA). HPLC-grade n-hexane, isopropanol, and methanol, as well as ammonium formate (purity ≥99.995 %), were obtained from Sigma-Aldrich (St. Louis, MO, USA). Purified water was acquired from Hangzhou Wahaha Group Co., (Zhejiang, China). All lipid standards were purchased from Avanti Polar Lipids Inc. (Alabaster, AL, USA), which are listed in the Electronic Supplementary Material (ESM).
Collection of plasma samples
Six patients with benign breast tumor and five patients with breast cancer who underwent surgery at the Cancer Hospital and Institute of Chinese Academy of Medical Sciences and Peking Union Medical College were included in this study. All these patients were diagnosed and confirmed by pathology. Plasma samples of these patients were collected at the time of inclusion before surgery. In addition, nine healthy individuals were also included in this study as control subjects at the same period, and plasma samples were also collected from these individuals. Verbal consent was obtained from every patient and from every healthy individual, but a written consent form was not required because the plasma samples used in this study were the residual after clinical use. This study was approved by the Institutional Review Board of Cancer Institute and Hospital, Chinese Academy of Medical Sciences.
Extraction of lipids
Prior to analysis, samples were naturally thawed at 4 °C. Lipids were extracted from human plasma according to the method of Bligh and Dyer [21] with some modification. Briefly, 100 μL of plasma was extracted with Folch solution (chloroform/methanol, 2:1, 6 mL) after adding an internal standard mixture (50 ng of each standard). The samples were sonicated for 3 min and centrifuged at 10,000 rpm for 30 min (LG 10-2.4A, China). Then 1.2 mL of water was added to the supernatant. After centrifuging at 5000 rpm for 15 min, the lower layer was filtered and dried by evaporation under nitrogen. Finally, the dried lipid extracts were redissolved in 0.5 mL Folch solution for HPLC injection.
Lipidomics analysis
Lipid profiling was performed on a non-stop-flow NP/RP 2D LC system [19], which was built based on a ten-port, two-position valve (Agilent Technologies, CA, USA) vacuum evaporation interface. As two loops were used to trap and transfer the first-dimensional elute to the second dimension separately, this system could suppress the sample band broadening in the first-dimensional column, increasing the recovery and repeatability of 2D LC interface. The first-dimensional separation was carried out on a Rx-SIL silica column (2.1 i.d. × 150 mm, 5 μm; Agilent Technologies, CA, USA) using an Agilent 1100 system consisting of a quaternary pump with an online degasser, an Agilent 1200 series autosampler, and a thermostated column compartment (Agilent Technologies, Waldbronn, Germany). In the second dimension, an Agilent 1200 series binary pump with an online degasser (Agilent Technologies, Waldbronn, Germany) delivered the mobile phase through a Poroshell 120 EC C8 column (2.1 i.d. × 50 mm, 2.7 μm; Agilent Technologies, CA, USA). Lipids were detected, and their structures were verified by an Agilent 6530 Accurate-Mass Quadrupole Time-of-Flight (QToF) mass spectrometer equipped with an Agilent Jet Stream ESI source (Agilent Technologies, CA, USA).
MS data were processed using the Mass Hunter Qualitative Analysis B.04.00 software (Agilent Technologies, CA, USA). All extracted ion chromatograms (EICs) were obtained with ±10 ppm m/z expansion. Peak areas of the validation standards and identified lipids were integrated from EICs.
Statistical analysis
Prior to principal component analysis (PCA), MS data of all samples were changed to compound exchange format (CEF) by MassHunter Qualitative Analysis B.04.00 (Agilent Technologies, CA, USA) software. Then the data was imported to the Mass Profiler Professional software. After mass filtering, alignment, internal standard normalization, the statistical results were analyzed by using PCA method. Differences were considered as significant if fold-change was larger than 2.0 and p value was less than 0.05.
Further statistical analysis was performed using IBM SPSS software, version 20 (SPSS Inc., Chicago, IL). Peak area data (after correction with corresponding internal standards) of each lipid species from all samples were analyzed by Mann–Whitney U test. Differences were considered to be statistically significant if fold-change was larger than 2.0 and p value was less than 0.05. Box plots were also generated from this software.
Result and discussion
Plasma lipid profiling
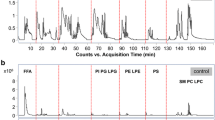
Lipid extracts from human plasma samples were first analyzed by NP/RP 2D LC-MS method mentioned above under both positive and negative ion modes for the identification of lipid species. The base peak chromatograms (BPCs) of control, benign breast tumor patient, and breast cancer patient obtained by this method are shown in Fig. 1, indicating that there were obvious differences among these groups. Besides, sphingolipids, including Cer, LacCer, GalCer, and GluCer, and glycerolipids, including MG, DG, and TG, were characterized at positive ion mode, while FFA and phospholipids, including PI, PG, LPG, PS, PE, LPE, PC, LPC, and SM, were characterized at negative ion mode.
a Comparison of base peak chromatograms (BPCs) of human plasma lipid extracts from a control sample, a benign breast tumor patient sample, and a breast cancer patient sample obtained by NP/RP 2D LC-MS at positive ion mode. b Comparison of base peak chromatograms (BPCs) of human plasma lipid extracts from a control sample, a benign breast tumor patient sample, and a breast cancer patient sample obtained by NP/RP 2D LC-MS at negative ion mode
Identification of lipid species was according to the accurate m/z, retention time, relative retention time of the species in the same class, and the spectra of tandem mass spectrometry (MS/MS) as described previously [19]. In the end, 512 lipid species were determined. The complete list of these lipid species was provided in ESM Table S1. This large database gave us a chance to fully investigate the influence of each lipid species other than lipid classes on breast cancer.
Multivariate statistical analysis
To obtain a preliminary understanding of the lipid profiling data, unsupervised PCA was used after picking out lipids from CEF files (based on the profiling data). The 2D and 3D PCA score plots of the three groups (healthy controls, benign breast tumor, and breast cancer) are shown in Fig. 2a (positive ion mode) and Fig. 2b (negative ion mode). As seen in Fig. 2, three distinctive clusters were formed at both positive and negative ion modes, which indicated the existence of lipid species with significant differences.
a 2D and 3D score plot of principal component analysis (PCA) of healthy control group (red), benign breast tumor group (brown), and breast cancer group (blue) at positive ion mode. b 2D and 3D score plot of principal component analysis (PCA) of healthy control group (red), benign breast tumor group (brown), and breast cancer group (blue) at negative ion mode
Quantitative analysis of lipids
For the quantitative analysis of identified lipid molecules, all samples were examined by the NP/RP 2D LC-MS method at both positive and negative precursor ion scan modes to measure the peak areas of detected species. The control group and patient groups were alternately injected to reduce systemic error. In order to compensate for the fluctuations in MS intensities during different runs, peak areas of each individual lipid species were corrected by that of an internal standard corresponding to the same lipid class. Then, the corrected peak areas of all identified lipid species were used for further analysis.
Comparison among three groups indicated candidate biomarkers for breast tumor and breast cancer
We first calculated the corrected peak areas representing the average value of the control samples (n = 9), the benign breast tumor samples (n = 6), and the breast cancer samples (n = 5) along with the standard deviation values, p values of the Mann–Whitney U test accomplished for each two of the three groups, and the ratio of the average corrected peak area values between each two of the three groups. Comparison of this part was based on these data.
Among the detected lipid species, four PGs, one LPE, one SM, three Cers, one GalCer, three MGs, ten DGs, and two TGs have shown significant differences between control group and either one of the two patient groups (p ≤ 0.05). Besides, there was no significant difference between breast cancer group and benign breast tumor group. Although these species could differentiate healthy controls and breast cancer patients (fold change ≥2), the similarities between benign breast tumor and breast cancer groups made them excluded from cancer biomarkers. To the best of our knowledge, many lipidomics studies which aim at revealing novel cancer biomarkers just compared healthy control group with breast cancer patient group. However, our results showed that introducing benign tumor group was vital for searching cancer biomarkers, as it could eliminate the compounds with similarities between the two patient groups. Thus, these species were considered to be candidate breast tumor biomarkers. All the results are listed in Table 1.
To select the candidate breast cancer biomarkers, the healthy control group and benign breast tumor group were taken as two control groups, and then compared with breast cancer group, respectively. Lipid species which showed significant differences between breast cancer group and both of the control groups (p ≤ 0.05) with more than two fold change between breast cancer group and healthy control group, and no significant difference between the two control groups, were picked out. As a result, PI (32:1) [PI (16:0/16:1)] and PI (38:4) [PI (18:0/20:4)] were selected as candidate breast cancer biomarkers. In addition, as the p value of PI (34:2) [PI (16:0/18:2)] was 0.052 between breast cancer group and benign breast tumor group, it was also considered as a candidate breast cancer biomarker initially. Details of the data are also listed in Table 2.
Apart from the results above, we found that four species, including PI (34:1) [PI (16:0/18:1)], PG (36:3), GluCer (33:2) [GluCer (d18:1/15:1)], and MG (16:0), showed significant differences in each two of the three groups (p = 0.052 was included initially), as indicated in Table 3. Thus, whether they were cancer biomarkers or not needed further investigation.
Validation of the candidate biomarkers for breast tumor
To further verify the candidate breast tumor biomarkers, the benign breast tumor and breast cancer groups were treated as one group, which was named as patient group. Then this new group was compared with the healthy control group. New parameters, including the corrected peak areas representing the average value of the control samples (n = 9) and the patient samples (n = 11) along with the standard deviation values, p values, and the ratio of the average relative peak area values between the two groups, were calculated for the 25 candidate breast tumor biomarkers. Lipid species with p ≤0.05 and fold-change ≥2.0 were further selected as potential breast tumor biomarkers, as shown in Fig. 3a. In total, 23 candidate breast tumor biomarkers have been verified. Among these species, PGs, LPE, SM, Cers, DGs, and TGs increased in patient group compared with control group, while GalCer and MGs showed distinct decreases. To further investigate this trend, Fig. 3b displays the average corrected peak areas of these lipid species in the three groups. As indicated in this figure, PG (32:0), PG (36:1), LPE (24:0), Cer (38:1), DG (32:0), DG (34:4), DG (34:0), DG (35:3), DG (35:0), DG (36:0), DG (37:0), DG (38:0), TG (50:0), and TG (54:0) showed a consistent increase from healthy control group to benign breast tumor group, then to breast cancer group, while others did not. We suppose these species are more important in the progression of breast tumor. In addition, we noticed that most of these species were DGs, and DG (34:4) has shown the largest increase in patient group compared to healthy control group. Previous studies have shown that DG signaling played an important role in cancer progression through protein kinase C (PKC) [22]. Changes in specific DG species could influence PKC activation and consequently the regulation of cell proliferation [23]. As DGs were important intermediates of lipid metabolism and cellular signaling, and their concentrations were demonstrated to be altered in diabetes, cancer, as well as other disease states [24], our results might provide critical information for the underlying molecular mechanisms leading to tumor.
a Average corrected peak areas of 23 lipid species selected from statistical treatments between control group and patient group. The blue bars represent the control group, and red bars represent the patient group. The data are expressed as mean value ± SD. b Average corrected peak areas of 23 lipid species selected from statistical treatments among control group, benign breast tumor group, and breast cancer group. The blue bars represent the control group, red bars represent the benign breast tumor group, and green bars represent the breast cancer group. The data are expressed as mean value ± SD
Apart from this, three GluCers, including GluCer (35:1), GluCer (40:6), and GluCer (40:3), were detected only in most of the control samples (eight of nine samples), which also had potential to predict the existence of breast tumor.
Validation of the candidate biomarkers for breast cancer
As for the validation of the candidate biomarkers for breast cancer, we regarded the benign breast tumor group and healthy control group as a non-cancerous group, and compared it with breast cancer group. Similar parameters were also obtained as before. At last, two PI species, including PI (16:0/16:1) and PI (18:0/20:4), were considered as potential breast cancer biomarkers. As shown in Fig. 4a, they showed clear decreases in the breast cancer group compared with the non-cancerous group.
PIs are phospholipids comprising two fatty acid chains linked by a glycerol moiety to a water-soluble inositol head group. They are precursors of phosphatidylinositol-3,4,5-tris-phosphate (PIP3) [7], which is a signaling lipid and included in the phosphatidylinositol 3-kinase (PI3K)/AKT pathway. In this pathway, the phosphotyrosines of activated growth factor receptors or G protein-coupled receptors can interact with class 1A PI3Ks, which catalyzes the formation of PIP3. On the other hand, phosphatase and tensin homologue deleted on chromosome 10 (PTEN), which acts as the tumor suppressor, can dephosphorylate PIP3 at 3′ inositol position [25, 26]. In recent years, the PI3K/AKT pathway has been found to play a crucial role in the cell metabolism, survival, growth, proliferation, polarity, and apoptosis, including breast cancer cells [27]. Upregulation of AKT and its upstream PI3K and loss of negative regulation of PTEN have been discovered in various malignant tumors. Bachman et al. [28] and Isakoff et al. [29] identified somatic mutations in PIK3CA, which encoded the p110alpha catalytic subunit of PI3K, in approximately 18 to 50 % of breast cancer tissues and cell lines. Besides, the gene was also found to be amplified and overexpressed in breast cancer [30]. These genomic alterations cause increased, growth factor-independent lipid kinase activity of the catalytic subunit, which leads to the accumulation of PIP3. In other words, the consumption of PIs has been expedited. Due to promoter hypermethylation and loss of heterozygosity owing to gene deletion or mutation, loss of PTEN function also occurs frequently in breast cancer, which stimulates PI3K/AKT downstream signaling and tumor growth [31]. These events will undoubtedly lead to the decrease of PIs in cancer samples.
In our study, the concentrations of PI (16:0/16:1) and PI (18:0/20:4) in cancer patients were lower than non-cancerous controls, which were consisted with the activation of PI3K/AKT pathway. In addition, a decrease of PI (18:0/20:4) was also reported in a previous study focusing on the urinary phospholipids in patients with breast cancer [11], as well as breast cancer cells [7], which were in line with our findings. To further reveal the differences, box plots of the three groups were presented. As shown in Fig. 4b, differences were more significant between breast cancer group and healthy control or benign breast tumor group. Besides, we also noticed two extreme abnormal values in PI (16:0/16:1). After further investigation, dot 18 represented a patient with multiple tumors and dot 17 represented the youngest patient in the breast cancer group. However, whether these reasons contributed to the abnormal phenomenon should be further investigated.
PI (16:0/18:1), PG (36:3), and GluCer (d18:1/15:1) were potential biomarkers to evaluate the level of malignancy of breast tumor
At last, we paid our attention to the four lipid species that showed significant differences in each two of the three groups. To verify their classification, we conducted the comparisons both for the validation of candidate breast tumor and breast cancer biomarkers. Based on the restricted conditions, we found that PI (16:0/18:1), PG (36:3), and GluCer (d18:1/15:1) could be classified as biomarkers for breast tumor or breast cancer, while MG (16:1) was excluded. In Fig. 5, PI (16:0/18:1) revealed a downregulation along with the severity of the disease, while PG (36:3) and GluCer (d18:1/15:1) were on the contrary. Thus, these lipid species could be proposed as biomarkers to evaluate the level of malignancy of breast tumor, which had the potential to discriminate people among healthy controls, benign breast tumor patients, and breast cancer patients.
As the change of PI (16:0/18:1) was in line with the activation of PI3K/AKT pathway mentioned above, and the upregulation of PG in breast cancer has been reported in other work [11], GluCer (d18:1/15:1) attracted our attention for the clear distinction among the three groups. GluCers were produced relying on glucosylceramide synthase (GCS), an enzyme which could catalyze the transfer of glucose to ceramide [32]. Studies on GCS activity suggested that it could potentiate the development of multidrug resistance in cancer cells [33, 34]. As for breast cancer, GCS has been demonstrated to be overexpressed in metastatic breast carcinoma [35]. Besides, expression of GCS conferred adriamycin resistance in human breast cancer cells [36]. On the contrary, inhibition of the activity of GCS downregulated P-glycoprotein and resensitized breast cancer cells to anticancer drugs [37]. As a product of GCS, GluCers were expected to be higher in cancer tissues as a consequence of GCS activity, which has been demonstrated in a breast tumor specimen [38]. However, previous studies about GluCers were in the lipid class level, and few studies have investigated the role of GluCers in molecular species level. Our study revealed that GluCer (d18:1/15:1) might act as a potential biomarker to evaluate the level of malignancy of breast tumor.
Conclusions
This study profiled the plasma lipidomes of patients with benign breast tumor and breast cancer using a NP/RP 2D LC-MS platform. Although the present work is preliminary with a limited number of samples, it provides a guideline to screen potential biomarkers for benign and malignant breast tumors, which could be used as target molecules for high-speed scanning of a large number of samples. Based on this study, 26 lipid species were proposed as potential breast tumor biomarkers. Among them, GluCer (35:1), GluCer (40:6), and GluCer (40:3) were detected only in healthy control samples, while the others were observed in all samples with significant variations between healthy controls and two patient groups, and significant differences between the patient subgroups were rare. More importantly, PI (16:0/16:1) and PI (18:0/20:4) were considered as potential breast cancer biomarkers, which could differentiate between benign and malignant breast tumors. As for PI (16:0/18:1), PG (36:3), and GluCer (d18:1/15:1), they showed significant differences among the three groups, and consistent variation trends were observed along with the severity of the disease. They were proposed as potential biomarkers to evaluate the level of malignancy of breast tumor. For the confirmation of these lipid biomarkers, a systematic examination of the proposed biomarkers with a larger number of patient samples is expected.
Abbreviations
- BPCs:
-
Base peak chromatograms
- CEF:
-
Compound exchange format
- Cer:
-
Ceramide
- DG:
-
Diacylglycerol
- EICs:
-
Extracted ion chromatograms
- FFA:
-
Free fatty acid
- GalCer:
-
Galactosylceramide
- GCS:
-
Glucosylceramide synthase
- GluCer:
-
Glucosylceramide
- LacCer:
-
Lactosylceramide
- LPC:
-
Lysophosphatidylcholine
- LPE:
-
Lysophosphatidylethanolamine
- LPG:
-
Lysophosphatidylglycerol
- MG:
-
Monoacylglycerol
- MS/MS:
-
Tandem mass spectrometry
- NP/RP 2D LC-MS:
-
Normal-phase/reversed-phase two-dimensional lipid chromatography-mass spectrometry
- PC:
-
Phosphatidylcholine
- PCA:
-
Principal component analysis
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- PI:
-
Phosphatidylinositol
- PI3K:
-
Phosphatidylinositol 3-kinase
- PIP3:
-
Phosphatidylinositol-3,4,5-tris-phosphate
- PKC:
-
Protein kinase C
- PS:
-
Phosphatidylserine
- PTEN:
-
Phosphatase and tensin homologue deleted on chromosome 10
- QToF:
-
Quadrupole Time-of-Flight
- SCD1:
-
Stearoyl-CoA desaturase
- SM:
-
Sphingomyelin
- TG:
-
Triacylglycerol
References
Siegel R, Ma JM, Zou ZH, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64(1):9–29. doi:10.3322/caac.21208
Sasco AJ (2001) Epidemiology of breast cancer: an environmental disease? APMIS 109(5):321–332. doi:10.1034/j.1600-0463.2001.090501.x
Aghaei M, Karami-Tehrani F, Salami S, Atri M (2010) Diagnostic value of adenosine deaminase activity in benign and malignant breast tumors. Arch Med Res 41(1):14–18. doi:10.1016/j.arcmed.2009.10.012
Gross RW, Han XL (2011) Lipidomics at the interface of structure and function in systems biology. Chem Biol 18(3):284–291. doi:10.1016/j.chembiol.2011.01.014
Spener F, Lagarde M, Geloen A, Record M (2003) What is lipidomics? Eur J Lipid Sci Technol 105(9):481–482. doi:10.1002/ejlt.200390101
Oresic M, Hanninen VA, Vidal-Puig A (2008) Lipidomics: a new window to biomedical frontiers. Trends Biotechnol 26(12):647–652. doi:10.1016/j.tibtech.2008.09.001
Doria ML, Cotrim Z, Macedo B, Simoes C, Domingues P, Helguero L, Domingues MR (2012) Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res Treat 133(2):635–648. doi:10.1007/s10549-011-1823-5
Doria ML, Cotrim CZ, Simoes C, Macedo B, Domingues P, Domingues MR, Helguero LA (2013) Lipidomic analysis of phospholipids from human mammary epithelial and breast cancer cell lines. J Cell Physiol 228(2):457–468. doi:10.1002/jcp.24152
Hilvo M, Denkert C, Lehtinen L, Muller B, Brockmoller S, Seppanen-Laakso T, Budczies J, Bucher E, Yetukuri L, Castillo S, Berg E, Nygren H, Sysi-Aho M, Griffin JL, Fiehn O, Loibl S, Richter-Ehrenstein C, Radke C, Hyotylainen T, Kallioniemi O, Iljin K, Oresic M (2011) Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res 71(9):3236–3245. doi:10.1158/0008-5472.can-10-3894
Kim H, Min HK, Kong G, Moon MH (2009) Quantitative analysis of phosphatidylcholines and phosphatidylethanolamines in urine of patients with breast cancer by nanoflow liquid chromatography/tandem mass spectrometry. Anal Bioanal Chem 393(6–7):1649–1656. doi:10.1007/s00216-009-2621-3
Min HK, Kong G, Moon MH (2010) Quantitative analysis of urinary phospholipids found in patients with breast cancer by nanoflow liquid chromatography-tandem mass spectrometry: II. Negative ion mode analysis of four phospholipid classes. Anal Bioanal Chem 396(3):1273–1280. doi:10.1007/s00216-009-3292-9
Hilvo M, Oresic M (2012) Regulation of lipid metabolism in breast cancer provides diagnostic and therapeutic opportunities. Clin Lipidol 7(2):177–188. doi:10.2217/clp.12.10
Schiffmann S, Sandner J, Birod K, Wobst I, Angioni C, Ruckhaberle E, Kaufmann M, Ackermann H, Lotsch J, Schmidt H, Geisslinger G, Grosch S (2009) Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis 30(5):745–752. doi:10.1093/carcin/bgp061
Shah FD, Shukla SN, Shah PM, Patel HRH, Patel PS (2008) Significance of alterations in plasma lipid profile levels in breast cancer. Integr Cancer Ther 7(1):33–41. doi:10.1177/1534735407313883
Nie HG, Liu RR, Yang YY, Bai Y, Guan YF, Qian DQ, Wang T, Liu HW (2010) Lipid profiling of rat peritoneal surface layers by online normal- and reversed-phase 2D LC QToF-MS. J Lipid Res 51(9):2833–2844. doi:10.1194/jlr.D007567
Li M, Yang L, Bai Y, Liu HW (2014) Analytical methods in lipidomics and their applications. Anal Chem 86(1):161–175. doi:10.1021/ac403554h
Li M, Zhou ZG, Nie HG, Bai Y, Liu HW (2011) Recent advances of chromatography and mass spectrometry in lipidomics. Anal Bioanal Chem 399(1):243–249. doi:10.1007/s00216-010-4327-y
Li M, Feng BS, Liang Y, Zhang W, Bai Y, Tang W, Wang T, Liu HW (2013) Lipid profiling of human plasma from peritoneal dialysis patients using an improved 2D (NP/RP) LC-QToF MS method. Anal Bioanal Chem 405(21):6629–6638. doi:10.1007/s00216-013-7109-5
Li M, Tong XL, Lv P, Feng BS, Yang L, Wu Z, Cui XJ, Bai Y, Huang YN, Liu HW (2014) Lipid profiling reveals galactosylceramides increasing in plasma of atherosclerotic stroke patients by online 2D LC QToF-MS. J Chromatogr A 1372:110–119
Fabian H, Lasch P, Boese M, Haensch W (2003) Infrared microspectroscopic imaging of benign breast tumor tissue sections. J Mol Struct 661:411–417. doi:10.1016/j.molstruc.2003.07.002
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37(8):911–917
Nishizuka Y (1995) Protein kinase C and lipid signaling for sustained cellular-responses. FASEB J 9(7):484–496
Deacon EM, Pettitt TR, Webb P, Cross T, Chahal H, Wakelam MJO, Lord JM (2002) Generation of diacylglycerol molecular species through the cell cycle: a role for 1-stearoyl, 2-arachidonyl glycerol in the activation of nuclear protein kinase C-beta II at G2/M. J Cell Sci 115(5):983–989
Li YL, Su X, Stahl PD, Gross ML (2007) Quantification of diacylglycerol molecular species in biological samples by electrospray ionization mass spectrometry after one-step derivatization. Anal Chem 79(4):1569–1574. doi:10.1021/ac0615910
Grunt TW, Mariani GL (2013) Novel approaches for molecular targeted therapy of breast cancer: interfering with PI3K/AKT/mTOR signaling. Curr Cancer Drug Targets 13(2):188–204
Vara JAF, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M (2004) PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 30(2):193–204. doi:10.1016/j.ctrv.2003.07.007
Cui WL, Cai Y, Zhou XY (2014) Advances in subunits of PI3K class I in cancer. Pathology 46(3):169–176. doi:10.1097/pat.0000000000000066
Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH (2004) The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther 3(8):772–775
Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, Cantley LC, Brugge JS (2005) Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res 65(23):10992–11000. doi:10.1158/0008-5472.can-05-2612
Jiang BH, Liu LZ (2008) PI3K/PTEN signaling in tumorigenesis and angiogenesis. BBA Proteins Proteomics 1784(1):150–158. doi:10.1016/j.bbapap.2007.09.008
Ghayad SE, Cohen PA (2010) Inhibitors of the PI3K/Akt/mTOR pathway: new hope for breast cancer patients. Recent Patents Anti-Cancer Drug Discov 5(1):29–57
Ryland LK, Fox TE, Liu X, Loughran TP, Kester M (2011) Dysregulation of sphingolipid metabolism in cancer. Cancer Biol Ther 11(2):138–149. doi:10.4161/cbt.11.2.14624
Liu YY, Han TY, Giuliano AE, Cabot MC (2001) Ceramide glycosylation potentiates cellular multidrug resistance. FASEB J 15(3):719–730. doi:10.1096/fj.00-0223com
Morjani H, Aouali N, Belhoussine R, Veldman RJ, Levade T, Manfait M (2001) Elevation of glucosylceramide in multidrug-resistant cancer cells and accumulation in cytoplasmic droplets. Int J Cancer 94(2):157–165. doi:10.1002/ijc.1449
Liu YY, Patwardhan GA, Xie P, Gu X, Giuliano AE, Cabot MC (2011) Glucosylceramide synthase, a factor in modulating drug resistance, is overexpressed in metastatic breast carcinoma. Int J Oncol 39(2):425–431. doi:10.3892/ijo.2011.1052
Liu YY, Han TY, Giuliano AE, Cabot MC (1999) Expression of glucosylceramide synthase, converting ceramide to glucosylceramide, confers adriamycin resistance in human breast cancer cells. J Biol Chem 274(2):1140–1146. doi:10.1074/jbc.274.2.1140
Gouaze V, Liu YY, Prickett CS, Yu JY, Giuliano AE, Cabot MC (2005) Glucosylceramide synthase blockade down-regulates P-glycoprotein and resensitizes multidrug-resistant breast cancer cells to anticancer drugs. Cancer Res 65(9):3861–3867. doi:10.1158/0008-5472.can-04-2329
Lucci A, Cho WI, Han TY, Giuliano AE, Morton DL, Cabot MC (1998) Glucosylceramide: a marker for multiple-drug resistant cancers. Anticancer Res 18(1B):475–480
Acknowledgment
This study was financially supported by the National Natural Science Foundation of China (grant no. 21175005).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Published in the topical collection Lipidomics with guest editor Michal Holčapek.
Li Yang and Xinge Cui contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 52 kb)
Rights and permissions
About this article
Cite this article
Yang, L., Cui, X., Zhang, N. et al. Comprehensive lipid profiling of plasma in patients with benign breast tumor and breast cancer reveals novel biomarkers. Anal Bioanal Chem 407, 5065–5077 (2015). https://doi.org/10.1007/s00216-015-8484-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-015-8484-x