Abstract
The purpose of this work was to fabricate an electrochemical DNA biosensor for detecting hepatitis B virus. Gold nanorods (GNRs), which are known for their conductivity, were used to increase surface area and consequently increase the immobilization of single-stranded DNA (ss-DNA) on the modified gold electrode. The GNRs were characterized via transmission electron microscopy. The morphology of the gold electrode before and after modification with GNRs was characterized by scanning electron microscopy. Atomic-force microscopy was used to evaluate the morphology of the GNR electrode surface before and after interaction with ss-DNA. Cyclic voltammetry was used to monitor DNA immobilization and hybridization, using [Co(phen)3]3+ as an electrochemical indicator. The target DNA sequences were quantified at a linear range from 1.0 × 10−12 to 10.0 × 10−6 mol L−1, with a detection limit of 2.0 × 10−12 mol L−1 by 3σ. The biosensor had good specificity for distinguishing complementary DNA in the presence of non-complementary and mismatched DNA sequences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of DNA biosensors has attracted much attention for many applications, including detecting gene sequences [1], identifying pathogens in product and food industries [2], and forensic applications [3]. Numerous techniques, including quartz-crystal microbalance [4], electrochemical [5], optic [6], and surface plasmon resonance [7] methods, have been widely studied for DNA detection. Among them, electrochemical biosensors have become very popular because of their great advantages, including simplicity, rapidity, portability, low cost, and high sensitivity [8].
An important problem in the development of DNA hybridization biosensors is enhancing the immobilization amount and accessibility of the DNA probe for hybridization with target DNA, to improve the DNA biosensor performance. Many kinds of nanomaterial, including metal [9], oxide [10], and semiconductor [11] nanoparticles, can be used to improve immobilization of biomolecules. Combining nanoparticles with biomolecules improves biosensor performance because of their large surface area. Cai et al. used colloidal gold with a 16 nm diameter to increase the immobilization of DNA on a gold electrode and improve an electrochemical DNA biosensor. Oligonucleotides with a mercaptohexyl group at the 5′-phosphate end were immobilized on gold nanoparticles self-assembled on a cysteamine-modified gold electrode, which increased immobilization of single-stranded DNA (ss-DNA) on the modified electrode in comparison with a bare gold electrode [12]. Zhang et al. reported immobilization of DNA on a silica-nanoparticle-modified electrode, resulting in increased immobilization of ss-DNA brought about by the large surface area [13]. Fu et al. reported fabrication of a DNA biosensor by self-assembling colloidal Ag to a thiol-containing sol–gel network. Silver nanoparticles could be assembled into the sol–gel network and mercapto oligonucleotides bound to the Ag nanoparticles. The results indicated that the Ag nanoparticles increased the surface area of the electrode, which led to an improved DNA anchor [14]. More research on electrochemical genosensors has focused on spherical nanoparticles [9] than on nanorods. However, GNRs have several advantages over spherical gold nanoparticles, including faster electron transfer [15], improved light-scattering properties [6]. These properties lead to higher sensitivity with lower detection limits. More studies on the use of GNRs in DNA detection have focused on localized surface plasmon resonance [16] and optical biosensors [17, 18] than on electrochemical ones. For example, HJ. Parab et al. [6] reported an optical biosensor for detecting target DNA, based on GNRs as molecular probes, and X. Wang et al. [16] designed a GNR biosensor using localized surface plasmon resonance behavior for detecting the hepatitis B surface antigen.
Viral hepatitis caused by hepatitis B virus (HBV) is regarded as a serious public health problem of worldwide importance, and causes other harmful consequences including cirrhosis and hepatocellular carcinoma. There are an estimated 400 million people worldwide who live with chronic HBV infection [19], and mortality as a result of liver disease caused by HBV infection is estimated to be approximately one million people annually [20]. Therefore, there have been great efforts to develop methods for detecting and quantifying HBV. Serum HBV DNA level can be regarded as a criterion for the efficacy of treatment for chronic hepatitis B and used as an effective marker for antiviral therapy, and this is performed via molecular hybridization assays [21, 22].
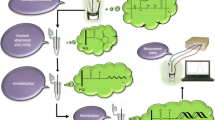
In this paper, we describe an electrochemical DNA biosensor for detecting HBV. The DNA biosensor surface was made from sequential modification of 1,6-hexanedithiol (HDT) and GNRs. First, HDT was assembled on the gold electrode surface, and GNRs were then placed on the HDT-modified electrode. The GNRs were used to increase the surface area of the electrode to enable the assembly of a greater amount of ss-DNA. Surface morphology of the gold electrode before and after modification with GNR was characterized by scanning electron microscopy (SEM), and atomic force microscopy (AFM) was used to evaluate the morphology of the GNR-modified electrode surface before and after interaction with ss-DNA. ss-DNA immobilization and hybridization on the GNR-modified electrode was studied by [Co(phen)3]3+ as an electrochemical indicator. The results revealed that GNRs improve the electrode surface in comparison with a bare gold electrode, and that the amount of DNA probe and the sensitivity of the DNA biosensor increase. The target DNA could be quantified in a linear range from 1.0 × 10−12 to 10.0 × 10−6 mol L−1, with a detection limit of 2.0 × 10−12 mol L−1. In addition, the biosensor had good specificity for distinguishing complementary DNA from non-complementary and mismatched DNA sequences.
Materials and methods
Materials
Gold (III) chloride trihydrate, sodium borohydride, L-ascorbic acid, potassium chloride, sodium chloride, sodium phosphate dibasic, and potassium phosphate monobasic were purchased from Sigma (USA). Hexadecyltrimethylammonium bromide (CTAB), silver nitrate, sodium hydroxide, absolute ethanol, 6-mercapto-1-hexanol (99 %), potassium ferrocyanide, and sulfuric acid were from Merck (Germany). 1,6-Hexanedithiol (HDT) (97 %) was bought from Acros (USA).
All electrochemical experiments were performed using a computer-controlled μAutolab type III Potentiostat (Eco Chemie, Utrecht, The Netherlands). Gold electrode, Ag–AgCl wire, and platinum wire electrode were used as working electrode, auxiliary reference, and counter electrodes, respectively.
The sequence of oligonucleotides was selected using Basic Local Alignment Search Tool (BLAST) to have the least similarity to the human serum genome. The oligonucleotides used in this work were:
Sequence name | Sequence of oligonucleotides | Company and country |
Thiolated-probe | 5′ - HS (CH2)6 TAC CGT CCC CTT CTT CAT CTG CCG T - 3′ | Vivantis, Malaysia |
Target | 5′ - ACG GCA GAT GAA GAA GGG GAC GGT A - 3′ | Vivantis, Malaysia |
One-point mismatch | 5′ - ACG CCA GAT GAA GAA GGG GAC GGT A - 3′ | Sinagen, Iran |
Non-complementary | 5′ - TAC CGT CCC CTT CTT CAT CTG CCG T - 3′ | Sinagen, Iran |
Stock solutions of oligonucleotides were prepared in PBS solution (100 mmol L−1, pH =7.4) and stored in a freezer (−20 °C). All solutions were prepared using double deionized water (DDW).
Preparation of GNRs
The GNRs were prepared using a seed-mediated growth method described in the literature [23]. For preparation of seed solution, CTAB solution (5 mL, 0.2 mol L−1) was mixed with HAuCl4 (5 mL, 0.5 mmol L−1). To the stirred solution, 0.6 mL 0.01 mol L−1 NaBH4 solution (kept in ice-cold conditions) was added as reducting agent, and stirring of the seed solution was then continued for 2 min. After stirring, the solution was kept at 25 °C.
For preparation of growth solution, 5 mL CTAB (0.2 mol L−1) was added to 0.25 mL AgNO3 solution (4 mmol L−1), and 22.5 mL HAuCl4 (1 mmol L−1) was then added to this solution during stirring. After mixing, 70 μL ascorbic acid (0.0788 mol L−1) was added as reducing agent. Finally, 12 μL seed solution was added to the growth solution; the temperature was constant at 24–27 °C, and the gold-nanorod solution was produced during 20–30 min. The GNRs were characterized by transmission electron microscopy (TEM) (Fig. 1). Before use of the gold nanorods for the next steps in the experiment, spherical nanoparticles and excess CTAB were removed by two rounds of centrifugation at 12,000 rpm, each round lasting 7 min, and the supernatant was decanted. GNRs were then suspended in an equal amount of DDW.
Preparation of gold electrode
The gold electrode was mechanically polished with alumina slurry on a micro cloth pad and washed with DDW. The electrode was then sonicated into ethanol and DDW, respectively, for 15 min, and dried. The electrode was then cycled in 0.5 mol L−1 sulfuric acid solution between −0.5 V and +1.3 V (versus Ag–AgCl); the scan rate was 0.15 V s−1 and 50 scans were performed.
GNR-modified gold electrode
Immobilization of GNRs on the gold electrode was performed using HDT. The monolayer of HDT was placed on the gold electrode by dipping a clean gold electrode into a 0.5 % (v/v) solution of HDT in ethanol for 1 h. The electrode was then washed by ethanol and dried with nitrogen stream. The electrode was next immersed into the aqueous goldnanorod solution for 1 h. The modified electrode was washed with DDW. SEM imaging indicated the presence of GNRs on the electrode surface.
Immobilization of thiolated single-stranded DNA probe
PBS (5 μL) (pH 4.5) containing 20 μmol L−1 ss-DNA probe was dropped on the modified electrode for 5 h, and the electrode was then washed with PBS (pH 7.4) and DDW, and dried with nitrogen stream. To remove unabsorbed ss-DNA from the electrode surface, the electrode was immersed into 2 mmol L−1 6-mercapto-1-hexanol solution for 30 min and then washed with PBS and DDW.
Hybridization of DNA probe with target DNA
The ss-DNA-probe-modified electrode was immersed in PBS (pH 7.4) containing 10 μmol L−1 ss-DNA target for 1 h; the DNA target was part of the DNA sequence of the hepatitis B virus and complementary to the DNA probe. Afterwards, the electrode was washed with DDW and dried with nitrogen stream.
Results and discussion
Surface study of GNR-modified electrode by SEM
Figure 2 shows the SEM images of a planar gold electrode after deposition of gold nanorods: (a) bare gold electrode, and (b) GNR-modified gold electrode. As seen in Fig. 2a, before deposition of GNRs the surface of the electrode was relatively smooth. After modification with GNR, the electrode surface is occupied by a large quantity of GNRs in random orientations (Fig. 2b), which have self-assembled on the electrode surface.
Surface study of GNR-modified electrode by AFM
AFM images of GNR are shown in Fig. 4. The GNR have ordered shapes before interaction with the DNA probe, as shown by 3D micrographs (Fig. 3a). However, the ordered, organized shapes of GNR were changed to disordered and disorganized shapes after interaction with the DNA probe (Fig 3b). This may be the result of destruction of longer GNRs to shorter ones after interaction with the DNA probe. However, GNRs remained on the surface of the gold electrode after interaction with the DNA probe, increasing the surface-to-volume ratio.
Cyclic-voltametric behavior of [Fe(CN)6]3−/4− at modified electrodes
HDT contains two thiol functionalities which can act as a mediator to anchor GNRs onto a gold electrode. A self-assembling monolayer of HDT forms as the bare gold electrode is immersed in the HDT solution. A second functional group at the end of the self-assembled monolayer was used for the attachment of GNRs. Figure 4a shows the cyclic voltametric behavior of the electrode after each modification step. Compared with the bare gold electrode (curve a), the peak current reduced after self-assembly of HDT. This may indicate that density on the electrode surface increased, and the electron-transfer reaction between electroactive species and the gold-electrode surface decreased. However, with Au–HDT (1 h)–GNR (curve c), the current increased after GNR attachment to the HDT-modified electrode. This may be because GNR have an important effect, acting like a conducting wire facilitating electron transfer. X. Han et al. constructed a simple electrochemical sensor for DNA detection based on GNR-modified graphene-oxide sheets (GNR–graphene-oxide nanocomposite) [24]. However, we used HDT (1 h)–GNR to fabricate the electrochemical DNA biosensor for HBV.
(a) Cyclic-voltametric behavior of (a) bare Au electrode, (b) Au–HDT (1 h), and (c) Au–HDT (1 h)–GNR; and of (b) (a) bare Au electrode, (b) Au–HDT (1 h)–GNR (1 h)–DNA probe, and (c) Au–HDT (1 h)–GNR (1 h)–DNA probe–DNA target, for electrolyte solution of 2.5 mmol L−1 [Fe(CN)6]3−/4− aqueous solution containing 0.1 mol L−1 KCl, at scan rate 50 mV s−1
Figure S1 (Electronic Supplementary Material, ESM) depicts the current–concentration relationship as the bare gold electrode was immersed in HDT solutions of different concentrations. As shown in ESM Fig. S1, the current of the HDT-modified electrodes reduced as the HDT concentration increased. This curve can be divided into two parts. The current reduced rapidly as the HDT concentration in ethanol solution increased to 0.5 % (v/v), whereas the reduction in current become slower from 0.5 % to 2 %. The fact the current reduced at a slower rate may indicate that most of the bare-electrode surface was covered by HDT, which formed a stable self-assembled monolayer on the gold-electrode surface by one of the two thiol functionalities of HDT. This decrease in the slope of the current is a reason for hypothesizing the formation of a self-assembled monolayer and not a multilayer: if a self-assembled multilayer had formed the current would have decreased at a constant rate.
Figure S2 (see ESM) shows the relationship of current with incubation time of the bare gold electrode in HDT solution. As seen in ESM Fig. S2, the current reduced as the incubation time increased. This indicates that assembly of HDT monolayers on the electrode surface increased with increasing incubation time. Therefore, both concentration of HDT and incubation time of the bare gold electrode in HDT affect self-assembly of HDT on the electrode surface.
Figure 4b shows the cyclic voltametric behavior for different modified electrodes in 2.5 mmol L−1 [Fe(CN)6]3−/4− aqueous solution containing 0.1 mol L−1 KCl, at a scan rate of 50 mV s1. The optimized experimental conditions for the DNA probe were 20 μmol L−1 for 5 h, and those for the DNA target were 10 μmol L−1 for 1 h. The peak current of the bare Au electrode is shown in curve 4B(a). When the electrode surface was immobilized by the DNA probe, the peak current reduced. This may be caused by the repellence of [Fe(CN)6]3−/4− by the negatively-charged phosphate backbone of probe DNA immobilized on the electrode surface (curve 4B(b)). The reduction in the peak current was preceded by the hybridization of the DNA target with the DNA probe (curve 4B(c)). This can attributed to increased negative charges on electrode surface after hybridization of the DNA target with the DNA probe, resulting in repelling of [Fe(CN)6]3−/4−. Also, immobilization of the DNA probe and DNA target lead to increased density on the gold-electrode surface and prevent electron transfer between the electrode surface and electroactive species.
Cyclic-voltametric behavior of [Co(phen)3]3+ at DNA-modified electrode
Figure 5 shows the cyclic voltametric behavior for different modified electrodes in 100 mmol L−1 buffer solution after incubation in 4 mmol L−1 [Co(phen)3]3+ for 1 h. In the absence of DNA, the intensity of the peak current follows curve 5a. After immobilization of the DNA probe, the peak current increased (curve 5b). This may be attributed to non-specific adsorption of electrostatic interaction between ss-DNA and [Co(phen)3]3+. After hybridization of the DNA probe with the DNA target, the peak current increased further (curve 5c). This was caused by binding between ss-DNA and [Co(phen)3]3+ as intercalated interactions.
Cyclic-voltametric behavior of (a) bare Au electrode, (b) Au–HDT (1 h)–GNR (1 h)–DNA probe, (c) Au–HDT (1 h)–GNR (1 h)–DNA probe–DNA target (10 μmol L−1), (d)10 μmol L−1 one-point-mismatch DNA, and (e) 10 μmol L−1 non-complementary DNA in 100 mmol L−1 buffer solution after incubation in 4 mmol L−1 [Co(phen)3]3+ for 1 h, at scan rate 50 mV s−1
The selectivity of the biosensor was also studied using cyclic voltametric behavior of [Co(phen)3]3+ to probe different DNA sequences. Curves 5d,e represent the electrochemical response of the hybridized electrode, in 100 mmol L−1 buffer solution after incubation in 4 mmol L−1 [Co(phen)3]3+, for one-point-mismatch ss-DNA and for non-complementary ss-DNA, respectively. A reduced peak current was observed for hybridization of the ss-DNA-probe-modified electrode with a one-point-mismatch ss-DNA (curve 5d) in comparison with an ss-DNA target, and the peak current was much reduced for non-complementary ss-DNA (curve 5e). The constructed DNA biosensor had high selectivity for detecting the complementary target.
Detection limit for target DNA
The sensitivity of electrochemical behavior for hybridization was studied in the range 1 pmol L−1–1 μ mol L−1 ss-DNA target. As seen in Fig. 6, the peak current response reduced as the concentration of ss-DNA target was reduced. The regression equation obtained was y = 1.1417x + 0.9058 (x was log (target DNA concentration), mol L−1; y was I, μA), and R 2 was 0.97 %. To estimate the limit of detection (LOD), the formula 3σ/a (where σ is the standard deviation of the blank solution and a is the slope of the obtained line in Fig. 6) was used; the LOD obtained was 2 × 10−12 mol L−1.
Conclusion
We fabricated an electrochemical DNA biosensor based on GNRs on a gold-electrode surface for detecting HBV. The GNRs were used because of their large surface area, enabling increased immobilization and hybridization. Self-assembly of GNR on the modified gold electrode led to more attachment of DNA probes and increased hybridization. The construction process of the biosensor was studied by cyclic voltammetry (CV) of [Fe(CN)6]3−/4−, as an electrochemical redox probe, and of [Co(phen)3]3+, as an electrochemical indicator. The target DNA were quantified in the linear range from 1.0 × 10−12 to 10.0 × 10−6 mol L−1, with a detection limit of 2.0 × 10−12 mol L−1 by 3σ. The DNA biosensor had good specificity for distinguishing complementary DNA from non-complementary and mismatched DNA sequences. The sequence of oligonucleotides was selected using BLAST so that it had the least similarity to the human serum genome. This biosensor can be generalized to serum samples in future studies. It is also suggested that the difference in aspect ratio of GNRs may affect sensitivity of electrochemical biosensors.
References
Labuda J, Brett AMO, Evtugyn G, Fojta M, Mascini M, Ozsoz M, Palchetti I, Palecek E, Wang J (2010) Electrochemical nucleic acid-based biosensors: concepts, terms, and methodology (IUPAC Technical Report). Pure Appl Chem 82:1116–1187
Taylor AD, Ladd J, Yu QM, Chen SF, Homola J, Jiang SY (2006) Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel SPR sensor. Biosens Bioelectron 22:752–758
Frederickx C, Verheggen FJ, Haubruge E (2011) Biosensors in forensic sciences. Biotechnol Agron Soc Environ 15:449–458
Wang D, Tang W, Wu X, Wang X, Chen G, Chen Q, Li N, Liu F (2012) Highly selective detection of single-nucleotide polymorphisms using a quartz crystal microbalance biosensor based on the toehold-mediated strand displacement reaction. Anal Chem 84:7008–7014
Zhang D, Peng Y, Qi H, Gao Q, Zhang C (2010) Label-free electrochemical DNA biosensor array for simultaneous detection of the HIV-1 and HIV-2 oligonucleotides incorporating different hairpin-DNA probes and redox indicator. Biosens Bioelectron 25:1088–1094
Parab HJ, Jung C, Lee JH, Park HG (2010) A gold nanorod-based optical DNA biosensor for the diagnosis of pathogens. Biosens Bioelectron 26:667–673
Sipova H, Zhang S, Dudley AM, Galas D, Wang K, Homola J (2010) Surface plasmon resonance biosensor for rapid label-free detection of microribonucleic acid at subfemtomole level. Anal Chem 82:10110–10115
Wang L, Lu D, Wang J, Du D, Zou Z, Wang H, Smith JN, Timchalk C, Liu F, Lin Y (2011) A novel immunochromatographic electrochemical biosensor for highly sensitive and selective detection of trichloropyridinol, a biomarker of exposure to chlorpyrifos. Biosens Bioelectron 26:2835–2840
Du P, Li H, Mei Z, Liu S (2009) Electrochemical DNA biosensor for the detection of DNA hybridization with the amplification of Au nanoparticles and CdS nanoparticles. Bioelectrochemistry 75:37–43
Kaushik A, Khan R, Solanki PR, Pandey P, Alam J, Ahmad S, Malhotra BD (2008) Iron oxide nanoparticles–chitosan composite based glucose biosensor. Biosens Bioelectron 24:676–683
Swain MD, Octain J, Benson DE (2008) Unimolecular, soluble semiconductor nanoparticle-based biosensors for thrombin using charge/electron transfer. Bioconjugate Chem 19:2520–2526
Cai H, Xu C, He P, Fang Y (2001) Colloid Au-enhanced DNA immobilization for the electrochemical detection of sequence-specific DNA. J Electroanal Chem 510:78–85
Zhang D, Chen Y, Chen HY, Xia XH (2004) Silica-nanoparticle-based interface for the enhanced immobilization and sequence-specific detection of DNA. Anal Bioanal Chem 379:1025–1030
Fu Y, Yuan R, Xu L, Chai Y, Liu Y, Tang D, Zhang Y (2005) Electrochemical impedance behavior of DNA biosensor based on colloidal Ag and bilayer two-dimensional sol–gel as matrices. J Biochem Biophys Methods 62:163–174
Jia Z, Liu J, Shen Y (2007) Fabrication of a template-synthesized gold nanorod-modified electrode for the detection of dopamine in the presence of ascorbic acid. Electrochem Commun 9:2739–2743
Wang X, Li Y, Wang H, Fu Q, Peng J, Wang Y, Du J, Zhou Y, Zhan L (2010) Gold nanorod-based localized surface plasmon resonance biosensor for sensitive detection of hepatitis B virus in buffer, blood serum and plasma. Biosens Bioelectron 26:404–410
He W, Huang CZ, Li YF, Xie JP, Yang RG, Zhou PF, Wang J (2008) One-step label-free optical genosensing system for sequence-specific DNA related to the human immunodeficiency virus based on the measurements of light scattering signals of gold nanorods. Anal Chem 80:8424–8430
Lu XC, Dong X, Zhang KY, Han XW, Fang X, Zhang YZ (2013) A gold nanorods based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst 138:642–650
Cha BH, Lee SM, Park JC, Hwang KS, Kim SK, Lee YS, Ju BK, Kim TS (2009) Detection of Hepatitis B Virus (HBV) DNA at femtomolar concentrations using a silica nanoparticle-enhanced microcantilever sensor. Biosens Bioelectron 25:130–135
Mahoney FJ (1999) Update on diagnosis, management, and prevention of Hepatitis B Virus infection. Clin Microbiol Rev 12:351–366
Erdem A, Kerman K, Meric B, Akarca US, Ozsoz M (1999) DNA electrochemical biosensor for the detection of short DNA sequences related to the Hepatitis B virus. Electroanalysis 11:586–587
Wright TL, Lau JYN (1993) Clinical aspects of hepatitis B virus infection. Lancet 342:1340–1344
Nikoobakht B, El-Sayed MA (2003) Preparation and growth mechanism of gold Nanorods (NRs) using seed-mediated growth method. Chem Mater 15:1957–1962
Han X, Fang X, Shi A, Wang J, Zhang Y (2013) An electrochemical DNA biosensor based on gold nanorods decorated graphene oxide sheets for sensing platform. Anal Biochem 443:117–123
Acknowledgments
This study was supported by Tehran University of Medical Sciences and the Iranian nanotechnology initiative (INI).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 59.5 kb)
Rights and permissions
About this article
Cite this article
Shakoori, Z., Salimian, S., Kharrazi, S. et al. Electrochemical DNA biosensor based on gold nanorods for detecting hepatitis B virus. Anal Bioanal Chem 407, 455–461 (2015). https://doi.org/10.1007/s00216-014-8303-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8303-9