Abstract
Recently, diesel engine exhaust (DEE) was reclassified as a known carcinogen to humans. DNA methylation alterations in DNA damage response (DDR)-related genes have the potential to affect DEE exposure-related cancer risk. However, the evidence regarding the association between DEE exposure and methylation alterations in DDR-related genes is limited. In 117 DEE-exposed workers and 112 non-DEE-exposed workers, we measured urinary concentrations of six mono-hydroxylated polycyclic aromatic hydrocarbons (OH-PAHs). We also determined the methylation levels of three DDR-related genes (p16, RASSF1A, and MGMT) and LINE-1 by bisulfite-pyrosequencing assay. We found that DEE-exposed workers exhibited significantly lower mean promoter methylation levels of p16, RASSF1A, and MGMT than non-DEE-exposed workers (all p < 0.001). In all study subjects and non-smoking workers, increasing quartiles of urinary summed OH-PAHs was associated with hypomethylation of p16, RASSF1A, and MGMT (all p < 0.05). In non-smoking workers, methylation in p16, RASSF1A, and MGMT decreased by 0.36 % [95 % confidential interval (CI): −0.60, −0.11 %], 0.46 % (95 % CI: −0.79, −0.14 %), and 0.55 % (95 % CI: −0.95, −0.15 %), respectively, in association with highest versus lowest quartile of urinary summed OH-PAHs. In addition, p16, RASSF1A, MGMT, and LINE-1 methylation levels showed negative correlations with cytokinesis-block micronucleus cytome index which was previously measured in the same workers (all p < 0.05). In conclusion, our results clearly indicated that DEE exposure and increased genetic damage were associated with hypomethylation of p16, RASSF1A, and MGMT. Future studies with larger sample size are needed to confirm these associations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cohort studies have consistently showed that ambient air pollution exposure, especially fine particulate matter (particulate matter with an aerodynamic diameter <2.5 µm, PM2.5), is associated with increased lung cancer risk (Puett et al. 2014; Raaschou-Nielsen et al. 2013). In urban settings, the major contributor to PM2.5 is diesel engine exhaust (DEE). DEE, which is a mixture of gases and elemental carbon (EC) core adsorbed with organic carcinogenic compounds such as polycyclic aromatic hydrocarbons (PAHs), has been previously suspected to cause lung cancer (IARC 1989). Recently, the International Agency for Research on Cancer reevaluated the carcinogenicity of DEE and concluded that DEE was a cause of lung cancer (Benbrahim-Tallaa et al. 2012). Therefore, considering that DEE exposure is posing great health threat to a huge number of urban residents and occupational population who perform routine work with diesel-powered equipments, assessment of DEE exposure-related cancer risk is urgently needed. Further, understanding of the potential biological mechanisms that mediate DEE exposure-related cancer risk is also beneficial to develop strategies to reduce relevant cancer risk.
Genomic instability is a hallmark of cancer that accelerates mutation rate, thus enabling the acquisition of additional capabilities required for carcinogenesis (Pikor et al. 2013). Marker of genomic instability, such as increased micronucleus frequency, has been found to be prospectively associated with increased cancer risk (Bonassi et al. 2007). For population exposed to genotoxic substance, such as DEE, the extent of genomic instability is determined not only by exposure-induced DNA lesions, but also by DNA damage response (DDR). DDR is a sophisticated network of signaling pathways that aims at maintaining genomic stability (Rouse and Jackson 2002). On the cellular level, DDR involves triggering cell-cycle arrest and recruiting DNA damage repair protein to the damage sites (Jackson and Bartek 2009). The expression levels of DDR-related genes can directly indicate the extent of DDR. Evidence is mounting that DNA methylation variation can affect the expression levels of DDR-related genes (Shin et al. 2013; Vo et al. 2002). These changes, in turn, have the potential to affect the extent of genomic instability, hence cancer risk. Previous studies have reported associations of air pollutants exposure with either hypomethylation or hypermethylation of DDR-related genes (Duan et al. 2013; Hou et al. 2011). Therefore, alterations in methylation levels of DDR-related genes may act as a potential mechanism that mediates air pollutants exposure-related cancer risk. However, no population study has yet examined whether DEE exposure is associated with methylation alterations in DDR-related genes.
In the present study, we recruited 117 diesel engine testing workers with exclusive exposure to DEE and 112 non-DEE-exposed workers. To characterize DEE exposure, we determined urinary concentrations of six mono-hydroxylated PAHs (OH-PAHs) as well as obtained detailed exposure history. We employed a candidate gene approach and measured promoter methylation levels of three DDR-related genes, including cyclin-dependent kinase inhibitor 2A (p16), Ras association domain family 1 isoform A (RASSF1A), and O6-methylguanine methyltransferase (MGMT). It is well known that both p16 and RASSF1A play a crucial role in cell-cycle regulation by inhibiting cell progression from G1 phase to S phase (Liggett and Sidransky 1998; Shivakumar et al. 2002). MGMT is a mismatch repair protein that removes the premutagenic lesions in damaged DNA (Kaina et al. 2007). Besides, we also measured methylation level of long interspersed nucleotide element 1 (LINE-1). LINE-1 accounts for approximately 20 % of human genome and is heavily methylated to prevent expression. Methylation in LINE-1 can be used as a surrogate marker of global DNA methylation (Yang et al. 2004). Aberrant methylation of p16, RASSF1A, MGMT, and LINE-1 has been detected in DNA from lung cancer patients (Daskalos et al. 2009; Fujiwara et al. 2005; Hsu et al. 2007). We aimed to compare the differences in methylation levels of p16, RASSF1A, MGMT, and LINE-1 between DEE and non-DEE-exposed workers and further examine their associations with both urinary OH-PAHs and DEE exposure duration. Taking advantage of the cytokinesis-block micronucleus (CBMN) cytome index measured previously in the same population, we also examined the correlations between p16, RASSF1A, MGMT, and LINE-1 methylation levels and CBMN cytome index to explore whether epigenetic variations were related to DEE exposure-related genetic damage.
Materials and methods
Study population and biological sample collection
We recruited 117 male DEE-exposed workers and 112 male non-DEE-exposed workers as described previously (Zhang et al. 2015). The DEE-exposed workers were diesel engine testing workers who spent most time in close proximity to the running diesel engines from a diesel engine manufacturing plant located in Luoyang, Henan, China. The non-DEE-exposed workers were water pump management workers from a water factory in the same city. The included workers had to be working in the current job position for at least 1 year and free of chronic diseases such as cancer and acute infection. All participants provided informed consent before enrollment. Demographic characteristics, smoking history, alcohol consumption, occupational history of exposure, and personal medical history were determined for all participants by an occupational physician using a structured questionnaire. At the end of work shift (after at least four consecutive working days), each worker provided 50 mL urine for measurement of OH-PAHs and 4 mL venous blood for analysis of complete blood count and extraction of DNA. The protocol was approved by the Research Ethics Committee of the National Institute for Occupational Health and Poison Control, Chinese Center for Disease Control and Prevention.
Exposure assessment
To characterize the DEE exposure levels of workers, airborne concentrations of PM2.5, EC, and PAHs were determined from diesel engine manufacturing plant and water factory, respectively, as described in our previous study (Zhang et al. 2015). In brief, PM2.5 concentrations were determined using gravimetric method. EC in collected PM was determined according to National Institute for Occupational Safety and Health method 5040 (thermal-optical analysis). A total of 16 PAHs in collected PMs were determined based on the Occupational Safety and Health Administration method 58 (high-performance liquid chromatography with a fluorescence detector).
Urinary OH-PAHs measurement
The concentrations of six OH-PAHs 1-hydroxynaphthalene (1-OHNa), 2-hydroxynaphthalene (2-OHNa), 2-hydroxyfluorene (2-OHFlu), 2-hydroxyphenanthrene (2-OHPhe), 9-hydroxyphenanthrene (9-OHPhe), and 1-hydroxypyrene (1-OHP) in urine were determined by HPLC–MS/MS method as described previously (Zhang et al. 2015). Creatinine concentration was determined by Jaffe’s colorimetric method to correct for urinary dilution. Measured concentrations below the limit of detection (LOD) were replaced with LOD divided by the square root of 2.
DNA methylation analyses
Genomic DNA was extracted from whole blood using TIANamp Blood DNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturers’ instructions. One microgram of genome DNA was treated with sodium bisulfite using the EpiTect Bisulfite Kit (Qiagen, Valencia, USA) and subjected to DNA methylation analyses by pyrosequencing. The primers of pyrosequencing for p16, RASSF1A, MGMT, and LINE-1 were listed (Table 1 in Online Resource 1). Approximately 40 ng bilsulfite-treated and purified DNA was used in a PCR along with Pyromark PCR kit (Qiagen, Valencia, USA) and primers (forward and biotinylated reverse). After purification of PCR products using Sepharose beads on PyroMark Vacuum Prep Workstation (Qiagen, Valencia, USA), pyrosequencing was performed using the PyroMark Q96MD System (Qiagen, Valencia, USA). Non-CpG cytosine residues were used to verify bisulfite conversion. The percentage of methylated and unmethylated cytosines was quantified for 7 CpG sites in p16 promoter, 7 CpG sites in RASSF1A promoter, 9 CpG sites in MGMT promoter, and 3 sites in LINE-1, respectively. The percentage of methylation was expressed as the methylated cytosines over the sum of methylated and unmethylated cytosines. We pyrosequenced each marker in two replicates, and the average was used in the statistical analysis. The within-sample coefficients of variation were 7.4 % for p16, 9.4 % for RASSF1A, 7.5 % for MGMT, and 1.3 % for LINE-1, respectively.
Statistical analysis
Standard descriptive statistics were calculated for general characteristics, urinary OH-PAH concentrations, and DNA methylation outcomes. Statistical comparisons of general characteristics between groups were conducted using Student t test for age and body mass index (BMI) and using Chi-square test for smoking status and alcohol use. Percentages of different white blood cell types and exposure levels of PM2.5, EC, and PAHs were compared between groups by Mann–Whitney test. Urinary OH-PAH concentrations, which were not normally distributed and thus natural logarithmic transformed, were compared between groups by Student t test. The differences of methylation levels of each CpG site as well as corresponding average across all CpG sites in the p16, RASSF1A, and MGMT promoters and of mean methylation level of LINE-1 repetitive element between groups were evaluated by Mann–Whitney test. Three separate general linear models were used to test the associations of p16, RASSF1A, MGMT, and LINE-1 mean methylation levels with categorized urinary summed OH-PAHs in all study subjects. In model 1, we presented crude results. In model 2, we included age (continuous), BMI (continuous), smoking status (current smokers, non-current smokers), and alcohol use (yes/no) into the model to adjust for potential confounding. In model 3, we additionally included percentages of neutrophils (continuous), lymphocytes (continuous), and monocytes (continuous) into the model. Covariates-adjusted means and 95 % confidential intervals (CIs) of methylation levels of p16, RASSF1A, MGMT, and LINE-1 were examined in model 2 and model 3. Regression coefficients and 95 % CIs relative to the reference category were also estimated. Because smoking was an important confounding factor, we further repeated the analysis by smoking status. Next, the associations of methylation levels of p16, RASSF1A, MGMT, and LINE-1 with categorized urinary summed OH-PAHs by exposure status and with categorized DEE exposure duration were examined in non-smoking workers using the general linear models as above mentioned. In addition, the correlations of p16, RASSF1A, MGMT, and LINE-1 methylation levels with CBMN cytome index were determined in non-smoking workers by Spearman correlation test. All analyses were performed with SPSS software version 15.0 (Chicago, USA), and all statistical tests were two-sided with a significance level of 0.05.
Results
General characteristics and DEE exposure levels
The study population included 117 DEE-exposed workers and 112 non-DEE-exposed workers, and their general characteristics and exposure levels had been described previously (Zhang et al. 2015). In brief, the differences of age, BMI, smoking status, and alcohol use were not significant between DEE and non-DEE-exposed workers. The percentages of neutrophils and lymphocytes were similar between DEE and non-DEE-exposed workers (59.89 ± 7.44 vs. 58.57 ± 8.47, p = 0.299 and 33.51 ± 7.18 vs. 34.30 ± 8.25, p = 0.641), and the percentage of monocytes was moderately lower in DEE-exposed workers (4.40 ± 1.12 vs. 5.01 ± 1.72, p = 0.014). The geometric means of PM2.5, EC, and PAH exposure levels of DEE-exposed workers were all significantly higher than those of non-DEE-exposed workers. Likewise, the urinary concentration of each OH-PAH was significantly higher in DEE-exposed workers than non-DEE-exposed workers (all p < 0.001) (Table 2 in Online Resource 1). When the urinary concentrations of six OH-PAHs were added together, the median of urinary summed OH-PAH concentration in the DEE-exposed workers was 2.7 times higher than that in the non-DEE-exposed workers (12.96 vs. 4.75 μg/g creatinine, p < 0.001).
DDR-related genes and LINE-1 methylation by exposure group
We compared the methylation levels of p16, RASSF1A, MGMT, and LINE-1 in DEE-exposed workers with those in non-DEE-exposed workers (Table 1). Of the seven CpG sites in the promoter region of p16, methylation levels decreased significantly at CpG sites 2, 3, 4, 5, and 7 in DEE-exposed workers than those in non-DEE-exposed workers (all p < 0.05). When the seven CpG sites were combined, the mean promoter methylation level of p16 was significantly lower in DEE-exposed workers than that in non-DEE-exposed workers (2.03 % vs. 2.21 %, p < 0.001). The promoter methylation in RASSF1A in DEE-exposed workers showed significantly lower levels at CpG sites 2, 4, 5, 6, and 7 than that in non-DEE-exposed workers (all p < 0.05). The mean promoter methylation level of the combined seven CpG sites was significantly lower in DEE-exposed workers than that in non-DEE-exposed workers (2.25 % vs. 2.48 %, p < 0.001). The methylation levels of all CpG sites except sites 6 and 9 in the promoter of MGMT decreased significantly in DEE-exposed workers than those in non-DEE-exposed workers (all p < 0.05). The mean methylation level of all nine CpG sites evaluated in the promoter of MGMT was significantly lower in DEE-exposed workers than that in non-DEE-exposed workers (2.22 % vs. 2.57 %, p < 0.001). No significant difference in the methylation level of LINE-1 was observed between DEE and non-DEE-exposed workers (p = 0.369). We also examined the correlations of age, BMI, smoking status, and alcohol use with p16, RASSF1A, MGMT, and LINE-1 mean methylation levels, and we found a negative correlation between smoking status (current smokers vs. non-current smokers) and MGMT methylation level (r = −0.181, p = 0.006) and a positive correlation between alcohol use (yes vs. no) and LINE-1 methylation level (r = 0.131, p = 0.049) (Table 2).
Associations of DDR-related genes methylation with DEE exposure
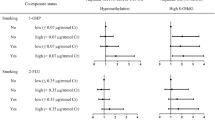
As summed OH-PAHs could provide more comprehensive and stable estimate of internal dose than individual OH-PAH and most individual CpG sites in the p16, RASSF1A, and MGMT promoters showed significant and consistent alterations in DEE-exposed workers, the association analyses including urinary summed OH-PAHs and mean methylation of p16, RASSF1A, MGMT, and LINE-1 were considered as the main results. Among all study subjects, increasing quartiles of urinary summed OH-PAHs was associated with hypomethylation of p16, RASSF1A, and MGMT (all p trend < 0.05) (Table 3). Because smoking was an important confounding factor, we further repeated the analyses by smoking status. We found that the above associations showed better dose–response trends in non-smoking workers but became nonsignificant in smoking workers, suggesting an apparent confounding effect of smoking. To avoid such confounding effect, the following analyses were based on non-smoking workers only. In non-smoking workers, the methylation levels of p16, RASSF1A, and MGMT decreased in the highest quartile of summed OH-PAHs compared with the lowest quartile (β = −0.355, SE = 0.125, p = 0.005 for p16; β = −0.464, SE = 0.162, p = 0.005 for RASSF1A; and β = −0.550, SE = 0.201, p = 0.007 for MGMT), with adjustment for age, BMI, alcohol use, and percentages of neutrophils, lymphocytes, and monocytes (Fig. 1 in Online Resource 2). No significant association was observed for mean methylation level of LINE-1. When the associations observed in non-smoking workers were separately analyzed by exposure group, no significant association was found (Table 3 in Online Resource 1).
Furthermore, the associations of DEE exposure duration with mean methylation levels of p16, RASSF1A, MGMT, and LINE-1 were examined (Table 4). The p16, RASSF1A mean promoter methylation levels were significantly or borderline significantly lower in workers exposed to DEE for <4 and 4–8 years compared with non-DEE-exposed workers, although they showed nonsignificant associations with categorized DEE exposure duration (Fig. 2 in Online Resource 2). The MGMT mean promoter methylation level was significantly lower across workers exposed to DEE for <4, 4–8, and >8 years and was associated with categorized DEE exposure duration (p trend = 0.031).
DDR-related genes and LINE-1 methylation and CBMN cytome index
We previously examined the effect of DEE exposure on CBMN cytome index in this study population and found that DEE exposure was associated with increased CBMN cytome index (Zhang et al. 2015). In this study, we observed negative correlations of p16, RASSF1A, MGMT, and LINE-1 mean promoter methylation levels with CBMN cytome index in non-smoking workers (r = −0.314, p = 0.002 for p16; r = −0.281, p = 0.006 for RASSF1A; r = −0.346, p = 0.001 for MGMT; and r = −0.216, p = 0.034 for LINE-1) (Fig. 1).
Discussion
In this study, the DEE-exposed workers showed significantly lower mean promoter methylation levels of p16, RASSF1A, and MGMT than non-DEE-exposed workers, and there were significant associations between increasing quartiles of urinary summed OH-PAHs and hypomethylation of p16, RASSF1A, and MGMT in all study subjects and non-smoking workers. In addition, non-smoking workers with hypomethylation of p16, RASSF1A, and MGMT showed increased CBMN cytome index. To our knowledge, this is the first study that reports the associations between DEE exposure and alterations in methylation levels of DDR-related genes in an occupational population who are exclusively exposed to DEE.
Exposure to DEE has been shown to be associated with increased lung cancer risk (Benbrahim-Tallaa et al. 2012), as well as early health effects related to carcinogenesis, such as chromosome damage (Zhang et al. 2015). However, the biological pathways that act as intermediates and/or consequences of these associations remain poorly understood. One of the commonly studied mechanisms through which environmental exposure exerts influence on biological pathways is DNA methylation. Nevertheless, there is a paucity of data on the association between DEE exposure and DNA methylation alteration mainly due to prevalent existence of mixed exposure (i.e., DEE coexists with other air pollutants in the setting). Recently, Jiang et al. (2014) investigated the association of DEE exposure with DNA methylation in a controlled exposure study wherein volunteers were exposed to pure DEE for 2 h and they found that DNA methylation at CpG sites residing in genes involved in inflammation and oxidative stress and overlapping with LINE-1 exhibited significant changes. In the present study, we extended the investigation to DEE-exposed workers who had been exclusively exposed to DEE for a mean of 8 years and we found that the DEE-exposed workers exhibited significant decrease in mean promoter methylation levels of DDR-related genes including p16, RASSF1A, and MGMT and nonsignificant change in LINE-1 methylation. The hypomethylation of p16, RASSF1A, and MGMT promoters might lead to increased expression levels of these genes and consequent activation of DDR. Therefore, the reason why Jiang et al. found nonsignificant associations with methylation at sites residing in genes involved in DDR might be that the exposure duration in their study was too short to cause DDR. As for LINE-1 methylation, two points should be considered when interpreting the differences between the studies. First, the study population were asthmatics in their study while were healthy subjects in our study. The asthmatics are more susceptible to exposure-induced oxidative stress and inflammation (Klumper et al. 2015), which have been shown to affect DNA methylation (Fratelli et al. 2005). Second, in their study, all study subjects are their own control, and thus, the analyses are immune to the effects of most confounding factors. Although we had adjusted common confounding factors in this study, there might be uncontrolled confounding (e.g., nutritional factors). As LINE-1 methylation is the marker of global methylation, it is more likely to be affected by confounding factors than gene-specific sites.
Considering that smoking was an important confounding factor, we further compared the p16, RASSF1A, and MGMT methylation levels between DEE-exposed and non-DEE-exposed workers in non-smoking and smoking workers, respectively. The results obtained in non-smoking workers were in line with those in all study subjects, thus providing more unequivocal evidence to the associations between DEE exposure and hypomethylation of p16, RASSF1A, and MGMT. In smoking workers, however, RASSF1A and MGMT methylation levels did not differ significantly between DEE and non-DEE-exposed workers (p = 0.123, p = 0.066), which were substantiated by the nonsignificant associations between urinary summed OH-PAHs and p16, RASSF1A, and MGMT methylation levels. Since we observed negative correlations of smoking status with methylation levels of p16, RASSF1A, and MGMT, the confounding effect of smoking could be explained by the possibility that the effect of DEE exposure on methylation was, to some extent, masked by that of smoking. Previous studies also examined the associations between exposure to air pollutants and alterations in methylation levels of DDR-related genes. Our group reported that coke oven workers who were exposed to higher levels of PAHs showed significantly lower mean promoter methylation level of MGMT than controls (Duan et al. 2013), which was in line with the results presented herein. Because PAH is one of the major organic components adsorbed to the elemental carbon core of diesel exhaust particle, our results suggested a potential role of PAHs in linking DEE exposure with MGMT hypomethylation. Another study by Hou et al. (2011) examined the associations of ambient PM exposure with methylation levels of p16 and RASSF1A, and their results were partially concordant with ours. Although both studies found hypomethylation of RASSF1A in exposed workers, their study reported that ambient PM exposure was associated with hypermethylation of p16, while our study reported that DEE exposure was associated with hypomethylation of p16. One possible explanation for the difference was that the chemical compositions of PM differed between these two studies. An alternative explanation was that the methylation alterations found in our study were the results of relatively long-term exposure, not rapid exposure as was seen in their study.
It was worth noting that the significant associations between increasing quartiles of urinary summed OH-PAHs and hypomethylation of p16, RASSF1A, and MGMT became insignificant when confining the analyses to DEE-exposed workers only. The narrower exposure range was the most likely explanation for this phenomenon. It was also possible that the limited sample size in the subgroup did not provide us with sufficient statistical power to detect exposure-related change in DNA methylation. On the other hand, although the mean promoter methylation levels of p16 and RASSF1A were significantly lower in workers exposed to DEE for <4 and 4–8 years, they did not show a significant decrease in workers exposed to DEE for >8 years when comparing to non-DEE-exposed workers. Indeed, when excluding non-DEE-exposed workers, we found that the mean promoter methylation levels of p16 and RASSF1A gradually increased with DEE exposure duration, which was consistent with previous findings that continuous exposure could lead to accumulation of methylation changes (Dolinoy et al. 2007). Adjustment for age did not alter the trend. To further take into account the collinearity between age and DEE exposure duration, we generated the residual by regressing DEE exposure duration on age and found that the residual was positively associated with the mean promoter methylation levels of p16 (p = 0.009) and RASSF1A (p = 0.004). Based on these data, one might expect for hypermethylation of p16 and RASSF1A which were frequently observed in lung cancer (Fujiwara et al. 2005; Hsu et al. 2007), as the cumulative exposure level of DEE accumulated to a certain degree. Unlike p16 and RASSF1A, the mean methylation level of MGMT was still significantly lower in workers exposed to DEE for >8 years, which suggested that hypermethylation of p16 and RASSF1A might precede that of MGMT in DEE-induced lung carcinogenesis. Further studies that follow up the DEE-exposed workers with longer exposure duration for a couple of years or even longer and then reevaluate the methylation levels are warranted to clarify this problem. Finally, the different results between our study and the study by Hou et al. might also be explained by the fact that the DEE-exposed workers in our study had a relatively shorter exposure duration than that of the steel workers in their study.
Our previous study had demonstrated a significantly higher CBMN cytome index in the same workers (Zhang et al. 2015). Here we further explored the correlations between CBMN cytome index and methylation levels of p16, RASSF1A, MGMT and found negative correlations between the indicators. Although CBMN cytome index was an integrated biomarker of micronucleus (MN), nucleoplasmic bridge (NPB), and nuclear bud (NBUD) and increased MN, NPB, and NBUD frequencies had been shown to be associated with increased cancer risk (Bonassi et al. 2007; El-Zein et al. 2006), the above negative correlations did not necessarily mean that hypomethylation of p16, RASSF1A, and MGMT contributed to increased cancer risk. The exposure duration of our exposed workers was relatively short. Previous studies had demonstrated that DDR was activated in the early precursor lesions of cancer while was attenuated in the cancer (Bartkova et al. 2005; Berwick and Vineis 2000), suggesting the regulation of DDR-related genes might be different in different stages of cancer. In addition, hypomethylation was typically associated with increased transcriptional levels, which led to cell-cycle arrest to facilitate DNA damage repair in the case of p16 and RASSF1A and directly strengthen DNA damage repair in the case of MGMT, thus mitigating genomic instability required for carcinogenesis. Although we could not directly determine the impact on RNA due to lack of RNA samples, we found positive correlations between methylation levels of p16 and RASSF1A which played a role in cell-cycle control and nuclear division index (r = 0.234, p = 0.021; and r = 0.250, p = 0.014), a biomarker indicative of mitogen response and cell proliferation (El-Zein et al. 2008), which suggested that hypomethylation of p16 and RASSF1A was associated with decreased cell proliferation. In this context, we speculated that the hypomethylation in the promoters of p16, RASSF1A, and MGMT associating with CBMN cytome index might represent the methylation-associated regulation pattern of DDR-related genes in DEE-exposed workers showing higher genetic damage, which acted as an adaptive response to confront the persistent genetic damage induced by DEE exposure, thus attenuating rather than exacerbating the increase in cancer risk among DEE-exposed workers with lower cumulative exposure level. This speculation was further supported by the results from epidemiology studies. Olsson et al. (2011) pooled 11 case–control studies and found that compared with never exposed workers, the odds ratios in lowest, second, and third quartile groups of cumulative DEE exposure only showed slight increase. Vermeulen et al. (2014) carried out a meta-regression to derive an exposure–response estimate for cumulative DEE exposure and lung cancer mortality, and they found that the relative risk showed a comparatively slow increase in the lower range of cumulative DEE exposure. Future studies that adjust for the methylation in DDR-related genes while examining the association between cumulative DEE exposure and lung cancer risk are warranted to gain deeper insight of the potential role of methylation alterations in DDR-related genes in mediating DEE exposure-related lung cancer risk. On the other hand, we also found a negative correlation of CBMN cytome index with LINE-1 methylation. This observation was consistent with previous results that showed both LINE-1 hypomethylation and increased MN, NPB, and NBUD frequencies were markers of genomic instability (Daskalos et al. 2009; Fenech 2006).
Although all of p16, RASSF1A, and MGMT engaged the DDR pathway and their hypomethylation were observed in DEE-exposed workers, whether the methylation alterations in these genes could act together in response to DEE was still largely unknown. To shed some light on this problem, we conducted a preliminary analysis among non-smoking workers in which we tested the possibility that DEE-exposed worker could simultaneously exhibit hypomethylation of two or more genes of p16, RASSF1A, and MGMT. Specifically, the methylation value of corresponding genes in DEE-exposed workers was rearranged from maximum to minimum and then was successively included into a test group. Once a new methylation value was incorporated, we repeated the comparison of methylation level between the test group and the reference group (i.e., non-DEE-exposed workers). Consequently, the cutoff methylation values that could be used to identify hypomethylation of corresponding genes at individual level were developed (1.72 for p16, 1.74 for RASSF1A, and 2.01 for MGMT) when the test group began to show significant decrease in methylation levels compared with the reference group. Using the cutoff methylation values, we indentified 4, 1, and 3 non-DEE-exposed workers and 9, 9, and 17 DEE-exposed workers who exhibited hypomethylation of p16, RASSF1A, and MGMT, respectively. The percentage of workers with hypomethylation of p16, RASSF1A, or MGMT in DEE-exposed workers was 2.4, 10.0, and 6.1 times higher than that in non-DEE-exposed workers, respectively. Among the DEE-exposed workers with hypomethylation of corresponding gene, there were five workers with hypomethylation of p16 and RASSF1A (38.5 %), seven workers with hypomethylation of p16 and MGMT (36.8 %), six workers with hypomethylation of RASSF1A and MGMT (30.0 %), and four workers with hypomethylation of p16, RASSF1A, and MGMT (14.8 %). Taken together, these results suggested that RASSF1A hypomethylation might be more sensitive than those of p16 and MGMT in discriminating DEE-exposed workers from non-DEE-exposed workers, and hypomethylation of p16 and RASSF1A was more likely to arise together in DEE-exposed workers. A combination of p16 and RASSF1A is also encouraged in future studies investigating the association between DEE exposure and methylation alterations in DDR-related genes.
In most exposure scenarios, exposure to DEE occurs concurrently with other air pollutants. On the contrary, the DEE-exposed workers in our study performed their duties in the indoor setting and there was no other major occupational exposure source except diesel engines. Further analyses confined to non-smoking workers showed consistent results, thus minimizing the possibility that the observed effects resulted from exposures other than DEE. In addition, we comprehensively evaluated urinary OH-PAHs, DNA methylation in DDR-related genes, and genetic damage, which linked epigenetic variation with both environmental exposure and genetic variation. We also recognize a number of limitations in this work. First, we used DNA extracted from whole blood for methylation analysis. Whole blood contained different white blood cell types that displayed specific methylation patterns (Reinius et al. 2012), and we noted that the percentage of monocytes in DEE-exposed workers was significantly lower than that in non-DEE-exposed workers. However, the percentages of neutrophils, lymphocytes, and monocytes were included into the model to account for the potential confounding effect of DEE-induced shift in the distribution of white blood cell types. Moreover, additional adjustment for lymphocyte subsets (i.e., T cell, B cell, and NK cell) resulted in similar results (data not shown). Second, we did not measure nutrition factors in this study. Previous studies had shown the impact of nutrition factors on DNA methylation (Fenech 2012). Therefore, we could not dismiss the possibility that the observed effects were attributable to the discrepancy of nutrition status between the two groups. Third, the methylation changes and associations with urinary OH-PAHs and CBMN cytome index we observed were tissue-dependent. Whether these findings extended to the target tissue (i.e., lung) remains to be explored in future studies.
In summary, our results provided clear evidence that DEE exposure and increased CBMN cytome index were associated with hypomethylation of p16, RASSF1A, and MGMT, suggesting that such methylation alterations might reflect the changes in the regulation of p16, RASSF1A, and MGMT under the circumstance of DEE-induced DNA damage. Because CBMN cytome index was an integrated biomarker indicative of cancer risk, our findings further suggested that the alterations in methylation levels of p16, RASSF1A, and MGMT could serve as potential mechanism that mediated DEE exposure-related cancer risk. Prospective studies with a larger sample size that employ cumulative DEE exposure and collect multiple methylation measurements of the same individual are warranted to replicate our findings and better understand the associations between DEE exposure and methylation alterations in DDR-related genes.
References
Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K et al (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864–870
Benbrahim-Tallaa L, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V et al (2012) Carcinogenicity of diesel-engine and gasoline-engine exhausts and some nitroarenes. Lancet Oncol 13:663–664
Berwick M, Vineis P (2000) Markers of DNA repair and susceptibility to cancer in humans: an epidemiologic review. J Natl Cancer Inst 92:874–897
Bonassi S, Znaor A, Ceppi M, Lando C, Chang WP, Holland N et al (2007) An increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of cancer in humans. Carcinogenesis 28:625–631
Daskalos A, Nikolaidis G, Xinarianos G, Sawari P, Cassidy A, Zakopoulou R et al (2009) Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer 124:81–87
Dolinoy DC, Weidman JR, Jirtle RL (2007) Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol 23:297–307
Duan H, He Z, Ma J, Zhang B, Sheng Z, Bin P et al (2013) Global and MGMT promoter hypomethylation independently associated with genomic instability of lymphocytes in subjects exposed to high-dose polycyclic aromatic hydrocarbon. Arch Toxicol 87:2013–2022
El-Zein RA, Schabath MB, Etzel CJ, Lopez MS, Franklin JD, Spitz MR (2006) Cytokinesis-blocked micronucleus assay as a novel biomarker for lung cancer risk. Cancer Res 66:6449–6456
El-Zein RA, Fenech M, Lopez MS, Spitz MR, Etzel CJ (2008) Cytokinesis-blocked micronucleus cytome assay biomarkers identify lung cancer cases amongst smokers. Cancer Epidemiol Biomark Prev 17:1111–1119
Fenech M (2006) Cytokinesis-block micronucleus assay evolves into a “cytome” assay of chromosomal instability, mitotic dysfunction and cell death. Mutat Res 600:58–66
Fenech M (2012) Folate (vitamin B9) and vitamin B12 and their function in the maintenance of nuclear and mitochondrial genome integrity. Mutat Res 733:21–33
Fratelli M, Goodwin LO, Orom UA, Lombardi S, Tonelli R, Mengozzi M et al (2005) Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc Natl Acad Sci USA 102:13998–14003
Fujiwara K, Fujimoto N, Tabata M, Nishii K, Matsuo K, Hotta K et al (2005) Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res 11:1219–1225
Hou L, Zhang X, Tarantini L, Nordio F, Bonzini M, Angelici L et al (2011) Ambient PM exposure and DNA methylation in tumor suppressor genes: a cross-sectional study. Part Fibre Toxicol 8:25
Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, Lee HC et al (2007) Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer 110:2019–2026
IARC (1989) IARC monographs on the evaluation of carcinogenic risks to humans. Diesel and gasoline engine exhausts and some nitroarenes. International Agency for Research on Cancer. IARC Monogr Eval Carcinog Risks Hum 46:1–458
Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461:1071–1078
Jiang R, Jones MJ, Sava F, Kobor MS, Carlsten C (2014) Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol 11:71
Kaina B, Christmann M, Naumann S, Roos WP (2007) MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst) 6:1079–1099
Klumper C, Kramer U, Lehmann I, von Berq A, Berdel D, Herberth G et al (2015) Air pollution and cytokine responsiveness in asthmatic and non-asthmatic children. Environ Res 138:381–390
Liggett WH Jr, Sidransky D (1998) Role of the p16 tumor suppressor gene in cancer. J Clin Oncol 16:1197–1206
Olsson AC, Gustavsson P, Kromhout H, Peters S, Vermeulen R, Bruske L et al (2011) Exposure to diesel motor exhaust and lung cancer risk in a pooled analysis from case-control studies in Europe and Canada. Am J Respir Crit Care Med 183:941–948
Pikor L, Thu K, Vucic E, Lam W (2013) The detection and implication of genome instability in cancer. Cancer Metastasis Rev 32:341–352
Puett RC, Hart JE, Yanosky JD, Spiegelman D, Wang M, Fisher JA et al (2014) Particulate matter air pollution exposure, distance to road, and incident lung cancer in the nurses’ health study cohort. Environ Health Perspect 122:926–932
Raaschou-Nielsen O, Andersen ZJ, Beelen R, Samoli E, Stafoggia M, Weinmayr G et al (2013) Air pollution and lung cancer incidence in 17 European cohorts: prospective analyses from the European study of cohorts for air pollution effects (ESCAPE). Lancet Oncol 14:813–822
Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D et al (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE 7:e41361
Rouse J, Jackson SP (2002) Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547–551
Shin DY, Sung Kang H, Kim GY, Kim WJ, Yoo YH, Choi YH (2013) Decitabine, a DNA methyltransferases inhibitor, induces cell cycle arrest at G2/M phase through p53-independent pathway in human cancer cells. Biomed Pharmacother 67:305–311
Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA (2002) The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol 22:4309–4318
Vermeulen R, Silverman DT, Garshick E, Vlaanderen J, Portengen L, Steenland K (2014) Exposure-response estimates for diesel engine exhaust and lung cancer mortality based on data from three occupational cohorts. Environ Health Perspect 122:172–177
Vo QN, Geradts J, Boudreau DA, Bravo JC, Schneider BG (2002) CDKN2A promoter methylation in gastric adenocarcinomas: clinical variables. Hum Pathol 33:1200–1204
Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32:e38
Zhang X, Duan H, Gao F, Li Y, Huang C, Niu Y et al (2015) Increased micronucleus, nucleoplasmic bridge, and nuclear bud frequencies in the peripheral blood lymphocytes of diesel engine exhaust-exposed workers. Toxicol Sci 143:408–417
Acknowledgments
This work was supported by the Key Program of National Natural Science Foundation of China (NSFC 81130050) and the National Key Technology Research and Development Program (2014BAI12B02).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Xiao Zhang and Jie Li have been contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, X., Li, J., He, Z. et al. Associations between DNA methylation in DNA damage response-related genes and cytokinesis-block micronucleus cytome index in diesel engine exhaust-exposed workers. Arch Toxicol 90, 1997–2008 (2016). https://doi.org/10.1007/s00204-015-1598-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-015-1598-2