Abstract
Circular RNAs (circRNAs) have been confirmed to be an important modulator and therapeutic target of cervical cancer (CC). The aim of this study is to explore the role and mechanism of circ_0081723 in CC progression. Circ_0081723, microRNA-545-3p (miR-545-3p), and CREB3 regulatory factor (CREBRF) levels were detected using quantitative real-time PCR (qRT-PCR) assay. CREBRF, ki-67, Bcl-2 related X protein (Bax), and E-cadherin expression levels were determined using western blot (WB) and immunohistochemistry (IHC) assays. Cell proliferation was assessed using Cell Counting Kit-8 (CCK-8), cell colony formation, and 5-ethynyl-2’-deoxyuridine (EdU) assays. Flow cytometry was used to measure cell apoptosis. Cell migration and invasion were examined using Transwell assay. Interaction between miR-545-3p and circ_0081723 or CREBRF was verified using dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assays. The biological role of circ_0081723 on CC growth was examined using the xenograft tumor model in vivo. Circ_0081723 and CREBRF were increased, and miR-545-3p was decreased in CC tissues and cells. Circ_0081723 silencing suppressed CC cell growth and motility whereas boosted CC cell apoptosis. Besides, circ_0081723 acted as a molecular sponge for miR-545-3p, and circ_0081723 knockdown-induced effects were largely reversed by miR-545-3p downregulation in CC cells. Moreover, miR-545-3p repressed CC progression by targeting CREBRF. Circ_0081723 absence blocked xenograft tumor growth in vivo. Circ_0081723 stimulated CC cell malignant behaviors by regulating the miR-545-3p/CREBRF pathway, providing a possible circRNA-targeted therapy for CC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer (CC) is a gynecological malignant tumor, with the second-highest incidence of female malignant tumors in the world (Siegel et al. 2019; Conesa-Zamora 2013). The good news is that human papillomavirus (HPV) vaccination can effectively prevent HPV infection and reduce the incidence of CC (Ferrall et al. 2021; O’Leary 2021). However, due to economic factors, diagnostic technology, and other factors, about two-thirds of CC patients are in advanced stages at the time of diagnosis and have a poor prognosis (Marquina et al. 2018; Kwong et al. 2021). In addition, various factors such as postoperative recurrence, tumor metastasis, and chemotherapy resistance greatly increase the difficulty of CC treatment (McMellen et al. 2020). Therefore, to improve the current status of CC treatment, an in-depth study of the mechanism of the progression of CC and finding effective therapeutic targets are essential.
Different from other non-coding RNAs, circular RNAs (circRNAs) are formed by reverse splicing of pre-RNA (Wang et al. 2021). Because of their wide expression, tissue diversity, structural stability, sequence conservation, and other characteristics, circRNAs have become a type of potential disease diagnosis and prognostic markers (Kristensen et al. 2019). Many circRNAs, such as hsa_circ_0003195 (Ma et al. 2022), hsa_circ_001787 (Chalbatani et al. 2021), has_circ_0006470 (Cui et al. 2021), and hsa_circ_0140271 (Chen et al. 2021) are dysregulated in tumor samples compared to the controls, and they can participate in cancer progression through different pathways, like affecting cell proliferation, metastasis, metabolism and autophagy. Circ_0082723 is elevated in CC tissues (Qu et al. 2020), but its role in CC formation remains to be explored.
MicroRNAs (miRNAs) are small RNAs that can target the 3ʹ untranscribed region (3’UTR) of mRNA and negatively regulate gene expression (Fabian et al. 2010). According to reports, circRNA can indirectly regulate gene expression and participate in tumor progression via sponging miRNA (Cui et al. 2018). For instance, Feng et al. proved that circ-ABCB10 contributed to CC progression and enhanced ZEB1 expression via sponging miR-128-3p (Feng et al. 2021). Here, we discovered that microRNA-545-3p (miR-545-3p) was a candidate miRNA for circ_0081723 based on the bioinformatics website. Meanwhile, there were binding sites for CREB3 regulatory factor (CREBRF) on the sequence of miR-545-3p. Accordingly, whether circ_0081723 might regulate CC development via the miR-545-3p/CREBRF axis was further explored. Then, we designed relevant experiments to verify the above hypothesis.
Materials and methods
Clinical specimens
CC tumor tissue samples (n = 60) and adjacent no-tumor tissues (n = 60) were obtained from Gongli Hospital of Shanghai Pudong New Area. Sample inclusion criteria are that the patients have no radiation or chemotherapy prior to surgery. This project was authorized by the ethics committee of Gongli Hospital of Shanghai Pudong New Area and written informed consent was signed by all subjects.
Cell culture
CC cell line SiHa and normal ectocervical cell line (Ect1/E6E7, ATCC, Manassas, VA, USA) were incubated in DMEM (Gibco, Grand Island, NY, USA). Besides, CC cell line Caski (ATCC) was cultured in RPMI-1640 medium (Gibco) in an incubator with 5% CO2 at 37 °C. Besides, 10% FBS (Gibco) was added to all media.
Quantitative real-time PCR (qRT-PCR)
Total RNAs were extracted from tissue samples and cell lines using TRIZOL reagent (Dingguo, Guangzhou, China). After detecting the RNA concentration by Nanodrop 2000 C, total RNA (2 µg) was reverse transcribed into cDNA using a Super Reverse Transcript PCR Kit (Dingguo). qRT-PCR was carried out using SYBR Green (Invitrogen, Paisley Scotland, UK). Finally, results were calculated according to the 2−ΔΔCT method and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6. Primer sequences are shown in Table 1.
RNase R assay
RNase R experiment was implemented to identify circ_0081723 stability. Part of the RNA isolated from CC cells was co-incubated with RNase R (Invitrogen). Then, qRT-PCR was conducted to determine the RNA expression of circ_0081723 and linear RNA Polymerase II Subunit J2 (POLR2J2).
Subcellular fractionation assay
To identify circ_0081723 localization, the PARISTM Kit (Invitrogen) was applied to separate RNA from nuclear and cytoplasmic extracts. At last, qRT-PCR assay was used to detect the expression levels of circ_0081723, GAPDH (cytoplasm control), and U6 (nucleus control).
Cell transfection
Generally, shRNA lentiviruses encoding sequence targeting circ_0081723 (sh-circ_0081723) or non-target control (sh-NC) were purchased from HANBIO (Shanghai, China). For lentiviral transduction, SiHa and Caski cells of 50% confluence were infected by virus particles in media containing 8 µg/mL of polybrene. 24 h later, the vector-positive cells were chosen using 1 µg/mL of puromycin over 72 h.
In addition, miR-545-3p mimic and inhibitor (miR-545-3p and in-miR-545-3p), CREBRF overexpression plasmid (CREBRF), and controls (miR-NC, in-miR-NC, and pcDNA) from Sangon Biotechnology (Shanghai, China) were transfected into SiHa and Caski cells in this research. Lipofectamine 3000 (Invitrogen) was used for cell transfection.
Cell counting Kit-8 (CCK-8)
In short, 2 × 104 CC cells/well were plated in a 96-well plate. At different time points, 10 µL of CCK-8 reagent (Beyotime, Shanghai, China) was added for another 4 h. The absorbance at 450 nm was estimated by an enzyme immunoassay analyzer.
Colony formation assay
200 cells CC cells were grown in 6-well plates. After culturing in an incubator for two weeks, cell colonies were subjected to 0.1% crystal violet staining, followed by counting under a microscope.
5-ethynyl-2’-deoxyuridine (EdU)
Referring to the KeyFluor488 EdU Kit (keyGEN Biotech, Jiangsu, China), the DNA synthesis of CC cells was analyzed in this experiment. Briefly, transfected CC cells were incubated with an EdU working solution (50 µM) for 2 h. Subsequently, the nuclei were stained with Hoechst for 30 min, and EdU-positive cells were visualized using a microscope.
Flow cytometry
CC cells were fostered for 24 h. After harvesting by centrifugation, cells were re-suspended in binding buffer and then stained with 5 µL Annexin V-FITC and 5 µL PI solution (Beyotime, Beijing, China) in the dark for 15 min, followed by the detection using a flow cytometer.
Transwell assay
CC cell invasion and migration ability were respectively evaluated using transwell chambers pre-coated with or without Matrigel (Corning, Tewksbury, MA, USA). First of all, cells were mixed with FBS-free medium and then added into the upper chambers. Meanwhile, medium with 10% FBS was placed into the bottom chambers. 24 h later, the migrated or invaded cells were fixed and stained with 0.1% crystal violet (Phygene). Finally, the migratory and invasive abilities were determined by counting cell numbers with a microscope.
Western blot (WB)
RIPA buffer (Beyotime) was implemented to lyse CC tissues and cells to obtain protein samples, which were separated by SDS-PAGE gel and transferred onto PVDF membranes. After blocking with 5% nonfat milk, membranes were incubated with primary and secondary antibodies (Table 2). At last, protein signals were analyzed based on the BeyoECL Plus kit.
Dual-luciferase reporter assay
CircInteractome, circBank, and Starbase software (Dudekula et al. 2016; Liu et al. 2019; Yang et al. 2011) retrieved the binding sequences between miR-545-3p and circ_0081723 or CREBRF. WT or MUT circ_0081723 and CREBRF 3’UTR were introduced into the pmirGLO vector (Promega, Madison, WI, USA), generating circ_0081723 or CREBRF 3’UTR WT/MUT reporter plasmids. Then, these plasmids and miR-545-3p mimic or miR-NC were co-transfected into CC cells. Luciferase activities were measured using the Dual-Lumi™ II Luciferase Assay Kit (Beyotime).
RNA immunoprecipitation (RIP)
Magna RIP Kit (Millipore, Billerica, MA, USA) was conducted to confirm the associated relation between circ_0081723 and miR-545-3p. Briefly, the CC cell lysates were co-incubated with magnetic beads pre-coated with anti-Ago2 or anti-IgG. At length, qRT-PCR assay was utilized to analyze the RNA enrichment of circ_0081723 and miR-545-3p.
Xenograft assay
10 female BALB/c nude mice (4–6 weeks old) purchased from Vital River Laboratory (Beijing, China) were used to establish the Xenograft tumor model. All animal experiments were approved by the Animal Ethics Committee of Gongli Hospital of Shanghai Pudong New Area. After dividing mice into two groups (n = 5/group), Caski cells with sh-NC or sh-circ_0081723 were subcutaneously inoculated into mice. Method: V (mm3) = (L × W2)/2 was used to calculate the tumor volume. 28 days after injection, mice were sacrificed and tumors were weighed and photographed. Besides, the IHC assay was performed as described (Huang et al. 2017). Other paraffin-embedded tissue were sectioned at a 4 μm thickness and hatched with antibodies shown in Table 2. Then, images were photographed and analyzed under light microscopy,
Statistical analysis
Mean ± standard deviation was the display form of all data. P < 0.05 was a sign of obvious difference. Data analysis in this research was carried out based on Student’s t-test and ANOVA with Tukey’s tests in GraphPad Prism software.
Results
The expression of circ_0081723 was increased in CC
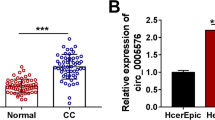
Circ_0081723 is a circular RNA derived from the exons 2–3 of the POlR2J2 gene (Fig. 1A). The expression of circ_0081723 was examined in 60 pairs of CC tissues and cells by qRT-PCR assay. As shown in Fig. 1B-C, circ_0081723 expression was evidently upregulated in CC tumor tissues (2.13 fold) and CC cell lines (SiHa, 2.81 fold; and Caski, 4.36 fold) when compared to the adjacent normal tissues and the control cell line Ect1/E6E7. These data suggested that circ_0081723 might be a biomarker of CC. Besides, we found that linear POLR2J2 expression was sharply digested by RNase R, but circ_0081723 expression had no obvious change after RNase R treatment (Fig. 1D-E), implying that the structure of circ_0081723 was more stable than its linear form POLR2J2. Then, we performed the nuclear-cytoplasmic fractionation and observed that circ_0081723 was mostly distributed in the cytoplasm rather than nuclear (Fig. 1F-G). These data suggested that circ_0081723 is indeed a circRNA and is mostly present in the cytoplasm in CC.
The expression of circ_0081723 was upregulated in the tissues and cell lines of CC. (A) Schematic diagram of circ_0081723 formation. (B) The circ_0081723 expression was measured by qRT-PCR in the adjacent normal and tumor tissues of CC patients. (C) The expression of circ_0081723 in CC cell lines (SiHa and Caski) and Ect1/E6E7 cells was determined by qRT-PCR. (D-E) The circular characteristic of circ_0081723 was confirmed by RNase R assay. (F-G) The subcellular localization of circ_0081723 was measured in cell nuclear and cytoplasm. ***P < 0.001
Repressing circ_0081723 caused suppressive effects on cell proliferation and motility of CC cells
Next, we performed loss-of-function experiments to explore the role of circ_0081723 in CC cells. First, SiHa and Caski cell lines stably expressing sh-NC or sh-circ_0081723 were established. The expression of circ_0081723 in CC cells in the sh-circ_0081723 group was notably reduced compared with the sh-NC group, while linear POLR2J2 expression was unaffected (Fig. 2A-B). Functionally, CCK-8 assay, colony formation assay, and EdU assay exhibited that circ_0081723 knockdown markedly reduced cell viability (with a 47.27% reduction in SiHa cells and a 52.94% decrease in Caski cells), colony number (with a 44.87% decrease in SiHa cells and a 52.96% reduction in Caski cells), and EdU positive rate (with a 47.95% decrease in SiHa cells and a 51.84% reduction in Caski cells) (Fig. 2C-F). Furthermore, flow cytometry analysis displayed that the downregulation of circ_0081723 might obviously induce cell apoptosis of CC cells (SiHa, 4.00 fold; and Caski, 4.31 fold) (Fig. 2G). In addition, the silencing of circ_0081723 notably decreased the number of migrated cells (with a 43.43% decrease in SiHa cells and a 53.85% reduction in Caski cells) and invaded cells (with a 41.85% decrease in SiHa cells and a 54.24% reduction in Caski cells) in CC cells (Fig. 2H-I). Consistently, WB assay revealed that circ_0081723 absence might reduce ki-67 protein level (proliferation-associated marker) (with a 43.67% reduction in SiHa cells and a 49.00% decrease in Caski cells), increase Bax protein level (pro-apoptotic marker) (SiHa, 1.89 fold; and Caski, 2.10 fold), and enhance E-cadherin protein level (EMT-associated marker) (SiHa, 1.73 fold; and Caski, 1.78 fold) (Fig. 2J-K). These findings demonstrated that circ_0081723 silencing suppressed CC cell malignant behaviors.
Circ_0081723 absence suppressed CC progressionin vitro. SiHa and Caski cells were transfected with sh-NC or sh-circ_0081723. (A-B) The expression of circ_0081723 was determined by qRT-PCR to assess the knockdown efficiency. (C-F) The cell proliferation ability of SiHa and Caski cells was explored using CCK-8 assay (C-D) cell colony formation assay (KeyFluor488 EdU Kit (keyGEN Biotech) and EdU assay (F). (G) The apoptotic rate of SiHa and Caski cells was tested by flow cytometry. (H-I) Transwell assay was used to evaluate the migration and invasion abilities of SiHa and Caski cells. (J-K) The protein expression of ki-67, Bax, and E-cadherin were detected by WB assay. **P < 0.01 and ***P < 0.001
Circ_0081723 was interacted with mir-545-3p via target binding
Furthermore, the mechanism by which circ_0081723 regulated the function of CC cells was further explored. Based on all three online websites (CircInteractome, circBank, and starBase) prediction, miR-545-3p was the only miRNA that was common among these websites (Fig. 3A) and the binding sites were presented in Fig. 3B. After confirming the overexpression efficiency of miR-545-3p (SiHa, 9.02 fold; and Caski, 12.55 fold) (Fig. 3C), a dual-luciferase reporter assay was performed in CC cells. As shown in Fig. 3D-E, miR-545-3p mimic significantly decreased the luciferase activity of circ_0081723 WT reporter plasmid, but had no effect on that of circ_0081723 MUT reporter plasmid. Moreover, the RIP assay revealed that the enrichment of circ_0081723 and miR-545-3p on Ago2 immunoprecipitates was greatly higher than that on IgG immunoprecipitates (Fig. 3F-G). In addition, miR-545-3p levels were notably decreased in CC tumor tissues (with a 36.21% reduction) and cells relative to their respective corresponding controls (adjacent normal tissues and Ect1/E6E7) (Fig. 3H-I). Beyond that, we observed that the level of circ_0081723 was negatively correlated with miR-545-3p expression in CC tumor tissues (Fig. 3J). Overall, these results illustrated that circ_0081723 was a sponge for miR-545-3p in CC.
Circ_0081723 acted as a molecular sponge for miR-545-3p in CC cells. (A) Venn diagram showed the number of miRNAs that were predicted as targets for circ_0081723 in circInteractome, circBank, and starBase online software. (B) The binding sites between circ_0081723 and miR-545-3p were shown. (C) SiHa and Caski cells were transfected with miR-NC or miR-545-3p, and the miR-545-3p expression was measured by qRT-PCR. (D-G) Dual luciferase reporter assay (D-E) and RIP assay (F-G) were performed to verify the interaction between circ_0081723 and miR-545-3p. (H-I) The miR-545-3p expression was measured by qRT-PCR in paired CC tissues and CC cells. (J) Pearson’s correlation analysis was used to analyze the linear correlation between the level of circ_0081723 and miR-545-3p in CC tumor tissues. **P < 0.01 and ***P < 0.001
Circ_0000463 silencing restrained CC cell malignant behaviors by upregulating mir-545-3p in vitro
Then, to confirm the functional relevance between circ_0081723 and miR-545-3p on CC development, rescue experiments were conducted in CC cells. At first, qRT-PCR data showed that miR-545-3p expression was effectively reduced in in-miR-545-3p-transfected CC cells (with a 56.33% reduction in SiHa cells and a 70.00% decrease in Caski cells) (Fig. 4A), suggesting that the knockdown was significant. Besides, qRT-PCR results displayed that circ_0081723 knockdown notably increased miR-545-3p expression, which was reverted by the co-transfection of in-miR-545-3p (Fig. 4B). The functional analysis presented that the suppressive effects of circ_0081723 silencing on CC cell proliferation were significantly attenuated by miR-545-3p downregulation (Fig. 4C-F). Furthermore, miR-545-3p inhibition strikingly reversed the sh-circ_0081723-induced increase in apoptotic rate in CC cells (Fig. 4G). Moreover, circ_0081723 absence-mediated CC cell migration and invasion repression were apparently restored by the knockdown of miR-545-3p (Fig. 4H-J). Meanwhile, the downregulation of miR-545-3p could partially relieve circ_0081723 knockdown-triggered decrease in ki-67 protein level and increase in Bax and E-cadherin protein levels in CC cells (Fig. 4K-L). Collectively, these outcomes indicated that the deficiency of circ_0081723 might hinder CC cell malignant behaviors via targeting miR-545-3p.
Circ_0000463 depletion restrained CC progression by targeting miR-545-3pin vitro. (A) SiHa and Caski cells were transfected with in-miR-NC or in-miR-545-3p, and the knockdown efficiency was detected by qRT-PCR. (B-L) SiHa and Caski cells were transfected with sh-NC, sh-circ_0081723, sh-circ_0081723 + in-miR-NC, and sh-circ_0081723 + in-miR-545-3p. (B) The expression of miR-545-3p was determined by qRT-PCR. (C-F) CCK-8 assay (C-D) cell colony formation assay (E) and EdU assay (F) were conducted to assess the cell proliferation ability of SiHa and Caski cells. (G) The apoptotic rate of SiHa and Caski cells was examined by flow cytometry. (H-J) Transwell assay was used to measure the migrated (H) and invaded (I-J) cells. (K-L) WB assay was applied to detect the protein levels of ki-67, Bax, and E-cadherin. *P < 0.05, **P < 0.01 and ***P < 0.001
CREBRF was a target of miR-545-3p
Based on StarBase online software analysis, we found that there were many target mRNAs of miR-545-3p. Among them, some mRNAs (CPEB4, PAK2, KDM2A, AKT2, RRMW2, ATF2, ERO1A, RPP25, and CREBRF) that were highly expressed in CC and promoted the progression of CC using literature research. For further selection, these mRNAs were subjected to qRT-PCR analysis responding to miR-545-3p upregulation. As a result, CREBRF presented the highest fold change (Figure S1). Hence, CREBRF was selected for further research. Then, StarBase online software presented that the 3’UTR of CREBRF possessed the complementary sites with miR-545-3p (Fig. 5A). Subsequently, dual-luciferase reporter assays showed that the luciferase activity of CREBRF 3’UTR WT vector was dramatically reduced by miR-545-3p overexpression in CC cells, rather than CREBRF 3’UTR MUT vector (Fig. 5B-C), indicating that miR-545-3p could interact with the 3’UTR of CREBRF. Additionally, qRT-PCR and WB assays showed that the mRNA (2.18 fold) and protein (2.29 fold) expression of CREBRF was highly expressed in NSCLC tissues in contrast to adjacent normal tissues (Fig. 5D-E). Meanwhile, the protein level of CREBRF was also upregulated in SiHa and Caski cells in comparison with Ect1/E6E7 cells (Fig. 5F). Interestingly, we discovered that circ_0081723 silencing markedly reduced the protein level of CREBRF, and this effect was partly rescued by the knockdown of miR-545-3p (Fig. 5G-H). Meanwhile, Pearson’s correlation analysis revealed that CREBRF mRNA level was negatively correlated with miR-545-3p expression and positively correlated with circ_0081723 expression in CC tumor tissues (Fig. 5I-J). Taken together, the above results proved that CREBRF was a direct target of miR-545-3p, and CREBRF was regulated by circ_0081723/miR-545-3p axis in CC.
MiR-545-3p interacted with CREBRF in CC cells. (A) The binding sites between CREBRF 3’UTR and miR-545-3p were shown. (B-C) SiHa and Caski cells were transfected with miR-NC or miR-545-3p, and dual-luciferase reporter assay was adopted to confirm the interaction between circ_0081723 and miR-545-3p. (D-E) The mRNA and protein expression of CREBRF were measured by qRT-PCR and WB assay in paired CC tissues. (F) The CREBRF protein expression was examined by WB assay CC cell lines (SiHa and Caski) and Ect1/E6E7 cells. (G-H) The protein expression of CREBRF was detected by WB assay in SiHa and Caski cells transfected with sh-NC, sh-circ_0081723, sh-circ_0081723 + in-miR-NC and sh-circ_0081723 + in-miR-545-3p. (I-J) Pearson’s correlation analysis was used to analyze the linear correlations between the level of CREBRF and miR-545-3p or circ_0081723 in CC tumor tissues. **P < 0.01 and ***P < 0.001
MiR-545-3p overexpression suppressed CC cell malignant behaviors by downregulating CREBRF in vitro
Subsequently, reversed function experiments were employed to investigate the functional role of miR-545-3p and CREBRF in CC progression. After confirming the overexpression efficiency of CREBRF (SiHa, 3.93 fold; and Caski, 5.48 fold) (Fig. 6A), CC cells were co-transfected with miR-NC or miR-545-3p and pcDNA or CREBRF. After that, WB data revealed that miR-545-3p overexpression effectively decreased the level of CREBRF protein in CC cells, and this effect was almost restored after the co-transfection of pcDNA-CREBRF (Fig. 6B). Then, CCK-8 assay, colony formation assay, and EdU assay presented that miR-545-3p overexpression remarkably hampered CC cell proliferation, which was clearly overturned by CREBRF overexpression (Fig. 6C-G). In parallel, flow cytometry results exhibited that the upregulation of CREBRF might significantly abrogate the positive effects of miR-545-3p on CC cell apoptosis rate (Fig. 6C-H). In addition, miR-545-3p-mediated CC cell migration and invasion inhibition were apparently attenuated by CREBRF overexpression (Fig. 6I-K). Moreover, downregulated ki-67 protein expression and upregulated Bax and E-cadherin protein expression caused by miR-545-3p mimic were mostly recovered by the introduction of the CREBRF overexpression plasmid (Fig. 6L-M). Overall, these outcomes validated that the upregulation of miR-545-3p might repress CC cell malignant behaviors via targeting CREBRF.
MiR-545-3p overexpression-mediated effects were largely reverted by CREBRF overexpressionin vitro. (A) SiHa and Caski cells were transfected with pcDNA or CREBRF, and the overexpression efficiency was detected by WB assay. (B-M) SiHa and Caski cells were transfected with miR-NC, miR-545-3p, miR-545-3p + pcDNA and miR-545-3p + CREBRF. (B) The CREBRF protein expression was measured by WB assay. (C-G) CCK-8 assay (C-D) cell colony formation assay (E-F) and EdU assay (G) were conducted to assess the cell proliferation ability of SiHa and Caski cells. (H) The apoptotic SiHa and Caski cells were examined by flow cytometry. (I-K) The migrated (I) and invaded (J-K) SiHa and Caski cells were examined by transwell assay. (L-M) The protein levels of ki-67, Bax, and E-cadherin were detected by WB assay. *P < 0.05, **P < 0.01 and ***P < 0.001
Circ_0081723 silencing repressed tumor growth in vivo
Additionally, the role of circ_0081723 in CC tumor growth in vivo was explored via animal experiments. Caski cells stably expressing sh-circ_0081723 or sh-NC were inoculated into mice to establish the mice model. As shown in Fig. 7A-C, tumor volume and weight in the sh-circ_0081723 group were evidently smaller than that in the sh-NC group. The knockdown of circ_0081723 resulted in the downregulation of circ_0081723 level and CREBRF protein level and the upregulation of miR-545-3p level (Fig. 7D-F). IHC assay data presented that the ki-67-positive cells were decreased and the Bax-positive cells and E-cadherin-positive cells were increased in the sh-circ_0081723 group when compared with the sh-NC group (Fig. 7G). Collectively, these findings implied that circ_0081723 served as a cancer-promoting factor in tumor growth in vivo.
Circ_0081723 knockdown notably suppresses xenograft tumor growth. Caski cells transfected with sh-NC or sh-circ_0081723 were injected into nude mice to establish a xenograft tumor model. (A-C) The volume and weight of xenograft tumors were measured. (D-E) The expression of circ_0081723 and miR-545-3p in the tumor tissues was determined by qRT-PCR. (F) WB assay was adopted to detect the protein expression of CREBRF. (G) The positive cells of ki-67, Bax, E-cadherin, and CREBRF were evaluated by IHC assay. ***P < 0.001
Discussion
This study tried to confirm the biological function of circ_0081723 in CC. Results presented that circ_0081723 abundance in CC tumor tissues and cells was significantly increased, and its silencing notably hindered the growth and metastasis of CC cells. Further mechanism studies demonstrated that the miR-545-3p/CREBRF axis was the key signaling pathway for circ_0081723 to promote the development of CC.
Genetic factors are an important factor in the pathogenesis of CC (Ramachandran and Dork 2021). Furthermore, studies have confirmed that the occurrence of HPV infection significantly increases the risk of CC (Schiffman et al. 2016). Although HPV vaccination can effectively prevent the occurrence of CC, medical conditions have resulted in lower vaccination rates in low- and middle-income countries (Cohen et al. 2019). Therefore, it is imperative to develop biomarkers with high specificity, good safety, and economical price. CircRNAs are a type of biomarker with high development value (Fontemaggi et al. 2021). The abnormally expressed circRNAs have been confirmed to be biomarkers for the diagnosis of many cancers (Tian et al. 2021; Harper et al. 2021). In addition, the role of circRNAs in the pathogenesis of CC has been gradually revealed. Hsa_circ_0043280 contributed to PAQAR3 expression and restrained the growth and motility of CC cells via absorbing miR-203a-3p, which suggested that hsa_circ_0043280 was a tumor suppressor in CC (Zhang et al. 2021a). Highly expressed circ-ACACA was found in CC, and circ-ACACA could sponge miR-582-5p to release the inhibitory effect on ERO1A expression, which exhibited that circ-ACACA acted as a tumor-promoting factor (Huang and Li 2021). In this study, we found that circ_0081723 was overexpressed in CC tissue samples (2.13 fold), which was consistent with the previous finding (Qu et al. 2020). Meanwhile, we confirmed that circ_0081723 had the same expression pattern in CC cells (SiHa, 2.81 fold; and Caski, 4.36 fold) as in CC tissues. The results verified that circ_0081723 knockdown had an inhibitory impact on CC cell proliferation and metastasis, implying circ_0081723 could facilitate the malignant process of CC.
CircRNAs could act as sponges of miRNA to regulate cell functions through an endogenous competitive RNA (ceRNA) mechanism (Zhong et al. 2018). According to bioinformatics predictions and related verification experiments, miR-545-3p could bind with circ_0081723. MiR-545-3p has been reported to exert anti-tumor effects in CC (Hu et al. 2019), colorectal cancer (Zhang et al. 2021b), oral squamous cell carcinoma (Ning et al. 2021), and non-small-cell lung cancer (Chen et al. 2020). However, it was still unclear whether miR-545-3p was involved in circ_0081723-mediated CC regulation. This study proved miR-545-3p partially abolished circ_0081723-mediated CC cell growth and metastasis repression via performing rescue experiments, implying that circ_0081723 played a tumor-promoting role in CC progression via targeting miR-545-3p.
MiRNAs negatively regulate gene expression by targeting the mRNA sequence of the gene (Jonas and Izaurralde 2015). Therefore, we further explored and confirmed that CREBRF was a target gene of miR-545-3p. As reported, CREBRF had the pro-cancer effect in gastric cancer (Han et al. 2018) and CC (Tang et al. 2021), and played an anti-cancer role in glioblastoma (Xue et al. 2016), acute myeloid leukemia (Han et al. 2020), and gallbladder carcinoma (Wu et al. 2019), suggesting that the function of CREBRF was disease-specific. Herein, a high level of CREBRF was presented in CC samples. Moreover, CREBRF expression was regulated by the circ_0081723/miR-545-3p axis. Additionally, miR-545-3p mediated suppressive effects on CC cell malignant behaviors were partially relieved by CREBRF overexpression.
All in all, our data confirmed that the abnormally upregulated circ_0081723 in CC tumor tissue, and its deficiency might hinder CC progression via targeting miR-545-3p/CREBRF (Fig. 8), providing a promising candidate target for CC treatment.
Data availability
No datasets were generated or analysed during the current study.
References
Chalbatani GM, Gharagouzloo E, Memari F, Dana H, Hadloo MHM, Zainalinia N et al (2021) Integrative and in-vitro analysis reveal hsa_circ_001787 can act as a diagnostic biomarker for colorectal cancer. Saudi J Biol Sci 28(11):6230–6238. https://doi.org/10.1016/j.sjbs.2021.06.071
Chen S, Lu S, Yao Y, Chen J, Yang G, Tu L et al (2020) Downregulation of hsa_circ_0007580 inhibits non-small cell lung cancer tumorigenesis by reducing mir-545-3p sponging. Aging 12(14):14329–14340. https://doi.org/10.18632/aging.103472
Chen Y, Xu X, Li X, Zhong J, Wu B, Shang J et al (2021) Identification of circular RNAs hsa_circ_0140271 in peripheral blood mononuclear cells as a novel diagnostic biomarker for female rheumatoid arthritis. J Orthop Surg Res 16(1):647. https://doi.org/10.1186/s13018-021-02794-8
Cohen PA, Jhingran A, Oaknin A, Denny L (2019) Cervical cancer. Lancet 393(10167):169–182. https://doi.org/10.1016/s0140-6736(18)32470-x
Conesa-Zamora P (2013) Immune responses against virus and tumor in cervical carcinogenesis: treatment strategies for avoiding the HPV-induced immune escape. Gynecol Oncol 131(2):480–488. https://doi.org/10.1016/j.ygyno.2013.08.025
Cui X, Wang J, Guo Z, Li M, Li M, Liu S et al (2018) Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer 17(1):123. https://doi.org/10.1186/s12943-018-0877-y
Cui Y, Cao J, Huang S, Ye J, Huang H, Liao D et al (2021) circRNA_0006470 promotes the proliferation and migration of gastric cancer cells by functioning as a sponge of miR-27b-3p. Neoplasma 68(6):1245–1256. https://doi.org/10.4149/neo_2021_210222N235
Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M (2016) CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 13(1):34–42. https://doi.org/10.1080/15476286.2015.1128065
Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79:351–379. https://doi.org/10.1146/annurev-biochem-060308-103103
Feng W, Guo R, Zhang D, Zhang R (2021) Circ-ABCB10 knockdown inhibits the malignant progression of cervical cancer through microRNA-128-3p/ZEB1 axis. Biol Proced Online 23(1):17. https://doi.org/10.1186/s12575-021-00154-8
Ferrall L, Lin KY, Roden RBS, Hung CF, Wu TC (2021) Cervical Cancer immunotherapy: facts and hopes. Clin Cancer Res 27(18):4953–4973. https://doi.org/10.1158/1078-0432.CCR-20-2833
Fontemaggi G, Turco C, Esposito G, Di Agostino S (2021) New Molecular mechanisms and Clinical Impact of circRNAs in Human Cancer. Cancers (Basel) 13(13):3154. https://doi.org/10.3390/cancers13133154
Han J, Zhang L, Zhang J, Jiang Q, Tong D, Wang X et al (2018) CREBRF promotes the proliferation of human gastric cancer cells via the AKT signaling pathway. Cell Mol Biol (Noisy-le-grand) 64(5):40–45
Han F, Zhong C, Li W, Wang R, Zhang C, Yang X et al (2020) hsa_circ_0001947 suppresses acute myeloid leukemia progression via targeting hsa-miR-329-5p/CREBRF axis. Epigenomics 12(11):935–953. https://doi.org/10.2217/epi-2019-0352
Harper KL, Mottram TJ, Whitehouse A (2021) Insights into the evolving roles of circular RNAs in Cancer. Cancers (Basel) 13(16):4180. https://doi.org/10.3390/cancers13164180
Hu C, Wang Y, Li A, Zhang J, Xue F, Zhu L (2019) Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J Cell Physiol 234(6):9225–9232. https://doi.org/10.1002/jcp.27601
Huang D, Li C (2021) circ-ACACA promotes proliferation, invasion, migration and glycolysis of cervical cancer cells by targeting the miR-582-5p/ERO1A signaling axis. Oncol Lett 22(5):795. https://doi.org/10.3892/ol.2021.13056
Huang DW, Huang M, Lin XS, Huang Q (2017) CD155 expression and its correlation with clinicopathologic characteristics, angiogenesis, and prognosis in human cholangiocarcinoma. Onco Targets Ther 10:3817–3825. https://doi.org/10.2147/OTT.S141476
Jonas S, Izaurralde E (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16(7):421–433. https://doi.org/10.1038/nrg3965
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20(11):675–691. https://doi.org/10.1038/s41576-019-0158-7
Kwong GA, Ghosh S, Gamboa L, Patriotis C, Srivastava S, Bhatia SN (2021) Synthetic biomarkers: a twenty-first century path to early cancer detection. Nat Rev Cancer 21(10):655–668. https://doi.org/10.1038/s41568-021-00389-3
Liu M, Wang Q, Shen J, Yang BB, Ding X (2019) Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol 16(7):899–905. https://doi.org/10.1080/15476286.2019.1600395
Ma Y, Li Z, Ma D, Guo J, Sun W (2022) Hsa_circ_0003195 as a biomarker for diagnosis and prognosis of gastric cancer. Int J Clin Oncol 27(2):354–361. https://doi.org/10.1007/s10147-021-02073-w
Marquina G, Manzano A, Casado A (2018) Targeted agents in Cervical Cancer: Beyond Bevacizumab. Curr Oncol Rep 20(5):40. https://doi.org/10.1007/s11912-018-0680-3
McMellen A, Woodruff ER, Corr BR, Bitler BG, Moroney MR (2020) Wnt signaling in gynecologic malignancies. Int J Mol Sci 21(12):4272. https://doi.org/10.3390/ijms21124272
Ning B, Guo S, Mei Y (2021) Long non-coding RNA CASC9 promotes tumor progression in oral squamous cell carcinoma by regulating microRNA-545-3p/laminin subunit gamma 2. Bioengineered 12(1):7907–7919. https://doi.org/10.1080/21655979.2021.1977103
O’Leary K (2021) HPV vaccines beat cervical cancer. Nat Med. https://doi.org/10.1038/d41591-021-00068-8
Qu X, Zhu L, Song L, Liu S (2020) Circ_0084927 promotes cervical carcinogenesis by sponging miR-1179 that suppresses CDK2, a cell cycle-related gene. Cancer Cell Int 20:333. https://doi.org/10.1186/s12935-020-01417-2
Ramachandran D, Dork T (2021) Genomic risk factors for cervical Cancer. Cancers (Basel) 13(20):5137. https://doi.org/10.3390/cancers13205137
Schiffman M, Doorbar J, Wentzensen N, de Sanjose S, Fakhry C, Monk BJ et al (2016) Carcinogenic human papillomavirus infection. Nat Rev Dis Primers 2:16086. https://doi.org/10.1038/nrdp.2016.86
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69(1):7–34. https://doi.org/10.3322/caac.21551
Tang X, Wen X, Li Z, Wen D, Lin L, Liu J et al (2021) Hsa_circ_0102171 aggravates the progression of cervical cancer through targeting miR-4465/CREBRF axis. J Cell Physiol 236(7):4973–4984. https://doi.org/10.1002/jcp.30210
Tian T, Zhao Y, Zheng J, Jin S, Liu Z, Wang T (2021) Circular RNA: a potential diagnostic, prognostic, and therapeutic biomarker for human triple-negative breast cancer. Mol Ther Nucleic Acids 26:63–80. https://doi.org/10.1016/j.omtn.2021.06.017
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H et al (2021) Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer 20(1):13. https://doi.org/10.1186/s12943-020-01298-z
Wu K, Huang J, Xu T, Ye Z, Jin F, Li N et al (2019) MicroRNA-181b blocks gensenoside Rg3-mediated tumor suppression of gallbladder carcinoma by promoting autophagy flux via CREBRF/CREB3 pathway. Am J Transl Res 11(9):5776–5787
Xue H, Zhang J, Guo X, Wang J, Li J, Gao X et al (2016) CREBRF is a potent tumor suppressor of glioblastoma by blocking hypoxia-induced autophagy via the CREB3/ATG5 pathway. Int J Oncol 49(2):519–528. https://doi.org/10.3892/ijo.2016.3576
Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH (2011) StarBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and degradome-seq data. Nucleic Acids Res 39(Database issue):D202–209. https://doi.org/10.1093/nar/gkq1056
Zhang C, Liu P, Huang J, Liao Y, Pan C, Liu J et al (2021a) Circular RNA hsa_circ_0043280 inhibits cervical cancer tumor growth and metastasis via miR-203a-3p/PAQR3 axis. Cell Death Dis 12(10):888. https://doi.org/10.1038/s41419-021-04193-7
Zhang L, Yu R, Li C, Dang Y, Yi X, Wang L (2021b) Circ_0026416 downregulation blocks the development of colorectal cancer through depleting MYO6 expression by enriching miR-545-3p. World J Surg Oncol 19(1):299. https://doi.org/10.1186/s12957-021-02407-y
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F et al (2018) Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 17(1):79. https://doi.org/10.1186/s12943-018-0827-8
Acknowledgements
None.
Funding
This study was supported by the Doctoral Research Foundation of Affiliated Hospital of Nantong University (No.Tdb2012).
Author information
Authors and Affiliations
Contributions
Q.M. performed experiments and wrote the manuscript. W.Y. and Z.L. collected and analyzed data. X.Z. contributed the methodology and edited the manuscript. L.Z. conceived and designed research. All authors reviewed the manuscript. The authors declare that all data were generated in-house and that no paper mill was used.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed with the authorization of the ethics committee of Gongli Hospital of Shanghai Pudong New Area and the written informed consent signed by all patients.
All animal experiments were approved by the Animal Ethics Committee of Gongli Hospital of Shanghai Pudong New Area.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.

Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, Q., Yu, W., Li, Z. et al. Circ_0081723 enhances cervical cancer progression and modulates CREBRF via sponging miR-545-3p. Naunyn-Schmiedeberg's Arch Pharmacol (2024). https://doi.org/10.1007/s00210-024-03175-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00210-024-03175-8