Abstract
Human primary hepatocytes represent a gold standard in in vitro liver research. Due to their low availability and high costs alternative liver cell models with comparable morphological and biochemical characteristics have come into focus. The human hepatocarcinoma cell line HepG2 is often used as a liver model for toxicity studies. However, under two-dimensional (2D) cultivation conditions the expression of xenobiotic-metabolizing enzymes and typical liver markers such as albumin is very low. Cultivation for 21 days in a three-dimensional (3D) Matrigel culture system has been reported to strongly increase the metabolic competence of HepG2 cells. In our present study we further compared HepG2 cell cultivation in three different 3D systems: collagen, Matrigel and Alvetex culture. Cell morphology, albumin secretion, cytochrome P450 monooxygenase enzyme activities, as well as gene expression of xenobiotic-metabolizing and liver-specific enzymes were analyzed after 3, 7, 14, and 21 days of cultivation. Our results show that the previously reported increase of metabolic competence of HepG2 cells is not primarily the result of 3D culture but a consequence of the duration of cultivation. HepG2 cells grown for 21 days in 2D monolayer exhibit comparable biochemical characteristics, CYP activities and gene expression patterns as all 3D culture systems used in our study. However, CYP activities did not reach the level of HepaRG cells. In conclusion, the increase of metabolic competence of the hepatocarcinoma cell line HepG2 is not due to 3D cultivation but rather a result of prolonged cultivation time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The liver represents the main drug- and xenobiotic-metabolizing organ. For in vitro investigations primary human hepatocytes are considered to be the gold standard for liver-specific toxicity and metabolism of xenobiotics (Godoy et al. 2013; LeCluyse 2001). A limitation of primary hepatocytes is their low availability due to the lack of human liver samples, the short life span, inter-individual differences between the donors as well as high costs (Gomez-Lechon et al. 2003). As an alternative, liver cell lines cultured in two-dimensional (2D) monolayers are often used to investigate effects of xenobiotics on human hepatocytes. However, 2D cell cultures have several limitations regarding morphology, biochemical properties, and enzyme activities compared to primary hepatocytes as well as the whole organ itself. During 2D culture numerous biological functions like cell–cell and cell–matrix contacts are lacking resulting in decreased cell differentiation, proliferation, cell functions (Bissell et al. 2003; Weaver et al. 1997) and leading further to alterations in cell signaling pathways (Debnath and Brugge 2005). Additionally, cell functions such as secretion of plasma proteins and the high potential to metabolize xenobiotics are strongly decreased or even lost under 2D culture conditions (Bissell et al. 1987; Clayton and Darnell 1983; Clayton et al. 1985; Godoy et al. 2009). It has been shown that cells growing under 3D culture conditions are more similar in regard to morphology and biochemical properties to the native tissue than cells cultivated in 2D (Daus et al. 2011; Lin et al. 2006; Loessner et al. 2010). Therefore, 3D cultivation has been reported as an important feature to improve traditional 2D cell culture and in vitro studies due to an imitation of the in vivo conditions of the respective organ (Breslin and O’Driscoll 2013). 3D cell culture comprises a wide variety of cultivation techniques where cells can grow in vitro in a micro-structured three dimensional environment enhancing the biophysical and biochemical cell–cell interactions and provide a more in vivo relevant environment for differentiation. The basis for 3D culture methods is the use of artificial scaffolds serving as matrices for cell growth. The origin of 3D scaffolds can be natural (collagen, gelatin, elastin, fibrinogen, etc.) or synthetic polymers. A combination of both can also be used; the choice always depends on the cell type and purpose of the study. Nevertheless, the concept of using any matrix is to mimic the natural organization in porosity, fibers, permeability and mechanical stability (Ravi et al. 2015).
The hepatoma cell line HepG2 represents a well-established liver cell line often used as an in vitro model for human hepatocytes. The cell line derived from the hepatocellular carcinoma of a 15-year-old Caucasian American male. During cultivation, HepG2 cells secrete numerous liver-specific plasma proteins like albumin, transferrin, and α-fetoprotein (Bouma et al. 1988). However, the expression of xenobiotic-metabolizing enzymes as well as nuclear receptors is low compared to human primary hepatocytes or the HepaRG cell line (Aninat et al. 2006; Wilkening and Bader 2003) resulting in a limited use and suitability of HepG2 cells for toxicological and pharmacological applications when metabolism is required. In many studies HepG2 cells have been cultivated under 2D conditions. However, Ramaiahgari et al. (2014) developed a HepG2 3D in vitro model using Matrigel as extracellular matrix. Under these conditions and 28 days of growth HepG2 cells form spheroids which leads to the recovery of lost hepatic functions like the storage of glycogen, transport of bile salts and the formation of bile canaliculi-resembling structures. Additionally, gene expression of xenobiotic-metabolizing enzymes (cytochrome P450 monooxygenases CYP1A2, 2E1, 2C9, UDP-glucuronosytransferases UGT1A1 and 2A3) as well as the enzyme activity of CYP was increased in comparison to the conventional HepG2 2D cultivation (Ramaiahgari et al. 2014). Based on these results, we aimed to study the effects of additional 3D cultivation methods to further increase the metabolic quality of HepG2 cells to use them as a robust, immortalized and permanent cell line to investigate human xenobiotic metabolism and liver toxicity in vitro. The HepG2 3D cultivation in collagen, Matrigel and the Alvetex scaffolds was investigated with regard to cell morphology, albumin production, CYP activity and gene expression of xenobiotic-metabolizing enzymes and some liver specific markers in comparison to 2D cultivation.
Materials and methods
Cell culture
The human hepatocellular carcinoma cell line HepG2 was obtained from the European Collection of Cell Cultures (ECACC, Porton Down, UK). The cells were grown as monolayer in Roswell Park Memorial Institute medium (RPMI, PAN-Biotech GmbH, Aidenbach, Germany) supplemented with 10 % fetal calf serum (PAN-Biotech GmbH, Aidenbach, Germany), 100 U/ml penicillin and 100 µg/ml streptomycin (PAA Laboratories GmbH, Pasching, Austria) at 37 °C in a humidified atmosphere containing 5 % CO2. Cells were passaged every 2–4 days at a confluence of about 80–90 % and plated at a density of 5 × 104 cells/cm2. HepaRG cells were obtained from Biopredic International (Saint-Gregoire, France) and cultured in William’s E medium (PAN-Biotech GmbH, Aidenbach, Germany) supplemented with 10 % (v/v) fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin, 5 µg/ml insulin (PAA Laboratories GmbH, Pasching, Austria) and 5 × 10−5 M hydrocortisone hemisuccinate (Sigma-Aldrich, Taufkirchen, Germany). For HepaRG differentiation, the cells were cultured for 4 weeks. The first 2 weeks cells were grown in medium. For the following 2 weeks 2 % dimethyl sulfoxide (DMSO) was added to the medium.
3D cell culture methods
For comparing different 3D cultivation methods the following techniques were investigated: collagen sandwich, Matrigel, and Alvetex scaffold. For each condition a cell density of 5 × 104 HepG2 cell per cm2 and a volume of 250 µl cell medium per cm2 culture vessel were used. Cell biochemistry, morphology and expression status were analyzed 3, 7, 14, and 21 days after seeding. For each method the culture vessels and their area are summarized in Table 1. For all cultivation conditions (2D and 3D) the medium was renewed every 2 days within the first week. Afterwards, medium was changed daily.
The collagen sandwich was prepared according to the manufacturer’s protocol with a final concentration of 1.7 mg/ml Corning Collagen I from rat tail (Corning, Schiphol-Rijk, The Netherlands). However, instead of 30 min the collagen was gelled for 1 h. For the preparation of the collagen gel 10 × RPMI instead of 10 × PBS was used. For the upper and lower collagen layer equal volumes of 55 µl per cm2 culture vessel surface were used.
For the Matrigel matrix the thick gel method was applied due to an enhanced migration of cells into the scaffold resulting in spheroid formation. The preparation of the Matrigel matrix was carried out according to the manufacturer’s instructions (Corning Matrigel Growth Factor Reduced Basement Membrane Matrix, Phenol Red-Free, Corning, Schiphol-Rijk, The Netherlands) with a final concentration of 5 mg/ml and 150 µl Matrigel per cm2 culture vessel surface.
The Alvetex scaffold represents a highly porous polystyrene scaffold consisting of a 200 µm thick membrane which forms a 3D structure into which cells can migrate and proliferate. The cells were seeded and cultured as recommended by the manufacturer (Reinnervate, Sedgefield, UK).
Cell morphology
As a suitable format for staining and confocal microscopy, the flexiPERM slide and disc cell culture chamber systems (Sarstedt, Nümbrecht, Germany) were used to grow HepG2 cells in 2D and collagen sandwich on cover slips. Subsequent to staining, the culture chambers were carefully removed and the cover slips placed on object slides with mounting medium (Vectashield HardSet, Vector Laboratories, Burlingame, USA). To prevent mechanical flattening of the HepG2 spheroids due to the absence of additional embedding, the µ-Slide Angiogenesis culture system (ibidi, Martinsried, Germany) was used for Matrigel. For Alvetex scaffold, HepG2 cells were grown on Alvetex 12 well inserts. Tissue sections (8 µm) of the polystyrene matrix were prepared after staining.
To visualize the nuclei and the cytoskeleton the following staining methods were used. After washing with phosphate-buffered saline (PBS) the HepG2 cells were fixed with 3.7 % formaldehyde for 10 min. They were rinsed three times with PBS containing 0.1 % (v/v) Tween 20 (PBS-T), incubated with 0.2 % Triton X-100 for 10 min and rinsed again with PBS-T for three times. For DNA visualization, the cells were incubated for 30 min in the dark with 1 µM SYTOX Orange Nucleic Acid Stain (Invitrogen, Eugene, USA) diluted in PBS. To stain the F-actin filaments, cells were then incubated for 30 min in the dark with 5 units/ml Alexa Fluor 488 Phalloidin (Lonza, Walkersville, USA) diluted in PBS with 1 % bovine serum albumin. Subsequent to each staining, the cells were washed with PBS-T to remove unbound dye. Finally, cellular morphology was imaged after 3, 7, 14, and 21 days using a TCS SP5 Confocal microscope and HCX PL APO 63x/1,4 objective (Leica Microsystems, Wetzlar, Germany).
Immunostaining and confocal microscopy
For detection of the differentiation status of the cultivated HepG2 cells in Matrigel or Alvetex a recently published staining protocol was applied (Hammad et al. 2014). Briefly, the cells were fixed in 4 % Roti-Histofix (Carl-Roth, Karlsruhe, Germany) for 3 days at 4 °C and then moved to another paraformaldehyde (PFA) buffer containing 15 % glucose. After washing in 1xPBS, cells were permeabilized in 1 % Triton X-100 (Sigma-Aldrich, Taufkirchen, Germany) for 1 h at room temperature. The cells were washed three times in 1 × PBS. Then the cells were incubated in PBS containing 3 % bovine serum albumin (BSA), 1 % Tween 80 for 60 min at room temperature to block unspecific bindings. Subsequently, the cells were incubated overnight with goat anti-human DPPIV (1:100; R&D systems, AF1180, Minneapolis, USA) and rabbit anti-human albumin (1:100, Abcam, Ab2406, Cambridge, UK) or rabbit anti-ZO1 (1:100, Invitrogen, 617300, Carlsbad, USA) in PBS containing 0.3 % BSA and 1 % Tween 80 at 4 °C. The next day, cells were washed and labelled with Cy3 conjugated donkey anti-goat (1:100, Dianova, 705-166-147, Hamburg, Germany), and Alexa Fluor 647 conjugated donkey anti-rabbit IgG (1:200, Dianova, 711-606-152, Hamburg, Germany) and Alexa Fluor 488 Phalloidin (1:250; Invitrogen, Carlsbad, USA) for 2 h on a shaker at room temperature. After three washing steps, nuclei were counterstained with 0.5 mg/l 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, USA) for 90 min and mounted on microscope glass slides. The stained cells were imaged using a FluoView FV100 (Olympus, Hamburg, Germany) confocal microscope with an UPlanSApo 60x Objective.
Detection of cell viability for normalization of albumin secretion and CYP activities to cell number
Viability measurements were used for normalization of data to cell number under the different culture conditions. Cell viability was measured by using the CellTiter-Blue Cell Viability Assay (Promega, Madison, USA), a test system detecting mitochondrial reductase activity which is proportional to the cell number. CellTiter-Blue reagent was diluted 1:10 with culture medium and pre-warmed to 37 °C. After washing the cells with PBS the CTB/medium solution was added in equal volume of culture medium into the cell culture vessel for 15 min. 100 µl of the supernatant were transferred in technical replicates into a 96 well plate and the fluorescence was measured at 590 nm (excitation at 540 nm). As background the pure CTB/medium solution was used and subtracted from the fluorescence signals.
Albumin secretion
To compare the different 2D and 3D cell culture methods the albumin secretion was chosen as a liver-specific marker. HepG2 cells were cultured as summarized in Table 1. For the measurement of secreted albumin the medium of each cell culture sample was renewed 24 h prior to sample taking. Medium was collected after 3, 7, 14, and 21 days and stored at −20 °C until albumin detection using the Human Albumin ELISA Quantitation kit (BethylLaboratories, Montgomery, USA) following the manufacturer’s protocol. Albumin secretion of each sample was determined in three adequate dilutions measured in two technical replicates. Data were normalized to the respective CTB values and day 3 of the corresponding culture condition as described in “Detection of cell viability for normalization of albumin secretion and CYP activities to cell number” section.
CYP activity
To elucidate CYP activities under the different culture conditions HepG2 cells were cultured as summarized in Table 1. As control for CYP activity differentiated HepaRG cells were used. After 4 h of cell treatment with CYP substrates 50 µl supernatant of HepaRG and HepG2 cells were taken in replicates at defined times as indicated in Fig. 5. The oxidation of 50 µM phenacetin (CYP1A2), 25 µM bupropion (CYP2B6), 5 µM amodiaquine (CYP2C8), 100 µM tolbutamide (CYP2C9), 100 µM S-mephenytoin (CYP2C19), 5 µM propafenone (CYP2D6) was measured as described previously (Feidt et al. 2010). As a substrate for CYP3A4, atorvastatin was exchanged with 10 µM midazolam (Klein and Zanger 2013). The detected metabolites are summarized in Table 2. Data were normalized to cell viability values as described in “Detection of cell viability for normalization of albumin secretion and CYP activities to cell number” section.
Isolation of RNA and gene expression analysis
For analysis of gene expression HepG2 cells were cultured in 2D and 3D (see Table 1) for 3, 7, 14 and 21 days. Prior to RNA isolation HepG2 cells were washed with PBS. For RNA isolation in general the RNeasy Mini Kit (Qiagen, Hilden, Germany) was employed according to the manufacturer’s protocol using a blunt 20-gauge needle for homogenization and an integrated on spin column DNA digestion (RNase-free DNase Set, Qiagen, Hilden, Germany). However, due to the different matrix characteristics cell lysis differed between the culture methods: Cells grown in conventional 2D or in Alvetex scaffolds were lysed directly using 600 µl RLT RNeasy lysis buffer. The cells grown in Alvetex scaffold were shaken 10 min at 150×g before homogenization. The RNA of HepG2 cells embedded into collagen sandwich was isolated using peqGold TRIfast (peqlab, Erlangen, Germany) according to the manufacturer’s instruction until phase separation into the lower DNA-containing phenol chloroform phase, the interphase, and the aqueous RNA containing phase. After taking the RNA phase, an equal volume of 70 % ethanol was added and cleaned-up with RNeasy Mini Kit following the protocol. For HepG2 cells grown in Matrigel cells were isolated from the matrix using the Cell Recovery solution (Corning, Schiphol-Rijk, The Netherlands) following Corning´s instructions prior to the RNeasy Mini Kit clean-up. Subsequent to the RNA isolation RNA concentration was determined by a spectrophotometer (Nano Drop 1000, Nanodrop Technologies, Wilmington, USA).
200 ng of RNA were reverse transcribed into single stranded cDNA by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, USA). According to the manufacturer’s protocol, thermal cycling conditions were chosen for two-step RT-PCR. Real-time quantitative PCR (qPCR) was performed on a LightCycler 96 system (Roche, Mannheim, Germany) using Maxima SYBR Green/ROX qPCR Master Mix (Fermentas, St. Leon Rot, Germany) with 300 nM of each primer (for primer sequences see Table 3) and 1 µl 1:5 diluted cDNA in an end volume of 10 µl. Thermal cycling conditions were selected as follows: initial denaturation at 95 °C for 15 min, 40 cycles of denaturation at 95 °C for 15 s, a combined annealing and elongation at 60 °C for 1 min, and a final elongation at 60 °C for 15 min. At the end of the run, a dissociation curve analysis was performed. For the relative quantification of mRNA content the 2−ΔΔCt method was used (Livak and Schmittgen, 2001), Ct values were normalized to the housekeeping genes β-actin (ACTB), ornithine decarboxylase antizyme 1 (OAZ1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and referred to expression values of HepG2 cells grown under 2D conditions.
Statistical analysis
SigmaPlot was used for the statistical analyses. Data are presented as mean ± standard deviation (SD). Differences between means were determined by one-way analysis of variance (ANOVA) followed by a Bonferroni post hoc test and considered significant at p < 0.05.
Results
HepG2 cell morphology in dependence of the extracellular matrix and cultivation time
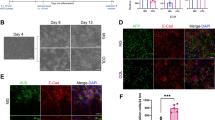
One of the limitations of the hepatocarcinoma cell line HepG2 is its low expression of phase I xenobiotic-metabolizing enzymes. However, a recently published study claimed that HepG2 cells grown under 3D conditions in Matrigel strongly improve liver-like functions such as expression of xenobiotic-metabolizing enzymes or albumin secretion (Ramaiahgari et al. 2014). For further elucidation and optimization of these characteristics in 3D culture different extracellular matrices were investigated in this study. In addition to the Matrigel matrix, the collagen sandwich and the Alvetex scaffold were in the focus of the morphological and biochemical characterization of HepG2 cells dependent on cultivation time. For morphological characterization of the cells, nuclei and the actin filaments were stained using SYTOX Orange Nucleic Acid Stain (red) and AlexaFluor 488 Phalloidin (green), respectively.
The conventional 2D cultivation resulted in a closed monolayer with a stacked growth at day 21 of cultivation (Fig. 1a, b). The morphology and expansion of cells grown in the collagen sandwich showed the highest similarity to 2D cultivation among the investigated 3D methods. The proliferation of cells grown on collagen followed a pattern where expansion initially occurred along the horizontal plane, and only after confluence was almost reached increased migration occurred along the vertical axis (Fig. 1b, day 7 and day 14). In contrast to 2D cultivation, even while planar growth seemed to prevail, the cells made use of the available space offered by the gel matrix to expand in all dimensions.
a Confocal microscopy and b Z-stack images of HepG2 cells cultured under conventional 2D and different 3D conditions (collagen sandwich, Matrigel and Alvetex scaffold). HepG2 cells were seeded at a density of 5 × 104 cells per cm2 in cell culture vessels as summarized in Table 1. Nuclei and actin cytoskeleton of HepG2 cells were stained as described in the “Materials and methods” section with Sytox Orange Nucleic Acid Stain (red) and Alexa Fluor 488 Phalloidin (green), respectively, after 3, 7, 14, and 21 days. Images were recorded with the HCX PL APO 63x/1.4 objective with an enlargement of 630x (scale as indicated in the Figure). d days, Col collagen sandwich, Mg Matrigel, Alv Alvetex scaffold
In contrast, HepG2 cells grown in Matrigel time-dependently formed spheroids (Fig. 1a, b). Especially the optical cross sections obtained by confocal microscopy (Fig. 1b) show the process of piling up finally leading to spheroid formation in the Matrigel matrix within 7 days. These structures were maintained until the end of the study period at day 21. The migration of HepG2 cells into the Alvetex scaffold increased time-dependently.
Cell proliferation was studied by analyzing the proliferation marker Ki-67. Ki-67 was strongly down-regulated under all cell culture conditions after 21 days of cultivation (Fig. 2).
Expression of the proliferation marker Ki-67 in HepG2 cells cultured under 2D and different 3D conditions (collagen sandwich, Matrigel, and Alvetex scaffold). HepG2 cells were seeded at a density of 5 × 104 cells per cm2 in cell culture vessels as summarized in Table 1. RNA was isolated after 3, 7, 14, and 21 days after seeding. After reverse transcription into cDNA, gene expression was analyzed by qPCR. Data were normalized to the housekeeping genes β-actin (ACTB), OAZ1, GAPDH and referred to day 3 in each cultivation method. Col, collagen sandwich; Mg, Matrigel; Alv, Alvetex scaffold. Shown are the mean ± SD of two independent experiments (three replicates each) (*p < 0.05, **p < 0.01, One Way ANOVA followed by a Bonferroni t test against day 3)
Bile canaliculi formation
To examine the effect of the culture systems on establishment of bile canaliculi DPPIV was used as a marker. These analyses focused on HepG2 cells in Matrigel, because this culture system allows the formation of relatively deep spheroid-like structures (Fig. 1b), and on the Alvetex culture system that to our knowledge has not yet been investigated for bile canaliculi formation. Co-staining of DPPIV together with the actin cytoskeleton, albumin and nuclear counterstaining illustrates a more intensive albumin staining and a more distinct canalicular network of HepG2 cells in Matrigel compared to Alvetex scaffolds (Fig. 3). Nevertheless, it should be considered that even in Matrigel HepG2 cells did not reach a state comparable to primary human hepatocytes that were included as a positive control (see supplemental data for further information Figure S1). To compare bile canaliculi formed by HepG2 cells in Matrigel and primary human hepatocytes three dimensional confocal reconstructions were performed. Human hepatocytes showed a more distinct canalicular network, while in HepG2 cells larger parts of the cellular surface were covered by DPPIV (Figure S1).
Expression of differentiation markers in HepG2 cells cultivated in Matrigel and Alvetex scaffold. Immunofluorescence staining of 3D HepG2 cells with DPPIV, and albumin counterstained with Alexa Fluor 488 Phalloidin for F-actin and DAPI for nuclei. No necrosis of cell death was reported and the cells maintained to some extent the polarity status particularly in Mg. Alv Alvetex, DPPIV Dipeptidyl-peptidase IV, Mg Matrigel. Scale bars as indicated in the figure
Albumin secretion
Albumin secretion was used as a liver-specific marker to compare the different cell culture models. Albumin protein content was determined in cell culture supernatant under conventional 2D and the different 3D conditions in dependence of the cultivation time.
To analyze albumin secretion time-dependently within one cultivation method data were normalized to day 3 of the respective cultivation method (Table 4). Overall, the time-dependent increase of albumin secretion by HepG2 cells during the 21 days incubation period was highest in conventional 2D culture. In all 3D cell culture methods HepG2 cells produced the highest amount of albumin after 7 days and afterwards the amount decreased until the end of the cultivation period.
Further, the increase of absolute albumin production within a cultivation method is shown in Fig. 4. High differences were observed after day 3 of cultivation. Cells grown in Matrigel and collagen secreted approximately fivefold (95 pg/ml/RFU) and threefold (54 pg/ml/RFU) higher amounts of albumin compared to HepG2 cells cultured in 2D (20 pg/ml/RFU), respectively. However, after 3 weeks of cultivation in collagen sandwich the secretion of albumin was lower than of HepG2 cells cultivated in 2D. Also the relative albumin content in the medium of HepG2 cells cultivated in Matrigel decreased within 3 weeks of culture detecting only twofold (157 pg/ml/RFU) higher albumin values compared to 2D (83 pg/ml/RFU) at the same time. The amounts of albumin secreted by HepG2 cells grown in Alvetex scaffold were comparable to the 2D culture but decreased under the level of 2D culture after 3 weeks of cultivation.
Albumin secretion in dependence of the cultivation time of HepG2 cells under 2D and different 3D (collagen sandwich, Matrigel and Alvetex scaffold) cultivation conditions. HepG2 were seeded at a density of 5 × 104 cells per cm2 in cell culture vessels as summarized in Table 1. 24 h prior to sample taking cell medium was refreshed. Values were normalized to the cell number, as indirectly determined by cell viability measurement. Values are given as mean ± SD (n = 6) (p < 0.05, OneWay ANOVA followed by a Bonferroni t test *against d3 Col collagen sandwich, Mg Matrigel, Alv Alvetex scaffold
CYP activities
For elucidating and comparing the metabolic capacity between the cultivation methods CYP activities were determined using specific substrates and LC–MS/MS detection of the formation of specific metabolites. The oxidation of phenacetin, bupropion, amodiaquine, tolbutamide, S-mephenytoin, propafenone, and midazolam was determined for CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A4, respectively.
As a positive control, the metabolic conversion of the CYP substrates was investigated using HepaRG cells which represent a cell model with higher metabolizing activities than HepG2 cells. Figure 5 shows the metabolite formation for the different HepG2 2D/3D cell culture techniques and HepaRG cells. The conversion of the CYP1A2 substrate phenacetin after 4 h incubation was highest for the conventional 2D culture after 21 days. Substantially lower activities were observed for HepG2 cells cultured for 3 days under 2D and 3D conditions as well as for HepaRG cells. However, HepaRG cells exhibited a high oxidation rate of bupropion as substrate of CYP2B6 compared to the HepG2 cells. The oxidation rates were comparable between the cultivation of HepG2 cells in 2D for 21 days and the collagen aas well as Matrigel matrix. After 3 days of cultivation of the cells in 2D CYP2B6 activity was below the detection limit.
Activity of CYP1A1/1A2, 2B6, 2C8, 2C9, 2C19, and 3A4 dependent of the cultivation time of HepG2 cells under 2D and different 3D (collagen sandwich, Matrigel, and Alvetex scaffold) cultivation conditions. HepG2 were seeded at a density of 5 × 104 cells per cm2 in cell culture vessels as summarized in Table 1 and medium samples were taken after 4 h. As positive control the CYP activity of HepaRG cells was measured after 4 h. CYP activity was determined as described in “Material and methods”. Values were normalized to the CTB cell viability values, given in relative fluorescence units (RFU), representing the cell number. Values are given in fmol/ml/RFU as mean ± SD of 3 replicates (*p < 0.05; **p < 0.01, OneWay ANOVA followed by a Bonferroni t test against d21 of conventional 2D). d days, Col collagen sandwich, Mg Matrigel, Alv Alvetex scaffold
HepG2 cells grown in the Alvetex scaffold exhibited the lowest CYP2B6 activity amongst all cultivation methods tested. The same effects as for CYP2B6 were also observed for CYP2C8 and 2C9 activity determined by the conversation of the substrates amodiaquine and tolbutamide, respectively. CYP2C8 activity increased in HepG2 cells cultured in conventional 2D time-dependently from day 3 to day 21. CYP2C8 enzyme activities were comparable between 2D and 3D HepG2 culture conditions. The highest substrate conversion was observed in HepaRG cells. CYP2C9 activity was enhanced in the control cell line HepaRG as well as in the conventional HepG2 2D cultivation compared to all 3D cultures which were substantially decreased compared to 2D cultivation at day 21. Oxidation of the CYP2C19 substrate S-mephenytoin was only detected in HepaRG cells and in HepG2 cells cultured in Matrigel as well as Alvetex matrix. However, after 24 h of substrate incubation (data not shown), low amounts of 4-hydroxymephenytoin were found in all samples except in the 2D culture at day 3 with a significantly increased level in the Matrigel sample.
The highest oxidation rate of midazolam metabolized by CYP3A4 was detected in HepaRG cells. After 3 days of growth under conventional 2D conditions a very low conversion rate was detected compared to 21 days of cell culture in 2D. Highest values of CYP3A4 activity were received by cultivation in Matrigel and collagen matrix, lower levels occured in the 21 day 2D culture and the 3D Alvetex matrix.
Comparison of gene expression
For characterization of differences in gene expression patterns between the conventional 2D culture and HepG2 cells cultivated under 3D conditions mRNA levels of different phase I (CYP1A2, 2B6, 2C9, 2D6, 2E1, 3A4) and phase II (UGT1A1, 1A6) xenobiotic-metabolizing enzymes, nuclear receptors (AhR, CAR, PXR) and the liver marker albumin were determined. Figure 6 shows a heat map representing the mRNA expression of these target genes depending on different points of reference. After normalizing raw data to housekeeping gene expressions, values were calculated relative to expression levels of HepG2 cells cultivated under conventional 2D conditions for 3 days (Fig. 6a). A time-dependent increase in gene expression was observed for CYP1A2, CYP2E1, CYP3A4, UGT1A1, UGT1A6 for HepG2 cells grown in Matrigel and Alvetex scaffold. CYP2C9 and albumin expression were also enhanced in HepG2 cells cultured in Matrigel matrix after 21 days of cultivation. However, a similar time-dependent increase of gene expression of these targets was also observed in HepG2 cells cultured for 21 days in 2D. Therefore, raw data of each 3D condition at a defined time were normalized to the corresponding time point of HepG2 cells cultured in 2D (Fig. 6b). After that change of reference point, the formerly observed differences in gene expression between 2D and 3D cultivation largely disappeared. The cultivation of HepG2 cells in collagen sandwich and Alvetex scaffold resulted in similar or even decreased expression values of analyzed target genes after 21 days compared to HepG2 cells grown in 2D (Fig. 6b). This was also observed for the expression of albumin in all 3D cultures (collagen sandwich, Matrigel, Alvetex scaffold). However, gene expression of CYP1A2, CYP3A4, and UGT in cells grown for 7 days in Matrigel was enhanced compared to the corresponding 2D control on day 7. However, this increase disappeared after 21 days of cultivation resulting in a similar expression pattern of all target genes as observed under 2D condition. Thus, increased expression values were solely observed after 7 days of HepG2 cell culture in Matrigel compared to day 7 in 2D.
Differential gene expression of phase I as well as phase II xenobiotic-metabolizing enzymes, nuclear receptors and liver markers in HepG2 cells cultured under 2D and different 3D conditions (collagen sandwich, Matrigel, and Alvetex scaffold). HepG2 cells were seeded at a density of 5 × 104 cells per cm2 in cell culture vessels as summarized in Table 1. RNA was isolated 3, 7, 14, and 21 days after seeding. After reverse transcription into cDNA gene expression was analyzed by qPCR as described in “Material and methods” section. Data were normalized to the housekeeping genes β-actin (ACTB), OAZ1, GAPDH and referred to a day 3 of 2D culture or b to the corresponding day of 2D culture. d days, Col collagen sandwich, Mg Matrigel, Alv Alvetex scaffold. Shown are the means of 5 to 6 replicates (*p < 0.05; **p < 0.01, OneWay ANOVA followed by a Bonferroni t test against a day 3 of 2D culture or b against corresponding day of 2D HepG2 cultivation)
Discussion
In vitro human primary hepatocytes represent the gold standard for toxicological research on liver metabolism and mode of action analysis. Due to their high cost, low availability and inter-donor variability researchers are looking for alternative in vitro models representing an additional appropriate model for human liver. In the last decades several human hepatoma cell lines (HepG2, Huh-7, HepaRG) were established and commonly used to assess hepatotoxicity of xenobiotics. These cell lines combine the advantages of an unlimited life span, high availability, easy handling and high reproducibility of experimental results due to a stable phenotype (Godoy et al. 2013). However, the expression of liver markers and xenobiotic-metabolizing enzymes e.g. in HepG2 and Huh-7 cells is quite low compared to human primary hepatocytes (Donato et al. 2008). By adding dimethylsulfoxide (DMSO) to HepaRG cells a differentiation process is induced resulting in higher expression of phase I and phase II xenobiotic-metabolizing enzymes. However, these levels are still lower compared to human primary hepatocytes with the exception of CYP3A4, which reaches even higher expression and activity values (Kanebratt and Andersson 2008). A lot of research currently focuses on the improvement of these hepatoma cell lines. A recently published study by Ramaiahgari et al. (2014) describes the development of a 3D Matrigel culture of HepG2 cells. Under these cultivation conditions HepG2 cells stop proliferating and differentiate to form multiple polarized spheroids. They were described to regain normal hepatocyte functions like storage of glycogen, enhanced production of albumin, enhanced transport of bile salts and the development of structures resembling bile canaliculi. Additionally, increased gene expression and partially enhanced activities of CYP enzymes like 3A4, 2E1, 2C9, 2C19, and 2D6 were detected after 28 days of HepG2 cultivation in Matrigel matrix compared to traditional HepG2 2D culture for 3 days. Analysis of some phase II xenobiotic-metabolizing enzymes (UGT, SULT) also showed increased transcript levels under 3D cultivation conditions. All in all the results of this study showed increased hepatic functions and metabolic capacities regained by HepG2 cells after cultivation in 3D Matrigel hydrogel. Based on this study we aimed to investigate if a further increase of hepatic functions, metabolic capacity and gene expression can be reached by alternative 3D cultivation methods. Therefore, HepG2 cells were cultured on the one hand in Matrigel for 21 days to compare our results directly to the published data. Please note that, as no further substantial increase in metabolic capacity was seen after cultivation day 21 in the study of Ramaiahgari et al. (2014), we decided to culture the cells only for 21 days. Beside cultivation in Matrigel, cells were additionally cultivated in collagen sandwich as well as Alvetex scaffold for 21 days. To compare the effects of the 3D method HepG2 cells in conventional 2D conditions were also cultivated for 21 days.
Comparing our data of the 3D cell culture of HepG2 cells with conventional HepG2 cell culture (treatment before confluence, HepG2 in 2D at day 3) we detected a strong increase in albumin secretion, in gene expression of xenobiotic-metabolizing enzymes as well as CYP activities as already described in the study of Ramaiahgari et al. (2014). The formation of spheroids was also observed in Matrigel as recently described in the cited study. In our study, two different cell numbers of HepG2 cells were used for the Matrigel hydrogel: a lower (15,000 cells/cm2) number, based on the experimental design of Ramaiahgari et al. (2014), and a higher number with 50,000 cells/cm2 as routinely used in our laboratory. Independent of the cell number we observed the same effects on gene expression, albumin secretion as well as CYP activity induction for both cell numbers cultivated in 3D Matrigel (data not shown). However, for collagen and Alvetex scaffold, the lower cell number was insufficient to achieve evaluable results especially with regard to the morphological development of the cells. Therefore, we compared the 3D cultivation techniques using only the standardized higher cell seeding numbers.
Comparing the biochemical and morphological characteristics of HepG2 cells grown in both hydrogels Matrigel and collagen as well as in Alvetex scaffold we observed that the proliferation rate decreased under all cultivation conditions as shown by the down-regulation of the proliferation marker Ki-67. This may be associated with a time-dependent differentiation process as shown for HepG2 cells grown in Matrigel and Alvetex scaffold (Figs. 2, 3). However, when comparing all three 3D cultivation methods in our study, cultivation of HepG2 cells in the Matrigel matrix resulted in the highest expression levels and CYP activities. By embedding the cells into the collagen sandwich especially the expression of the analyzed xenobiotic-metabolizing enzymes were only marginally induced. This might be associated with the components of the hydrogels. The here applied collagen I isolated of rat tail consists mostly of only one protein component whereas in Matrigel numerous proteins were detected by proteomics analysis (Hughes et al. 2010). The extracellular matrix mix Matrigel is isolated from Engelbreth–Holm–Swarm tumors of mice and consists of a mixture of laminin, collagen IV, fibronectin, heparan sulfate, and a lot of other components (Hughes et al. 2010). Harma et al. (2010) showed with normal prostate and prostate cancer cell lines no formation of differentiated spheroids in purely collagen-rich extracellular matrix, whereas in contrast the same cell lines formed differentiated spheroids in Matrigel (Harma et al. 2010). These observations support the hypothesis that the gel components are mainly responsible for the development of cell morphology, differentiation and spheroid formation. Laminin, fibronectin, and heparan sulfate are described to exhibit a substantial influence on spheroid formation (Ivascu and Kubbies 2006; McClelland et al. 2008). Alvetex scaffold represents an artificial growth skeleton consisting of polystyrene (Knight et al. 2011). During cultivation we observed a continuous growth of the cells into the scaffold and a partial differentiation. However, the gene expression and CYP activity data revealed that levels of HepG2 cells cultivated in Alvetex scaffold are all in all lower than the expression levels and activities of HepG2 cells grown in the Matrigel hydrogel.
Comparing the results of HepG2 cells cultured in 3D at day 21 with HepG2 cells cultured in conventional 2D monolayer at day 21 revealed surprising results. Most effects of the formerly observed enhanced metabolic capacity and increased gene expression when comparing 3D at day 21 with 2D at day 3 were relativized. HepG2 cells cultured in 2D over 21 days showed the strongest increase in albumin secretion, a strong enhancement in gene expression of xenobiotic-metabolizing enzymes, as well as an increase in CYP activities partially comparable to or even higher (CYP2C9, CYP1A2) than those observed in Matrigel (with the exception of CYP2C19). This increase of the metabolic properties of HepG2 cells under conventional 2D conditions may be associated on the one hand with the induction of a partial differentiation process over the long-time cultivation for 21 days based on cell aging. On the other hand, piling up and growth into the third dimension may contribute to the increased metabolic capacity after a cultivation period of 21 days. Furthermore, the enhanced metabolic properties of HepG2 cells in 2D might be associated with a continuously increasing stress potential due to increasing cell numbers and limited space. Enhancement of metabolic capacities due to chemically induced stress response is also known for HepaRG cells. Here, the addition of DMSO to the culture medium forces hepatocyte-committed cells to activate their metabolic detoxifying systems to resist the toxic effects of DMSO (Guguen-Guillouzo and Guillouzo 2010). Taken together, the effects observed in HepG2 cells in 3D culture primarily seem to be more time-induced than 3D-induced.
Long-term cell cultivation of HepG2 cells leads to an enhancement of expression of liver-specific markers as shown for albumin or the formation of bile canaliculi. Independent of 2D or 3D cultivation, the elevated gene expression levels and metabolic capacities at day 21 became apparent in the increased conversion of CYP substrates which was clearly enhanced in all 2D and 3D cultures at day 21 compared to the conventional HepG2 cell culture at day 3. However, gene expression and enzyme activity were even more pronounced in the hepatocarcinoma cell line HepaRG. HepaRG differentiate into hepatocyte-resembling cells by adding 1.7 % DMSO to the growth medium for 14 days. As already described by Aninat et al. (2006) mRNAs of some nuclear receptors (AhR, PXR, CAR, peroxisome proliferator-activated receptor α), CYP (CYP1A2, 2C9, 2D6, 2E1, 3A4), and phase II enzymes (UGT1A1, GSTA1, GSTA4, GSTM1) were analyzed in differentiated HepaRG cells with partially comparable expression as observed in human primary hepatocytes. The mRNA pattern of confluent HepG2 cells was also analyzed showing lower expression of all CYP enzymes than HepaRG cells (Aninat et al. 2006). In our study we also detected highest CYP enzyme activities in HepaRG cells for CYP2B6, 2C8, 2C9, 2C19, and 3A4 compared to HepG2 cells in 2D and 3D at day 21. Only for CYP1A1/1A2 long-term HepG2 cultures reached a threefold higher activity compared to HepaRG or HepG2 culture for 3 days. However, it should be noted that, in contrast to HepaRG, HepG2 cells were grown in the absence of differentiation-inducing DMSO or the PXR ligand hydrocortisone hemisuccinate, which both positively influence CYP gene expression and activities in the HepaRG system and might lead to an under-estimation of HepG2 performance if the compounds would elicit comparable effect in HepG2 cells. However, it has been shown that HepG2 cells do not gain substantial benefit from DMSO treatment with regard to various metabolism- and differentiation parameters (Mowbray 2011).
In conclusion, our study shows that the effect of increased metabolic capacity of HepG2 cells in 3D culture, as reported by Ramaiahgari et al. (2014), is most likely not primarily an effect of the 3D cultivation but rather a consequence of the long-term cultivation of HepG2 cells. This may be associated with a partial differentiation of HepG2 cells due to the aging of cells under 2D cultivation, a secondary 3D differentiation due to piling up when confluency is reached, or with stress-related up-regulation of metabolic enzymes. It should be considered that the relatively high costs of 3D culture material do not necessarily reflect the benefit of increased hepatic functions of HepG2 cells in 3D compared to 2D cultivation.
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- ACTB:
-
β-Actin
- AhR:
-
Aryl hydrocarbon receptor
- Alv:
-
Alvetex
- CAR:
-
Constitutive androstane receptor
- Col:
-
Collagen
- CTB:
-
CellTiter-Blue
- CYP:
-
Cytochrome-P450 monooxygenase(s)
- DPPIV:
-
Dipeptidyl-peptidase IV
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- Mg:
-
Matrigel
- OAZ1:
-
Ornithine decarboxylase antizyme 1
- PXR:
-
Pregnane-X receptor
- qPCR:
-
Real-time quantitative PCR
- RFU:
-
Relative fluorescence units
- SD:
-
Standard deviation
- UGT:
-
UDP-glucuronosyltransferase(s)
- ZO-1:
-
Zona occludens-1
References
Aninat C, Piton A, Glaise D et al (2006) Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos 34(1):75–83. doi:10.1124/dmd.105.006759
Bissell DM, Arenson DM, Maher JJ, Roll FJ (1987) Support of cultured hepatocytes by a laminin-rich gel. Evidence for a functionally significant subendothelial matrix in normal rat liver. J Clin Invest 79(3):801–812. doi:10.1172/jci112887
Bissell MJ, Rizki A, Mian IS (2003) Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol 15(6):753–762
Breslin S, O’Driscoll L (2013) Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today 18(5–6):240–249. doi:10.1016/j.drudis.2012.10.003
Clayton DF, Darnell JE Jr (1983) Changes in liver-specific compared to common gene transcription during primary culture of mouse hepatocytes. Mol Cell Biol 3(9):1552–1561
Clayton DF, Weiss M, Darnell JE Jr (1985) Liver-specific RNA metabolism in hepatoma cells: variations in transcription rates and mRNA levels. Mol Cell Biol 5(10):2633–2641
Daus AW, Goldhammer M, Layer PG, Thielemann C (2011) Electromagnetic exposure of scaffold-free three-dimensional cell culture systems. Bioelectromagnetics 32(5):351–359. doi:10.1002/bem.20649
Debnath J, Brugge JS (2005) Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer 5(9):675–688. doi:10.1038/nrc1695
Donato MT, Lahoz A, Castell JV, Gomez-Lechon MJ (2008) Cell lines: a tool for in vitro drug metabolism studies. Curr Drug Metab 9(1):1–11
Feidt DM, Klein K, Hofmann U et al (2010) Profiling induction of cytochrome p450 enzyme activity by statins using a new liquid chromatography-tandem mass spectrometry cocktail assay in human hepatocytes. Drug Metab Dispos 38(9):1589–1597. doi:10.1124/dmd.110.033886
Godoy P, Hengstler JG, Ilkavets I et al (2009) Extracellular matrix modulates sensitivity of hepatocytes to fibroblastoid dedifferentiation and transforming growth factor beta-induced apoptosis. Hepatology 49(6):2031–2043. doi:10.1002/hep.22880
Godoy P, Hewitt N, Albrecht U et al (2013) Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch Toxicol 87(8):1315–1530. doi:10.1007/s00204-013-1078-5
Gomez-Lechon MJ, Donato MT, Castell JV, Jover R (2003) Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr Drug Metab 4(4):292–312
Guguen-Guillouzo C, Guillouzo A (2010) General review on in vitro hepatocyte models and their applications. Methods Mol Biol 640:1–40. doi:10.1007/978-1-60761-688-7_1
Hammad S, Hoehme S, Friebel A et al (2014) Protocols for staining of bile canalicular and sinusoidal networks of human, mouse and pig livers, three-dimensional reconstruction and quantification of tissue microarchitecture by image processing and analysis. Arch Toxicol 88(5):1161–1183. doi:10.1007/s00204-014-1243-5
Harma V, Virtanen J, Makela R et al (2010) A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS ONE 5(5):e10431. doi:10.1371/journal.pone.0010431
Hughes CS, Postovit LM, Lajoie GA (2010) Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 10(9):1886–1890. doi:10.1002/pmic.200900758
Ivascu A, Kubbies M (2006) Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J Biomol Screen 11(8):922–932. doi:10.1177/1087057106292763
Kanebratt KP, Andersson TB (2008) Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab Dispos 36(7):1444–1452. doi:10.1124/dmd.107.020016
Klein K, Zanger UM (2013) Pharmacogenomics of cytochrome P450 3A4: recent progress toward the “missing heritability” problem. Front Genet 4:12. doi:10.3389/fgene.2013.00012
Knight E, Murray B, Carnachan R, Przyborski S (2011) Alvetex(R): polystyrene scaffold technology for routine three dimensional cell culture. Methods Mol Biol 695:323–340. doi:10.1007/978-1-60761-984-0_20
LeCluyse EL (2001) Human hepatocyte culture systems for the in vitro evaluation of cytochrome P450 expression and regulation. Eur J Pharm Sci 13(4):343–368
Lin RZ, Chou LF, Chien CC, Chang HY (2006) Dynamic analysis of hepatoma spheroid formation: roles of E-cadherin and beta1-integrin. Cell Tissue Res 324(3):411–422. doi:10.1007/s00441-005-0148-2
Loessner D, Stok KS, Lutolf MP, Hutmacher DW, Clements JA, Rizzi SC (2010) Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 31(32):8494–8506. doi:10.1016/j.biomaterials.2010.07.064
McClelland R, Wauthier E, Uronis J, Reid L (2008) Gradients in the liver’s extracellular matrix chemistry from periportal to pericentral zones: influence on human hepatic progenitors. Tissue Eng Part A 14(1):59–70. doi:10.1089/ten.a.2007.0058
Mowbray CA (2011) In vitro systems to predict hepatotoxicity: models based on hepatocarcinoma cell lines. Newcastle University, Faculty of Medical Sciences, Institute of Cell and Molecular Biosciences, https://theses.ncl.ac.uk/dspace/bitstream/10443/1337/1/Mowbray%2011.pdf
Ramaiahgari SC, den Braver MW, Herpers B et al (2014) A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch Toxicol 88(5):1083–1095. doi:10.1007/s00204-014-1215-9
Ravi M, Paramesh V, Kaviya SR, Anuradha E, Solomon FD (2015) 3D cell culture systems: advantages and applications. J Cell Physiol 230(1):16–26. doi:10.1002/jcp.24683
Weaver VM, Petersen OW, Wang F et al (1997) Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol 137(1):231–245
Wilkening S, Bader A (2003) Influence of culture time on the expression of drug-metabolizing enzymes in primary human hepatocytes and hepatoma cell line HepG2. J Biochem Mol Toxicol 17(4):207–213. doi:10.1002/jbt.10085
Acknowledgments
This work was financed by the Federal Institute for Risk Assessment (grant number 1322-591) and by the Robert Bosch Foundation, Stuttgart. We thank Beatrice Rosskopp, Markus König (Stuttgart) and Igor Liebermann (Stuttgart) for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luckert, C., Schulz, C., Lehmann, N. et al. Comparative analysis of 3D culture methods on human HepG2 cells. Arch Toxicol 91, 393–406 (2017). https://doi.org/10.1007/s00204-016-1677-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-016-1677-z