Abstract
Benzophenones (BPs) and their hydroxylated derivatives exhibited low estrogenic activity in human breast cancer cell. However, the available reported data are insufficient to describe structure modification relationship of these molecules that can be developed as a potential breast cancer drug. In this work, we present a 240-ns molecular dynamics (MD) simulation analysis on the conformational flexibility and changes of both the monomer and dimer forms of human estrogen receptor α (hERα) upon binding with the newly designed morpholine ether benzophenone (BP). The structural stability and conformational changes of an apo conformation hERα with respect to Helix 12 (H12) upon binding were determined by analyzing the H-bond formation, radius of gyration, root mean square deviation (RMSD) and root mean square fluctuation (RMSF). Molecular mechanics Poisson–Boltzmann surface area (g_mmpbsa) method was used to predict binding free energies and to estimate the energy contribution per residue to the binding energy of the complexes. Our results revealed low energy values for Leu525 and Ala350 residues in both monomer and dimer forms of hERα complexes. On the other hand, the hydrogen bonding formation between the host and the ligand in the binding pocket involved Glu353, Arg394, Asp351 and Lys529 amino acid residues, indicating that morpholine ether BP has stable interaction with hERα by mimicking the behavior of 17β-estradiol. Furthermore, transition paths analysis of H12 reveals a new stable hERα apo conformation during the simulation time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
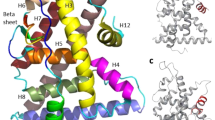
Nuclear hormone receptors (NRs), which are transcription regulators modulated by ligand binding, play important roles in ligand-activated transcription factors involved in a number of biological processes such as homeostasis, lipid metabolism and cell death [1,2,3]. Thus, they are significantly related to breast and prostate cancers [4]. NRs are composed of three domains. Starting from N-terminal, the principal domains are (1) the N-terminal domain (NTD); (2) DNA-binding domain (DBD); and (3) ligand-binding domain (LBD) (Fig. 1) [5]. LBD is composed of 12 helices pack into a three-layer sandwich motif, contains hormone binding pocket [6], and is responsible for ligand recognition and dimerization interface [7, 8]. Transcription behavior of NR differs when different ligands bind to its LBD, especially the C-terminal, Helix 12 (H12), orientation which changes when different ligands interact to NR. It is reported [6, 9] that the transcriptional activity changes with the location of H12. The position of H12 is far from the binding site in the apo form [10], whereas the H12 is located near the ligand binding site when the ligand is bound. The position of the H12 depends on the kind of ligand binds to the NR. When the agonist ligands bind to the LBD, the H12 is repositioned to cap ligand binding site, allowing the co-activator protein to bind and the transcription to take place [11, 12]. On the other hand, the H12 lay over the co-activator groove in antagonist state, thus preventing dimerization and transcription from occurring [13]. Agonist and antagonist forms have similar binding site region, but with a major difference in the H12 location [14].
The extended apo conformation of retinoic X receptor-α (RXRα) was mostly described in 1995 [15]; the H12 in RXRα is extended away from the LBD core. Similarly, the apo-form human estrogen receptor hERα (PDB ID: 1A52, Fig. 2a) also has unfolded H12 conformation [10]. The apo hERα 1A52 shows that H12 is highly flexible compared to known apo structures of other NRs [10, 15,16,17]. Many studies have considered the apo hERα 1A52 as the best available model of an apo ERα LBD form [18, 19].
a Backbone structure of apo ERα bound to 17β-estradiol (PDB ID: 1A52). b Molecular structure of newly designed morpholine ether BP ligand used in this study [29]
It has been shown that the apo dimer conformation of ER is more stable than monomer form and the stability increases when it interacts with various ligands [20]. Antagonist ligands such as 4-hydroxytamoxifen and raloxifene enhance the stability of ER dimer by providing greater kinetic stabilization more than agonists like 17β-estradiol, and final C-terminal encodes the LBD, which forms a hydrophobic binding pocket responsible for estrogen and antiestrogen binding [13]. This domain also contains a second ligand dependent activation factor, AF2, which activates ER in response to 17β-estradiol or synthetic agonists. Due to the implication of controlling ER activity through its modulation, the LBD is important for the development of synthetic agonists and antagonists [9].
Several computational studies were performed in order to further understand the molecular mechanism of the ERα. The fragment molecular orbital (FMO) method has been employed to study the binding energies between ERα and several ligands; the calculated binding energies were found in good agreement with the experimental values [21,22,23].
Also, 158 ns MD simulations were performed for apo, agonist and antagonist conformations of hERα describing the binding of the 17β-estradiol (E2) in the presence and absence of co-activator protein [24]. The results from MD studies did not show any transition for the apo monomer to either an agonist or an antagonist state. In addition, 2.2 μs MD runs were performed for ERα using the accelerated molecular dynamics (aMD) method [25]. The transition paths of unfolded apo state to either an agonist or an antagonist state were investigated. It was observed that the H12 transition occurs in the dimer form and is not governed only by the presence of a ligand. However, the dimerization state is essential to get separated landscapes for agonist and antagonist conformations of H12.
The present study aims to investigate the stability of hERα forming complex with the newly designed morpholine ether BP (Fig. 2b) through MD simulation. In addition, to describe the transition path of the extended H12 in the PDB ID: 1A52 structure during simulation time, we treated the apo structure as monomer and dimer forms, even though other MD studies proposed that the position of the H12 will not be influenced by the choice of monomer or dimer conformation [19, 24], but up to nine independent accelerated MD simulations of apo ERα showed the same behavior and reproduced H12 states from apo to agonist or even antagonist conformations only after dimer formation while no transition has been observed in the monomer complex [25]. In addition, the authors presumed that the ligands do not directly determine the H12 orientation but stabilize or destabilize substructural parts of the protein, which in turn affects the dimerization and H12 conformation. In this study, we applied normal MD simulations to understand the dynamics of apo ERα in dimer and monomer forms with or without the newly designed morpholine ether (BP) as a ligand [29].
2 Methods
2.1 Molecular dynamics simulation protocol
The X-ray crystal structure of apo human estrogen receptor hERα, PDB ID: 1A52, was retrieved from the Protein Data Bank (www.rcsb.org). The ligand, 17β-estradiol bound to apo 1A52, was removed from the crystal structure; the hydrogen atoms were added to the protein, and the missing residues were fixed. The protonation state of the key residue, His524 imidazole ring, was set as neutral in which the H atom was assigned to the epsilon (Nε) and His524 (Nδ) was pointing toward the ligand.
The starting ligand structure was generated and optimized using a semi-empirical PM3 method [27] available in Gaussian 2003 software [28] and was used as the initial ligand structure in the docking calculation with hERα receptor [29]. The lowest binding energy which corresponds to the most stable conformation complex from the docking calculation was used in the present MD simulation. Additional parameters were added to the ligand by using the ACPYPE in the Amber Tools package.
All MD simulations were carried out using the Gromacs 5.0.7 program with the AMBER FF99SB-ILDN force field for hERα receptor [26]. The TIP3P model [30] was used for the water molecules in a cubic box with the distance from the box to the surface of the protein set to be at least 12 Å. The general AMBER force field (GAFF) was used for the parameters of the BP ligand. To neutralize the systems, counter ions were added to balance the charge of the protein. The systems were minimized using the steepest descent method for 6000 steps followed by the Berendsen thermostat [31] equilibration run in NVT (constant number of particles, volume and temperature) ensemble for 200 ps at 300 K. Then the production runs were performed using Parrinello–Rahman barostat [32] in the NPT (constant number of particles, pressure and temperature) ensemble for 1 ns at 1 bar and 300 K. After the temperature and pressure adjustments, a 60-ns MD simulation was performed for the four systems at 1 bar and 300 K. The cutoff for Coulomb and van der Waals interactions was set to 12 Å and was updated every 2 fs. The particle mesh Ewald [33, 34] method was used to correct the electrostatic interactions. The LINCS [35, 36] algorithm was used to constrain the bonds with hydrogen atoms. A total of four different models of the system were investigated (see Table 1 for details).
The analysis of the computed trajectories were conducted using root mean square deviation (RMSD), radius of gyration (Rg) and root mean square fluctuation (RMSF), and the VMD 1.9.3 [37] software was used to visualize the interaction.
2.2 Free energy calculations
Free energy calculations were performed by the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method available in GROMACS software packages using g_mmpbsa tool [38]. In this study, the last 20 ns for simulation of complex was chosen as an equilibrium part of the trajectory for energy analysis. MM-PBSA was applied to predict average binding free energies using python script, MmPbSaStat.py, available in the g_mmpbsa package; two output files summary_energy.dat and full_energy.dat were obtained; summary_energy. dat contains average of all energetic components including the binding energy, polar solvation energy ΔGpolar, solvent accessible surface area (SASA) ΔGnonpolar, electrostatic interaction ΔEelec and van der Waals interaction EvdW, whereas full_energy.dat contains the values of energetic terms as a function of time which were plotted with Xmgrace software. On the other hand, to calculate average contribution of the residues to the binding energy [39], a python script MmPbSaDecomp.py was used, and the results including binding energy for each residue were plotted using file energyMapIn.dat with Xmgrace software. Furthermore, energy2bfac tool was used to visualize the contribution energy of residue with its structure using VMD program.
3 Results and discussion
3.1 Conformational stability of simulations
For all simulations, the calculated RMSDs reach stable values after approximately 10 ns. The RMSDs for MF and ML reach the average value of 4.4 Å (Fig. 3a) while the dimer ligand complex model DL is more dynamic and has the RMSD average value of 5.4 Å. The free dimer DF, on the other hand, has lower RMSD values, between 2 to 4 Å (Fig. 3b). While initially this may indicate unstable simulations of the apo dimer complex, DL, such high RMSD values which correspond to the conformational changes of H12 are not very surprising.
The larger RMSD values in the free and complex apo forms appear to have occurred due to the large fluctuation in the extended H12. This is in agreement with most apo hERα studies compared to the folding conformations of hERα in agonist and antagonist forms [24, 25]. For example, Fratev [25] also reported high conformational flexibility with respect to H12, thus affecting the stability of the overall hERα structure compared to the folding of H12 conformation. Similar results were also reported by Celik et al. [24]. The results from MD runs showed high RMSD value, around 4 Å for apo hERα (PDB: 1A52) bound to 17β-estradiol, whereas the RMSD values for the antagonist and agonist were around 3 and 2 Å, respectively. Therefore, the rather high RMSD values obtained with our morpholine ether benzophenone ligand with the apo dimer are in agreement with the previous reports.
Consistently, the RMSFs of the monomer model are smaller than those of the dimer complex DL. Further analysis reveals that the RMSFs of the beginning of C-terminal of H3, loops 330–340 between H1 and H3, the beginning of C-terminal of H9, loops 556–565 between H8 and H9, and the end of H11, loops 531–536 between H11 and H12, are relatively higher than other regions in all apo hERα LBD models. Thus, helix–loop–helix segments of H3 (loops 330–340), H9 (loops 556–565), H11 (loops 531–536) and H12 are the most flexible regions in the apo hERα LBD. We further analyzed the stability of the apo hERα LBD models during the simulation in terms of the radius of gyration (Rg). The plot of the variation of radius of gyration of each monomer and dimer models with time is shown in Fig. 4. Due to the orientational difference of H12, the hERα dimer DL complex has higher Rg values compared to the free dimer, DF. The Rg profile obtained from MD simulations reveals that the free receptors are more stable than the ligand-bound receptors with respect to both monomer and dimer forms.
The root mean square fluctuations (RMSF) per residue were also analyzed to identify the regions of high fluctuations. Results are summarized in Fig. 5. In general, hERα LBD dimer, DF, is more stable than its monomeric form, MF, and the upward movement of the linker region between H8 and H9 was observed for the monomer, but not for the dimer. This is due to the dimerization which constraints the movement of the linker region, thus making the dimer form more stable compared to the monomer. We found that the observed high RMSD fluctuations of the apo ERα/morpholine ether BP complex are mainly due to the long loop region between H1 and H3, H8 and H9 and H12. These regions are found to be highly flexible during the MD simulation in both the monomer and dimer forms. Analysis of the hydrogen bonding formation between the morpholine ether BP and the monomer and dimer forms reveals high similarity for both complexes. The hydrogen bonding formation between the hERα and morpholine ether BP in the binding pocket involved Glu353, Arg394, Asp351 and Lys529 amino acid residues (Fig. 6), indicating that morpholine ether BP has stable interaction with hERα by mimicking the behavior of 17β-estradiol (Fig. 7a, b). Both were observed to form hydrogen bonds mainly with Glu353 residue throughout the simulation period (Fig. 8a, b), thus suggesting that the stability of the complexes is due to hydrophobic interactions. Further analysis based on the free energy analysis results for individual contributions of the residues during ligand receptor interaction in Sect. 3.3 confirms this assumption.
3.2 Transition path analysis of H12 in dimer form
To illustrate the dynamics of H12 in the free and ligand bound dimer, distance analysis was also performed on eight key residues of H12 for chains A and B in the dimer models, DF and DL (Fig. 9). The minimum-distance analysis for DF showed distance between H12 in chain A and chain B was within the range of 4.5 nm, indicating that H12 swung up toward the binding pocket for both chains during the simulation (Fig. 10a). In DL, the dynamics of H12 is studied in the presence of morpholine ether BP ligand in both chains. Interestingly and in contrast to the free dimer form, this simulation reveals a new stable conformation of apo hERα complex after 15 ns to the end of the simulation time with dramatic changes in the behavior of the dimer. H12 is very dynamic and travels from chain A to interact with H12 in chain B within the range of 0.2 nm (Fig. 10b).
3.3 Free energy analysis
The MM-PBSA method has been applied to predict binding free energies and to evaluate the relative stabilities and molecular interactions of the whole apo ERα ligand binding domain with morpholine ether BP. The calculated energies including the total binding energy and the separate energy components for MD trajectories from 40 to 60 ns are listed in Table 2. As shown in Fig. 11, the calculated binding free energy of monomer complex ML (− 126.13 kJ/mol) is higher than dimer complex, DL (− 359.40 kJ/mol). The results demonstrated that the dimer form promoted stability to the apo ERα and morpholine ether BP complex more than the monomer conformation. In order to understand the molecular interaction of apo ERα/morpholine ether BP complex, it would be helpful to determine the ERα residues that interact with the morpholine ether BP. The g_mmpbsa tool decomposes the total binding energy into the contributions made by each residue, thus enabling the comparison of the relative residue contribution to the overall binding energy. We calculated per residue energy contributions for both monomer and dimer complexes in Fig. 12a, b, respectively. As it can be seen, during the initial transition to the stable forms, the energy contribution came mainly from the binding site residues of H3, H5 and H11. The hydrophobic residues strongly interacting with morpholine ether BP are Leu525, Ala350, Leu346 and Leu387 in both monomer and dimer forms. Weak interactions were observed between the morpholine ether BP and many surrounding charged/polarized residues, such as Glu353, Lys529, Glu419 and Gly521 for monomer (see Fig. 13a) and Glu353, Lys529, Arg394 and His524 for the dimer form (see Fig. 13b), suggesting that the stability of the apo ERα/morpholine ether BP complex is mainly due to hydrophobic interactions.
4 Conclusion
The interactions between the human ERα both in monomer and in dimer forms with morpholine ether BP were studied using MD simulation. Our results shed light on the structural fluctuation of the ERα apo state and its structural change upon morpholine ether BP binding. It was observed that the fluctuation of the H12 is much larger than other helices as determined from the MD trajectories. Furthermore, this study reveals a new stable dimer conformation during the simulation time as well as identifies that the motions of the H12 in the dimer form swing toward each other until the end of the simulation time. From g_mmpbsa analysis, the calculated binding free energy of dimer complex DL is more stable than the monomer complex ML. Furthermore, significant interactions of the ligand, morpholine ether BP, with the dimer form were found to occur with the same amino acid residues observed in the monomer complex. Strong interactions were observed between the morpholine ether BP and several hydrophobic residues, in particular for leucine (Leu525, Leu346, Leu387) and alanine Ala350. The interaction of ligand and surrounding charged/polar residues with the highest energy was found for Glu353, suggesting the stability of the apo ERα/morpholine ether via hydrophobic interaction.
References
Gronemeyer H, Gustafsson JA, Laudet V (2004) Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov 3(1):950–964
Moore JT, Collins JL, Pearce KH (2006) The nuclear receptor superfamily and drug discovery. ChemMedChem 1(5):504–523
Berkenstam A, Gustafsson JA (2005) Nuclear receptors and their relevance to diseases related to lipid metabolism. Curr Opin Pharmacol 5(2):171–176
Horwitz KB, McGuire W (1978) Estrogen control of progesterone receptor in human breast cancer: correlation with nuclear processing of estrogen receptor. J Biol Chem 253(7):2223–2228
Beato M (1989) Gene regulation by steroid hormones. Cell 56(3):335–344
Bourguet W, Germain P, Gronemeyer H (2000) Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci 21(10):381–388
Kumar R, Thompson EB (1999) The structure of the nuclear hormone receptors. Steroids 64(5):310–319
Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR (1998) Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev 12(21):3343–3356
Aranda A, Pascual A (2001) Nuclear hormone receptors and gene expression. Physiol Rev 81(3):1269–1304
Tanenbaum DM, Wang Y, Williams SP, Sigler PB (1998) Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains. Proc Natl Acad Sci 95(11):5998–6003
Wärnmark A, Treuter E, Gustafsson J-Å, Hubbard RE, Brzozowski AM, Pike AC (2002) Interaction of transcriptional intermediary factor 2 nuclear receptor box peptides with the coactivator binding site of estrogen receptor α. J Biol Chem 277(24):21862–21868
Heery DM, Kalkhoven E, Hoare S, Parker MG (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387(6634):733–736
Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95(7):927–937
Egner U, Heinrich N, Ruff M, Gangloff M, Mueller-Fahrnow A, Wurtz JM (2001) Different ligands–different receptor conformations: modeling of the hERα LBD in complex with agonists and antagonists. Med Res Rev 21(6):523–539
Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D (1995) Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature 375(6530):377–382
Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature 395(6698):137–143
Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D (1995) Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature 378(6558):681–689
Watanabe C, Watanabe H, Tanaka S (2010) An interpretation of positional displacement of the helix12 in nuclear receptors: preexistent swing-up motion triggered by ligand binding. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics 1804(9):1832–1840
Batista MR, Martínez L (2013) Dynamics of nuclear receptor Helix-12 switch of transcription activation by modeling time-resolved fluorescence anisotropy decays. Biophys J 105(7):1670–1680
Tamrazi A, Carlson KE, Daniels JR, Hurth KM, Katzenellenbogen JA (2002) Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol Endocrinol 16(12):2706–2719
Fukuzawa K, Kitaura K, Nakata K, Kaminuma T, Nakano T (2003) Fragment molecular orbital study of the binding energy of ligands to the estrogen receptor. Pure Appl Chem 75(11–12):2405–2410
Fukuzawa K, Kitaura K, Uebayasi M, Nakata K, Kaminuma T, Nakano T (2005) Ab initio quantum mechanical study of the binding energies of human estrogen receptor α with its ligands: an application of fragment molecular orbital method. J Comput Chem 26(1):1–10
Fukuzawa K, Mochizuki Y, Tanaka S, Kitaura K, Nakano T (2006) Molecular interactions between estrogen receptor and its ligand studied by the ab initio fragment molecular orbital method. J Phys Chem B 110(32):16102–16110
Celik L, Lund JDD, Schiøtt B (2007) Conformational dynamics of the estrogen receptor α: molecular dynamics simulations of the influence of binding site structure on protein dynamics. Biochemistry 46(7):1743–1758
Fratev F (2015) Activation helix orientation of the estrogen receptor is mediated by receptor dimerization: evidence from molecular dynamics simulations. Phys Chem Chem Phys 17(20):13403–13420
Piana S, Lindorff-Larsen K, Shaw DE (2011) How robust are protein folding simulations with respect to force field parameterization? Biophys J 100(9):L47–L49
Stewart JJ (1989) Optimization of parameters for semiempirical methods II. Applications. J Comput Chem 10(2):221–264
Frisch M, Trucks G, Schlegel H, Scuseria G, Robb M, Cheeseman J, Montgomery JR, Vreven T, Kudin K, Burant J (2004) Gaussian 03. Gaussian Inc, Wallingford
Shtaiwi A, Adnan R, Khairuddean M, Soib SF (2016) Investigation of newly designed human estrogen receptor inhibitors from benzophenones derivatives (BPs) by molecular docking and molecular dynamic simulation. Aust J Basic Appl Sci 10(18):49–59
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935
Berendsen HJ, Postma JP, van Gunsteren WF, Hermans J (1981) Interaction models for water in relation to protein hydration. Intermol Forces 14:331–342
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52(12):7182–7190
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N log (N) method for Ewald sums in large systems. J Chem Phys 98(12):10089–10092
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103(19):8577–8593
Hess B, Bekker H, Berendsen HJ, Fraaije JG (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472
Hess B (2008) P-LINCS: a parallel linear constraint solver for molecular simulation. J Chem Theory Comput 4(1):116–122
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Kumari R, Kumar R, Consortium OSDD, Lynn A (2014) g_mmpbsa-A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Meliciani I, Klenin K, Strunk T, Schmitz K, Wenzel W (2009) Probing hot spots on protein-protein interfaces with all-atom free-energy simulation. J Chem Phys 131(3):034114
Funding
The authors gratefully acknowledge the financial support from Universiti Sains Malaysia through USM Fellowship Scheme under the Institute of Postgraduate Studies and Ministry of Higher Education through FRGS Grant No. 203/PKIMIA/6711558.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shtaiwi, A., Adnan, R., Khairuddean, M. et al. Molecular dynamics simulation of human estrogen receptor free and bound to morpholine ether benzophenone inhibitor. Theor Chem Acc 137, 101 (2018). https://doi.org/10.1007/s00214-018-2277-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-018-2277-1