Abstract
Circadian clock genes influence biological processes and may be involved in tumorigenesis. We systematically evaluated genetic variants in the circadian clock pathway genes associated with colorectal cancer risk and survival. We evaluated the association of 119 single nucleotide polymorphisms (SNPs) in 27 circadian clock pathway genes with the risk of colorectal cancer in a case–control study (1150 cases and 1342 controls). The false discovery rate (FDR) method was applied to correct for multiple comparisons. Gene-based analysis was performed by the sequence kernel association test (SKAT). Cox proportional hazards regression was used to calculate the effects of SNPs on the overall survival of patients. We identified that compared to those with the G allele, individuals with the rs76436997 A allele in RORA had a significant 1.33-fold increased risk of colorectal cancer (P = 3.83 × 10− 4). Specifically, the GA/AA genotypes were related to an enhanced risk of colorectal cancer compared with that associated with the GG genotype, which was more common in patients with well and moderately differentiated tumors and Dukes A/B stages. The SNP rs76436997 significantly increased the overall survival time of colorectal cancer patients (P = 0.044). Furthermore, RNA-seq data showed that the mRNA levels of RORA were significantly lower in colorectal tumors than the paired normal tissues. Gene-based analysis revealed a significant association between RORA and colorectal cancer risk. These findings highlight the important roles of genetic variations in circadian clock pathway genes play in colorectal cancer risk and suggest that RORA is potentially related to colorectal carcinogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer is the third most common malignancy and has a high mortality rate worldwide (McGuire 2016). The incidence of colorectal cancer increased significantly in China with the improvement of the economy and changes in lifestyle. Colorectal cancer accounts for a large proportion of cancer morbidity and mortality in China (Chen et al. 2016). The etiology of colorectal cancer involves numerous risk factors, such as alcohol consumption, smoking, low levels of physical activity, unhealthy dietary habits and increased body mass index (BMI) (Hughes et al. 2017). However, these known risk factors cannot fully explain the high incidence of colorectal cancer. Genetic variations might also affect individual susceptibility and prognosis of colorectal cancer (Lemire et al. 2015).
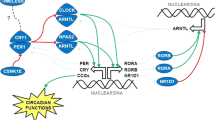
The circadian “clock” is located in the suprachiasmatic nucleus in the hypothalamus, and it plays an important role in driving many biologic processes in the body through the periodic transcription of genes (Yu and Weaver 2011). The circadian clock pathway genes are associated with the susceptibility and prognosis of several cancers through their involvement in regulating DNA damage and repair, carcinogen metabolism and detoxification, cell proliferation, apoptosis and the cell cycle (Zmrzljak and Rozman 2012). Several lines of evidence from previous studies suggest that single nucleotide polymorphisms (SNPs) in circadian clock genes are associated with the risk of developing prostate cancer (Chu et al. 2008; Markt et al. 2015), breast cancer (Grundy et al. 2013; Monsees et al. 2012; Rabstein et al. 2014; Truong et al. 2014; Zienolddiny et al. 2013), and glioma (Madden et al. 2014) as well as the likelihood of surviving gastric cancer (Qu et al. 2016). However, only a few studies have investigated the core circadian genes in relation to colorectal cancer risk, and a limited number of SNPs in each gene were demonstrated (Karantanos et al. 2013). These reported results have not been replicated by many studies, and no associations between gene variants in circadian clock genes and the survival of colorectal cancer have been found.
The primary circadian clock pathway genes in mammals have been proposed as the following: CLOCK, neuronal PAS domain protein 2 (NPAS2), ary1 hydrocarbon receptor nuclear translocator-like (ARNTL), period 1 (PER1), period 2 (PER2), period 3 (PER3), cryptochrome 1 (CRY1), cryptochrome 2 (CRY2) and casein kinase 1-epsilon (CSNK1E)(Kondratov et al. 2007). Melatonin, which regulates circadian rhythms, has been demonstrated to be related to cancer susceptibility (Blask et al. 2011). The levels of melatonin are regulated by its biosynthesis and signaling pathway, which is mediated by the arylalkylamine N-acetyltransferase (AANAT) protein, melatonin receptor MTNR1A, melatonin receptor MTNR1B and the retinoic acid-related orphan receptors RORA and RORB (Slominski et al. 2012). Momma et al. (Momma et al. 2017) demonstrated that PER1, PER2, CRY1 and CRY2 were frequently expressed in colorectal cancer but not in adenomas. The expression levels of PER1, PER2 and CLOCK were associated with tumor size, the depth of invasion and survival of patients with colorectal carcinomas.
In the present study, we systematically investigated the relationships between genetic variants of the circadian clock pathway genes and the risk of colorectal cancer in 1150 cases and 1342 healthy controls. We also examined whether these selected genetic variants are related to the prognosis of colorectal cancer in the Chinese population.
Materials and methods
Study population
Briefly, 1150 cases with newly and histologically diagnosed colorectal cancer were included in this case–control study from the Affiliated Nanjing First Hospital and the First Affiliated Hospital of Nanjing Medical University on September 2010. Patients with previous cancer, colorectal neoplasia, and inflammatory bowel disease were excluded from the study. A total of 1342 cancer-free controls were randomly selected from a pool of more than 25,000 cancer-free individuals on the basis of physical examinations and frequency matched to cases on age (± 5 years) and gender. A total of 344 colorectal cancer patients were followed up by telephone interviews, 57 patients were excluded due to incomplete overall survival information. The last follow up date was April 2, 2016. All the participants gave their written informed consent that their information could be used and published for research purposes and donated approximately 5 mL of whole blood. The study was approved by the Institutional Review Board of Nanjing University.
Selection of SNPs in the circadian clock pathway
A total of 63 circadian clock pathway genes were selected from the Cancer Genome Anatomy Project (CGAP) database (http://cgap.nci.nih.gov) and previously reported studies (Supplementary Table 1). We identified 27 genes that were differentially expressed in tumor tissues and normal tissues through the Cancer Genome Atlas (TCGA) database from among those 63 genes in the circadian clock pathway; the 27 selected genes met the following criteria: (a) P < 0.05, (b) fold change > 1.5, and (c) call rate > 80% (Supplementary Table 2). The selected key genes in the circadian clock pathway are shown in Supplementary Fig. 1.
The flow chart of SNP selection is shown in Fig. 1. First, we identified 5223 SNPs that met the criteria of minor allele frequency (MAF) ≥ 0.05, Hardy–Weinberg equilibrium (HWE) ≥ 0.05, and call rate > 95% from the 1000 Genomes Project. Then, we predicted the potential function of the SNPs using RegulomeDB, SNPinfo, and HaploReg. Next, we used HaploView 4.2 software to select the tagged SNPs (r2 ≥ 0.8) by pairwise linkage disequilibrium (LD) analysis. Finally, we included a total of 119 SNPs for genotyping in this study.
SNPs genotyping
Genomic DNA was isolated from the peripheral blood samples using the Qiagen Blood Kit (Qiagen). DNA was successfully extracted from the blood samples obtained from all subjects. A total of 119 candidate SNPs in the circadian clock pathway genes were included in the present analysis. Genotyping was performed using Illumina Human Omni ZhongHua Bead Chips. The genotyping success rate was at least 98% in cases and controls. Laboratory personnel were blind to the status of the case and control samples.
Statistical analysis
The differences in the distributions of demographic variables between cases and controls were compared by t test for continuous variables and Chi square test for categorical variables. Allele frequencies in the control groups were analyzed for HWE. The associations between genetic variants and colorectal cancer risk were evaluated with odds ratios (ORs) and their 95% confidence intervals (CIs) using an unconditional logistic regression model. The false discovery rate (FDR) method was applied to control for multiple comparisons. Gene-based analysis was performed by the sequence kernel association test (SKAT) (Wu et al. 2011). We performed the mRNA expression analysis using colorectal cancer data from TCGA database (http://cancergenome.nih.gov/) and the Gene Expression Omnibus (GEO) datasets. Significant differences in gene expression between colorectal tumors and normal tissues were compared by a two-sided Mann–Whitney test. In TCGA colorectal cancer database, differential gene expression was measure in 625 colorectal cancer tumors and 51 normal tissues. Expression quantitative trait loci (eQTL) analysis was performed based on TCGA database and the Genotype Tissue Expression (GTEx) project dataset to evaluate the genetic variant effects on the expression of genes (Cookson et al. 2009; Emilsson et al. 2008).
Unconditional univariate and multivariate Cox regression analyses were used to assess the hazard ratios (HRs) and 95% CIs for the associations between SNPs and the overall survival of patients with colorectal cancer. The associations between survival time and genetic variants were measured using the Kaplan–Meier method and the log-rank test. P value < 0.05 was the statistically significant threshold. R 3.3.3 and PLINK 1.09 were used for all the statistical analyses.
Results
Characteristics of the study population
This study was a case–control study with a total of 1150 colorectal cancer cases and 1342 controls. The demographic characteristics of the cases and controls are shown in Table 1. There were no significant differences in terms of the distributions of age, gender, smoking status and alcohol consumption between the cases and controls (P = 0.994, 0.738, 0.334, and 0.077, respectively). Of the patients, 51.0% had colon cancer. The most common tumor grade was moderately differentiated (77.3%). The percentages of Dukes stages A, B, C, and D were 5.7, 38.6, 37.3, and 18.4%, respectively.
SNPs in RORA and colorectal cancer risk
A total of 119 SNPs in 27 genes in the circadian clock pathway were analyzed for their relationships with colorectal cancer risk (data not shown). As shown in Table 2, we found that 12 SNPs (rs76436997, rs61815118, rs62576340, rs340023, rs1437551, rs1997644, rs3803479, rs11635975, rs1542178, rs2102928, rs2227631 and rs919000) were nominally associated with colorectal cancer risk in the additive genetic model (P < 0.05). After FDR correction, only rs76436997 in RORA was associated with an increased risk of colorectal cancer (P = 0.046). The gene-based analysis further revealed the most significant correlations existed between genetic variants in RORA and colorectal cancer risk (P = 4.60 × 10− 4) (Supplementary Table 3).
Association between rs76436997 and colorectal cancer risk
The genotype frequencies of RORA rs76436997 and their associations with colorectal cancer susceptibility according to four genetic models (additive, dominant, codominant and recessive model) are shown in Table 3. The frequencies of the GG, GA, and AA genotypes were 3.87, 29.09, and 67.04% in the cases and 1.71, 23.89, and 74.40% in the controls. In the additive model, we found that individuals with the A allele had a significantly increased risk of colorectal cancer compared to that of those with the G allele (OR = 1.33, 95% CI = 1.14–1.55). Compared with the GG genotype, the GA/AA genotypes were significantly linked with colorectal cancer susceptibility (OR = 1.41, 95% CI = 1.17–1.70). Further stratified analyses in the dominant model showed that there was no significant heterogeneity in the subgroup analyses according to age, gender, smoking, and alcohol consumption (Supplementary Table 4).
The associations between RORA rs76436997 and the clinicopathologic variables related to colorectal cancer risk stratified by tumor site, tumor grade and Dukes stage are shown in Table 4. Statistical analysis revealed a significant association between the GA/AA genotypes and the risk of tumors in the colon and rectum (OR = 1.47, 95% CI = 1.17–1.86 for colon tumors and OR = 1.35, 95% CI = 1.07–1.71 for rectal tumors). A subsequent stratification analysis by tumor grade showed that there was a significantly increased risk in well and moderately differentiated colorectal cancers (OR = 1.44, 95% CI = 1.18–1.75) but not in poorly differentiated tumors (OR = 1.23, 95% CI = 0.82–1.84). In addition, we observed a stronger significant association between the GA/AA genotypes and colorectal cancer risk in Dukes A and B stages (OR = 1.45, 95% CI = 1.15–1.83) than in Dukes C and D stages (OR = 1.37, 95% CI = 1.09–1.73).
Expression levels of RORA in colorectal tumor and normal tissues
In addition, we evaluated the mRNA expressions levels of RORA in 17 paired clinical samples from Nanjing University. The expression levels were further validated in 638 colorectal tumor tissues and 51 normal tissues from TCGA and two GEO datasets. As shown in Fig. 2, the mRNA expression levels of RORA were significantly lower in colorectal tumors than in the normal tissues (P = 3.48 × 10− 4 in in-house RNA-seq data, P = 2.20 × 10− 16 in TCGA data and P = 8.01 × 10− 24 in GSE21510 data, respectively). An insignificantly lower expression level was found in the GSE21510 data (P = 0.103). Based on the expression data and genotyping data from TCGA and GTEx, we found that rs76436997 was not an eQTL for RORA (Supplementary Fig. 2).
Effects of RORA on the overall survival of patients with colorectal cancer
According to the multivariate Cox regression analyses using age, sex, smoking and alcohol consumption, we evaluated the prognostic ability of rs76436997 in RORA for the overall survival of patients with colorectal cancer. Compared with the GG genotype, the GA/AA genotypes were significantly associated with longer overall survival of patients with colorectal cancer (HR = 0.69, 95% CI = 0.49–0.99, P = 0.044) (Supplementary Fig. 3a). We also detected significantly poorer overall survival in patients who smoked than in nonsmokers (HR = 1.53, 95% CI = 1.01–2.30, P = 0.043) (Supplementary Table 5). Furthermore, we compared the overall survival time between patients with low expression levels of RORA and those with high levels of expression in TCGA database. Individuals with low levels of expression of RORA had slightly longer overall survival times than those with high levels of RORA expression, although the P value was larger than 0.05 (Supplementary Fig. 3b).
Discussion
The circadian system plays a vital role in the regulation of various physiologic, metabolic and behavioral processes (Fu and Lee 2003). Disturbances of the circadian rhythm in humans may increase the risk of cancer (Costa et al. 2010). A previous meta-analysis demonstrated that the disruption of the natural circadian rhythm was a potential risk factor associated with an increased risk of colorectal cancer (Wang et al. 2015). The molecular mechanism of the circadian pathway is based on interlocking positive/negative transcriptional-translational feedback loops that are regulated by a series of core circadian clock genes (Lee et al. 2001). Genetic variation in genes involved in the circadian clock pathway has been the focus of attention in recent years. However, there have been few studies on the relationship between genetic variants and the risk of colorectal cancer. Only one study identified that genetic variants in the CLOCK1 gene significantly increased the risk of colorectal cancer, while they did not affect the prognosis of colorectal cancer patients (Karantanos et al. 2013). In this study, we evaluated the effect of genetic variants in 27 circadian clock pathway genes on colorectal cancer susceptibility and prognosis. We found that the rs37436997 SNP in RORA was significantly associated with an increased risk of developing colorectal cancer.
RORA, retinoic acid receptor-related orphan nuclear receptor A, is a member of the orphan nuclear receptor family, which is located at 15q21–q22 (Polakis 2000; Xiong et al. 2012). RORA is involved in lipid metabolism, the maintenance of circadian rhythm clock function, immune regulation and tumor progression (Boukhtouche et al. 2004; Kottorou et al. 2012). The mRNA and protein expression levels of RORA in colorectal cancer tissue were significantly downregulated compared to those in normal tissue and were related to the time to disease progression (Kottorou et al. 2012). Our study found that RORA was dramatically downregulated in colorectal cancer tumors in paired clinical samples and TCGA data, which is consistent with the findings of previous studies. However, the functional significance of RORA in the development of colorectal cancer has not yet been studied extensively. The role of RORA was proposed to be the inhibition of the proliferation and motility of colorectal cancer cells through the activation of RORA by cholesterol sulfate (Xiao et al. 2015), the inhibition of the Wnt/β-catenin signaling pathway to suppress colorectal cancer cell growth (Lee et al. 2010), and the promotion of apoptosis by enhancing the stability of the p53 gene (Kim et al. 2011). These studies have shown that RORA is a functional tumor suppressor gene, but further functional studies are still needed to confirm the effects of RORA.
We also evaluated the potential impact of RORA on clinicopathological parameters and the survival of patients with colorectal cancer. A previous study showed that the low level of expression of RORA was correlated with a high level of serum alpha fetoprotein (AFP), poor pathology grade, tumor recurrence, and vascular invasion in hepatocellular carcinoma (HCC). Fu et al. found that RORA was an independent predictor of overall and disease-free survival in HCC patients (Fu et al. 2014). Moreover, the reduction in methylation of the RORA promoter is associated with late stages (stages III and IV) of colorectal cancer (Kano et al. 2016). In addition, Li et al. demonstrated that RORA SNPs rs782917 and rs17204952 were associated with an increased risk of death due to cutaneous melanoma (Li et al. 2018). However, the associations between genetic variants of RORA and the clinical outcome of colorectal cancer have not been reported. This study demonstrated that compared with the GG genotype, the GA/AA genotypes of rs76436997 in RORA were significantly associated with better differentiation and the early stages of colorectal cancer. Our findings also suggest that colorectal cancer patients with the GA/AA genotypes may have a longer overall survival times. The possible mechanism is that genetic variation may affect the function of the gene, and studies with a larger sample size and further in-depth functional studies are needed to validate the results.
In conclusion, we identified genetic variants in RORA that may contribute to the risk and prognosis of colorectal cancer. The genetic associations, together with the differences in the expression levels of RORA, suggest that RORA might play important roles in colorectal tumorigenesis. Further studies are needed to confirm the role of circadian genes in the development and outcome of colorectal cancer.
References
Blask DE, Hill SM, Dauchy RT et al (2011) Circadian regulation of molecular, dietary, and metabolic signaling mechanisms of human breast cancer growth by the nocturnal melatonin signal and the consequences of its disruption by light at night. J Pineal Res 51(3):259–269. https://doi.org/10.1111/j.1600-079X.2011.00888.x
Boukhtouche F, Mariani J, Tedgui A (2004) The “CholesteROR” protective pathway in the vascular system. Arterioscler Thromb Vasc Biol 24(4):637–643. https://doi.org/10.1161/01.ATV.0000119355.56036.de
Chen W, Zheng R, Baade PD et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66(2):115–132. https://doi.org/10.3322/caac.21338
Chu LW, Zhu Y, Yu K et al (2008) Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis 11(4):342–348. https://doi.org/10.1038/sj.pcan.4501024
Cookson W, Liang L, Abecasis G et al (2009) Mapping complex disease traits with global gene expression. Nat Rev Genet 10(3):184–194. https://doi.org/10.1038/nrg2537
Costa G, Haus E, Stevens R (2010) Shift work and cancer - considerations on rationale, mechanisms, and epidemiology. Scand J Work Environ Health 36(2):163–179
Emilsson V, Thorleifsson G, Zhang B et al (2008) Genetics of gene expression and its effect on disease. Nature 452(7186):423–428. https://doi.org/10.1038/nature06758
Fu L, Lee CC (2003) The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer 3(5):350–361. https://doi.org/10.1038/nrc1072
Fu RD, Qiu CH, Chen HA et al (2014) Retinoic acid receptor-related receptor alpha (RORalpha) is a prognostic marker for hepatocellular carcinoma. Tumour Biol 35(8):7603–7610. https://doi.org/10.1007/s13277-014-2007-9
Grundy A, Schuetz JM, Lai AS et al (2013) Shift work, circadian gene variants and risk of breast cancer. Cancer Epidemiol 37(5):606–612. https://doi.org/10.1016/j.canep.2013.04.006
Hughes LAE, Simons C, van den Brandt PA et al (2017) Lifestyle, diet, and colorectal cancer risk according to (epi)genetic instability: current evidence and future directions of molecular pathological epidemiology. Curr Colorectal Cancer Rep 13(6):455–469. https://doi.org/10.1007/s11888-017-0395-0
Kano H, Takayama T, Midorikawa Y et al (2016) Promoter hypomethylation of RAR-related orphan receptor alpha 1 is correlated with unfavorable clinicopathological features in patients with colorectal cancer. Biosci Trends 10(3):202–209. https://doi.org/10.5582/bst.2016.01097
Karantanos T, Theodoropoulos G, Gazouli M et al (2013) Association of the clock genes polymorphisms with colorectal cancer susceptibility. J Surg Oncol 108(8):563–567. https://doi.org/10.1002/jso.23434
Kim H, Lee JM, Lee G et al (2011) DNA damage-induced RORalpha is crucial for p53 stabilization and increased apoptosis. Mol Cell 44(5):797–810. https://doi.org/10.1016/j.molcel.2011.09.023
Kondratov RV, Gorbacheva VY, Antoch MP (2007) The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol 78:173–216. https://doi.org/10.1016/S0070-2153(06)78005-X
Kottorou AE, Antonacopoulou AG, Dimitrakopoulos FI et al (2012) Altered expression of NFY-C and RORA in colorectal adenocarcinomas. Acta Histochem 114(6):553–561. https://doi.org/10.1016/j.acthis.2011.10.005
Lee C, Etchegaray JP, Cagampang FR et al (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107(7):855–867
Lee JM, Kim IS, Kim H et al (2010) RORalpha attenuates Wnt/beta-catenin signaling by PKCalpha-dependent phosphorylation in colon cancer. Mol Cell 37(2):183–195. https://doi.org/10.1016/j.molcel.2009.12.022
Lemire M, Qu C, Loo LWM et al (2015) A genome-wide association study for colorectal cancer identifies a risk locus in 14q23.1. Hum Genet 134(11–12):1249–1262. https://doi.org/10.1007/s00439-015-1598-6
Li B, Wang Y, Xu Y et al (2018) Genetic variants in RORA and DNMT1 associated with cutaneous melanoma survival. Int J Cancer. https://doi.org/10.1002/ijc.31243
Madden MH, Anic GM, Thompson RC et al (2014) Circadian pathway genes in relation to glioma risk and outcome. Cancer Causes Control 25(1):25–32. https://doi.org/10.1007/s10552-013-0305-y
Markt SC, Valdimarsdottir UA, Shui IM et al (2015) Circadian clock genes and risk of fatal prostate cancer. Cancer Causes Control 26(1):25–33. https://doi.org/10.1007/s10552-014-0478-z
McGuire S (2016) World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 7(2):418–419. https://doi.org/10.3945/an.116.012211
Momma T, Okayama H, Saitou M et al (2017) Expression of circadian clock genes in human colorectal adenoma and carcinoma. Oncol Lett 14(5):5319–5325. https://doi.org/10.3892/ol.2017.6876
Monsees GM, Kraft P, Hankinson SE et al (2012) Circadian genes and breast cancer susceptibility in rotating shift workers. Int J Cancer 131(11):2547–2552. https://doi.org/10.1002/ijc.27564
Polakis P (2000) Wnt signaling and cancer. Genes Dev 14(15):1837–1851
Qu F, Qiao Q, Wang N et al (2016) Genetic polymorphisms in circadian negative feedback regulation genes predict overall survival and response to chemotherapy in gastric cancer patients. Sci Rep 6:22424. https://doi.org/10.1038/srep22424
Rabstein S, Harth V, Justenhoven C et al (2014) Polymorphisms in circadian genes, night work and breast cancer: results from the GENICA study. Chronobiol Int 31(10):1115–1122. https://doi.org/10.3109/07420528.2014.957301
Slominski RM, Reiter RJ, Schlabritz-Loutsevitch N et al (2012) Melatonin membrane receptors in peripheral tissues: distribution and functions. Mol Cell Endocrinol 351(2):152–166. https://doi.org/10.1016/j.mce.2012.01.004
Truong T, Liquet B, Menegaux F et al (2014) Breast cancer risk, nightwork, and circadian clock gene polymorphisms. Endocr Relat Cancer 21(4):629–638. https://doi.org/10.1530/ERC-14-0121
Wang X, Ji A, Zhu Y et al (2015) A meta-analysis including dose-response relationship between night shift work and the risk of colorectal cancer. Oncotarget 6(28):25046–25060. https://doi.org/10.18632/oncotarget.4502
Wu MC, Lee S, Cai T et al (2011) Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89(1):82–93. https://doi.org/10.1016/j.ajhg.2011.05.029
Xiao L, Wang J, Li J et al (2015) RORalpha inhibits adipocyte-conditioned medium-induced colorectal cancer cell proliferation and migration and chick embryo chorioallantoic membrane angiopoiesis. Am J Physiol Cell Physiol 308(5):C385–C396. https://doi.org/10.1152/ajpcell.00091.2014
Xiong G, Wang C, Evers BM et al (2012) RORalpha suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res 72(7):1728–1739. https://doi.org/10.1158/0008-5472.CAN-11-2762
Yu EA, Weaver DR (2011) Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany NY) 3(5):479–493. https://doi.org/10.18632/aging.100323
Zienolddiny S, Haugen A, Lie JA et al (2013) Analysis of polymorphisms in the circadian-related genes and breast cancer risk in Norwegian nurses working night shifts. Breast Cancer Res 15(4):R53. https://doi.org/10.1186/bcr3445
Zmrzljak UP, Rozman D (2012) Circadian regulation of the hepatic endobiotic and xenobitoic detoxification pathways: the time matters. Chem Res Toxicol 25(4):811–824. https://doi.org/10.1021/tx200538r
Funding
This study was supported by the National Key R&D Program of China (2017YFC0908200), the National Natural Science Foundation of China (81773516), the Foundation of Six Talent Peaks Project of Jiangsu (YY-020), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Author information
Authors and Affiliations
Contributions
JC, ZFZ, and MW conceived and designed the experiments. DG and SL wrote the paper. SB and MD contributed reagents/materials/analysis tools. HC and ZZ recruited samples. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors made no disclosures.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gu, D., Li, S., Ben, S. et al. Circadian clock pathway genes associated with colorectal cancer risk and prognosis. Arch Toxicol 92, 2681–2689 (2018). https://doi.org/10.1007/s00204-018-2251-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-018-2251-7