Abstract
Recent studies have established the role of mid-chain hydroxyeicosatetraenoic acids (HETEs) in the development of cardiovascular disease. Mid-chain HETEs have been reported to have vasoconstrictive and pro-inflammatory effects. However, whether mid-chain HETEs can induce cardiac hypertrophy remains unclear. Therefore, the overall objective of the present study was to elucidate the potential hypertrophic effect of mid-chain HETEs in the human ventricular cardiomyocytes, RL-14 cells, and to explore the mechanisms involved. For this purpose, RL-14 cells were treated with increasing concentrations of mid-chain HETEs (2.5, 5, 10 and 20 µM). Thereafter, the cardiac hypertrophy markers and cell size were determined using real-time polymerase chain reaction and phase contrast imaging, respectively. Phosphorylated mitogen-activated protein kinase (MAPK) level and nuclear factor kappa B (NF-κB) binding activity were determined. Our results showed that mid-chain HETEs induced cellular hypertrophy in RL-14 cells as evidenced by the induction of cardiac hypertrophy markers, α- and β-myocin heavy chain and atrial and brain natriuretic peptide as well as the increase in cell size. Mechanistically, all mid-chain HETEs were able to induce the binding activity of NF-κB to its responsive element in a HETE-dependent manner, and they significantly induced the phosphorylation of ERK 1/2. The induction of cellular hypertrophy was associated with proportional increase in the formation of dihydroxyeicosatrienoic acids parallel to the increase of soluble epoxide hydrolase enzyme activity. In conclusion, our study provides the first evidence that mid-chain HETEs induce cellular hypertrophy in RL-14 cells through MAPK- and NF-κB-dependent mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mounting evidence is shedding the light on the role of arachidonic acid (AA) metabolites in the pathogenesis of cardiovascular disease (CVD) (Roman 2002). AA is known to be metabolized by cyclooxygenases, lipoxygenases (LOXs) and cytochrome P450 (CYP) enzymes to form biologically active eicosanoids (Capdevila et al. 1981). CYP, cysteinato-heme mixed function mono-oxygenases enzymes, oxidizes AA into epoxyeicosatrienoic acids (EETs) and hydroxyeicosatetraenoic acids (HETEs), which are known to play an important role in the maintenance of cardiovascular health (Zordoky and El-Kadi 2010). CYP ω-hydroxylases, namely CYP4 family, metabolize AA into its cardiotoxic form 20-HETE, whereas (Althurwi et al. 2013; Elshenawy et al. 2013; Fava et al. 2012; Gross et al. 2005; Schwartzman et al. 1996; Wu and Schwartzman 2011; Yousif et al. 2009; Zordoky et al. 2008, 2010) CYP epoxygenases, mainly CYP2B, CYP2C and CYP2J subfamilies, metabolize AA into four regioisomers of cardioprotective EETs, 14,15-EET, 11,12-EET, 8,9-EET and 5,6-EET metabolites (Roman 2002). EETs are further metabolized by soluble epoxide hydrolase (sEH) into their corresponding degradation products dihydroxyeicosatrienoic acids (DHETs) (Imig et al. 2002).
LOXs, non-heme iron dioxygenase enzymes, constitute a family of lipid-peroxidizing enzymes that insert molecular oxygen into free and esterified polyunsaturated fatty acids. AA is metabolized by LOXs into mid-chain HETEs, typified by 5-, 12- and 15-HETE. CYP1B1 can also metabolize AA in vitro by its bis-allylic oxidation reaction (LOX-like reaction) to generate mid-chain HETEs. Recent studies have established the role of mid-chain HETEs in the development of cardiovascular disease (Cyrus et al. 1999; Jenkins et al. 2009; Nozawa et al. 1990). For example, 5-HETE has been reported to have vasoconstrictive and pro-inflammatory actions (Burhop et al. 1988). 12-HETE has been reported to act as vasoconstrictors in small renal arteries and induce cellular hypertrophy and fibrosis (Kayama et al. 2009). In addition, an enhanced 12-HETE production has been reported in patients with essential hypertension and in the spontaneously hypertensive rat (SHR) (Gonzalez-Nunez et al. 2001; Sasaki et al. 1997). 15-HETE has been shown to increase the sensitivity of the isoproterenol-mediated β-adrenergic response in cardiomyocytes and has been proposed to be implicated in heart failure by induction of cardiac fibrosis (Kayama et al. 2009; Wallukat et al. 1994).
Several molecular mechanisms have been shown to play a role in cardiac hypertrophy such as mitogen-activated protein kinases (MAPKs) (Kayama et al. 2009). MAPKs are serine-/threonine-specific protein kinases that are involved in the regulation of various cellular responses such as gene expression, mitosis, differentiation, proliferation and survival/apoptosis (Kayama et al. 2009). MAPKs were found to exert control over those genes that stimulate protein synthesis and initiate hypertrophy (Thorburn et al. 1994). When hypertrophy stimulus is initiated at cell membrane, activated MAPKs move through large pores on the nuclear membrane, translocating into the nucleus, and activate transcription factors involved in cardiac hypertrophy (Pearson et al. 2001). MAPK signaling cascade consists of extracellular-regulated kinases (ERK), c-Jun NH2-terminal kinases (JNKs) and p38 MAPK (Sopontammarak et al. 2005). Previous studies analyzing MAPK activities in cardiac hypertrophy have demonstrated differential effects; in that, persistent activation of p38 and JNK can promote apoptosis, resulting in cardiac dilation and dysfunction (Pearson et al. 2001), whereas ERK1/2 has been proposed to regulate smooth muscle contraction and to promote cellular hypertrophy (Modesti et al. 2008; Pearson et al. 2001; Sopontammarak et al. 2005).
Cardiac hypertrophy is also regulated by several transcription factors such as nuclear factor kappa B (NF-κB), myocyte enhancer factor 2 (MEF2) and homeobox transcription factors Csx/Nkx 2-5 (Akazawa and Komuro 2003). Among these transcription factors, NF-κB plays a wide range of physiological and pathophysiological functions, such as B cell proliferation, cell cycle control, carcinogenesis and cardiac hypertrophy (Grabellus et al. 2002). Upon activation by inflammatory mediators and hypertrophy agonist, NF-κB binds to its responsive element sequences, κB, to initiate target gene transcription that is involved in cellular hypertrophy (Leychenko et al. 2011). Genetic NF-κB inhibition attenuates angiotensin II-induced hypertrophy, suggesting an important role of NF-κB in cardiac hypertrophy (Esposito et al. 2002).
Based on the information described above, the possibility that mid-chain HETEs would induce cardiac hypertrophy through MAPK- and NF-κB-dependent pathway has never been investigated before and could not be ruled out. Accordingly, we hypothesize that mid-chain HETEs induce cellular hypertrophy in the human ventricular cardiomyocytes through MAPK- and NF-κB-dependent mechanism. For this purpose, the current study was designed to (a) examine the potential cellular hypertrophy effect of mid-chain HETEs in the human ventricular cardiomyocytes, RL-14 cell line, and (b) explore the role of MAPKs and NF-κB signaling pathways. Our study provides the first evidence that mid-chain HETEs induce cellular hypertrophy in RL-14 cells through MAPK- and NF-κB-dependent mechanism.
Materials and methods
Materials
5-, 12- and 15-HETEs as well as the deuterated metabolites (internal standards) were purchased from Cayman Chemical (Ann Arbor, MI). Dulbecco’s Modified Eagle’s Medium/F-12 (DMEM/F-12), goat IgG peroxidase secondary antibody and CYP4F11 mouse monoclonal primary antibodies were purchased from Sigma Chemical Co. (St. Louis, MO). TRIzol reagent was purchased from Invitrogen Co. (Grand Island, NY). High Capacity cDNA Reverse Transcription kit and SYBR® Green PCR Master Mix were purchased from Applied Biosystem (Foster city, CA). Nitrocellulose membrane was purchased from Bio-Rad Laboratories (Hercules, CA). GAPDH mouse monoclonal primary antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), whereas CYP2B6, 2C8, 2C19 and 4F2 rabbit polyclonal primary antibodies and CYP2J2 mouse monoclonal primary antibody and MAPKs kit were purchased from abcam (abcam, CA). NF-κB Family EZ-TFA Transcription Factor Assay Chemiluminescent Kit was purchased from Millipore (Millipore, Schwalbach/Ts., Germany, #70-660). Chemiluminescence Western blot detection kits were obtained from GE Healthcare Life Sciences (Piscataway, NJ). All other chemicals were purchased from Fisher Scientific Co. (Toronto, ON).
Cell culture and treatments
Human cardiomyocyte RL-14 cells (American Type Cell Culture, Manassas, VA) were maintained in DMEM/F-12, with phenol red supplemented with 12.5 % fetal bovine serum, 20 µM l-glutamine, 100 IU/ml penicillin G and 100 µg/ml streptomycin. Cells were grown in 75 cm2 tissue culture flasks at 37 °C under a 5 % CO2 humidified environment.
The cells were seeded in 12- and 6-well cell culture plates in F12/DMEM culture media for RNA and protein assays, respectively. In all experiments, the cells were treated for the indicated time intervals in serum-free media with various concentrations of mid-chain HETEs as indicated.
Cytotoxicity of mid-chain HETEs
The effect of mid-chain HETEs on RL-14 cells viability was determined by measuring the capacity of reducing enzymes present in only viable cells to convert 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazoliumbromide (MTT) to colored formazan crystals as described previously (Maayah et al. 2013).
RNA extraction and cDNA synthesis
After treatment of RL-14 cells with the test compound for the specified time periods, total cellular RNA was isolated using TRIzol reagent (Invitrogen®) according to the manufacturer’s instructions and quantified by measuring the absorbance at 260 nm. RNA purity was determined by measuring the 260/280 ratio (>1.8). Thereafter, first-strand cDNA synthesis was performed using the High Capacity cDNA reverse transcription kit (Applied Biosystems), according to the manufacturer’s instructions as described previously (Zordoky et al. 2008). Briefly, 1.5 µg of total RNA from each sample was added to a mixture of 2.0 µl of 10× reverse transcriptase buffer, 0.8 µl of 25× dNTP mix (100 mM), 2.0 µl of 10× reverse transcriptase random primers, 1.0 µl of MultiScribe reverse transcriptase and 4.2 µl of nuclease-free water. The final reaction mixture was kept at 25 °C for 10 min, heated to 37 °C for 120 min, heated for 85 °C for 5 min and finally cooled to 4 °C.
Quantification of mRNA expression by real-time polymerase chain reaction (real-time PCR)
Quantitative analysis of specific mRNA expression was performed by real-time PCR by subjecting the resulting cDNA to PCR amplification using 96-well optical reaction plates in the ABI Prism 7500 System (Applied Biosystems) (Maayah et al. 2014). The 25-µl reaction mixture contained 0.1 µl of 10 µM forward primer and 0.1 µl of 10 µM reverse primer (40 nM final concentration of each primer), 12.5 µl of SYBR Green Universal Master mix, 11.05 µl of nuclease-free water and 1.25 µl of cDNA sample. Human primers sequences and probes for CYP2B6, 2C8, 2J2, 4F2, 4F11, brain natriuretic peptide (BNP), atrial natriuretic peptide (ANP), α-myocin heavy chain (α-MHC), β-myocin heavy chain (β-MHC) and β-actin are illustrated in Table 1. These primers were purchased from Integrated DNA Technologies (IDT, Coralville, IA). The fold change in the mRNA expression levels between treated and untreated cells was corrected by the levels of β-actin. The real-time PCR data were analyzed using the relative gene expression (i.e., ∆∆CT) method, as described and explained previously (Livak and Schmittgen 2001). Briefly, the fold change in the level of target genes between treated and untreated cells, corrected by the level of β-actin, was determined using the following equation: fold change = 2−Δ(ΔCt), where ΔCt = Ct(target) − Ct(β-actin) and Δ(ΔCt) = ΔCt(treated) −ΔCt(untreated).
Measurement of cell surface area
Relative changes in cell size, as indicator for hypertrophy, in response to treatments were measured using phase contrast imaging as described previously (Tse et al. 2013). Briefly, RL-14 cells were treated for 24 h with test compounds; thereafter, phase contrast images were taken with Zeiss Axio Observer Z1 inverted microscope using 20× objective lens. Surface area was then quantified by imaging to the complete boundary of individual cells with Zeiss AxioVision Software (Carl Zeiss Imaging Solutions). Five different images have been taken, and twenty cells were counted for each treatment group.
Protein extraction and western blot analysis
Twenty-four hours after incubation with the test compound, approximately 1.5 × 106 cells per 6-well culture plates were collected in 100 µl lysis buffer (50 mM HEPES, 0.5 M sodium chloride, 1.5 mM magnesium chloride, 1 mM EDTA, 10 % glycerol (v/v), 1 % Triton X-100 and 5 µl/ml of protease inhibitor cocktail). Total cellular proteins were obtained by incubating the cell lysates on ice for 1 h, with intermittent vortex mixing every 10 min, followed by centrifugation at 12,000×g for 10 min at 4 °C.
Western blot analysis was performed using a previously described method (Korashy and El-Kadi 2004). Briefly, 25 µg of protein from each treatment group was separated by 10 % sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) and then electrophoretically transferred to nitrocellulose membrane. Protein blots were then blocked overnight at 4 °C in blocking solution containing 0.15 M sodium chloride, 3 mM potassium chloride, 25 mM Tris-base (TBS), 5 % skim milk powder, 2 % bovine serum albumin and 0.5 % Tween-20. After blocking, the blots were washed several times with TBS–Tween-20 before being incubated with primary antibodies, 2B6, 2C8, 2J2, 4F2, 4F11 and GAPDH, for 2 h at room temperature in TBS solution containing 0.05 % (v/v) Tween-20 and 0.02 % sodium azide. Incubation with peroxidase-conjugated secondary antibodies was carried out in blocking solution for 1 h at room temperature. The bands were visualized using the enhanced chemiluminescence method according to the manufacturer’s instructions (GE Healthcare, Mississauga, ON). The intensity of different protein bands was normalized to the signals obtained for GAPDH protein and quantified using ImageJ® image processing program (National Institutes of Health, Bethesda, MD).
Preparation of nuclear extract
Nuclear extracts from RL-14 cells were prepared according to a previously described procedure with minor modifications (Andrews and Faller 1991). Briefly, RL-14 cells were grown on 100-mm petri dishes and treated with vehicle or mid-chain HETEs (20 μM) for 2 h. Thereafter, cells were washed twice with cold PBS, pelleted and suspended in cold buffer A [10 mM Hepes–KOH, 1.5 mm MgCl2, 10 mM KC1, 0.5 mM dithiothreitol and 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] pH 7.9, at 4 °C. After 15 min on ice, the cells were centrifuged at 6,500g and the pellets were suspended again in high salt concentration cold buffer C (20 mM Hepes–KOH, pH 7.9, 25 % glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol and 0.5 mM PMSF) to extract nuclear proteins. The cells were then incubated on ice with vigorous agitation for 60 min followed by centrifugation for 10 min at 12,000g at 4 °C. The nuclear extracts (supernatant) were stored at −80 °C till further use.
Determination of NF-κB binding activity
The NF-κB Family EZ-TFA Transcription Factor Assay Chemiluminescent Kit (Millipore, Schwalbach/Ts., Germany, #70-660) was used according to the manufacturer’s protocol (Bhattacharya et al. 2010).
Determination of MAPKs signaling pathway
MAPKs protein phosphorylation was determined in cytoplasmic protein extracts using the PhosphoTracer ERK1/2 (pT202/Y204) + p38 MAPK (pT180/Y182) + JNK1/2/3 (pT183/Y185) ELISA Kit (Abcam, Cambridge, UK). The kit detects ERK1 and 2, p38 and JNK1, 2 and 3 when phosphorylated on the indicated conserved threonine or tyrosine sites of each protein and was used according to manufacturer’s instructions. Fluorescent data were normalized against total protein concentration from the same sample.
Metabolism of AA by RL-14 cells
To determine whether CYP enzymes in RL-14 cells are metabolically active, cells were plated in 100-mm petri dishes and treated with 20 µM 5-, 12- and 15-HETE for 24 h, and then, the cells were incubated with 50 μM AA for 3 h. AA metabolites were extracted from the media by ethyl acetate and dried using speed vacuum (Savant, Farmingdale, NY). Extracted AA and metabolites were analyzed using liquid chromatography–electrospray ionization–mass spectrometry (LC–ESI–MS) (Waters Micromass ZQ 4000 spectrometer) method as described previously (Zordoky et al. 2010).
sEH catalytic activity
To determine sEH activity in RL-14 cells, cells were plated in 100-mm petri dishes and treated with 20 µM 5-, 12- and 15-HETE for 24 h, and then, the cells were incubated with 5 μM 14, 15-EET for 30 min. 14,15-EET and 14,15-DHET metabolites were extracted from the media by ethyl acetate and dried using speed vacuum (Savant, Farmingdale, NY). Extracted 14,15-EET and 14,15-DHET metabolites were analyzed using liquid chromatography–electrospray ionization–mass spectrometry (LC–ESI–MS) (Waters Micromass ZQ 4000 spectrometer) method as described previously (Zordoky et al. 2010).
Statistical analysis
The comparative analysis of the results from various experimental groups with their corresponding controls was performed using SigmaPlot® for Windows (Systat Software, Inc, CA). One-way analysis of variance (ANOVA) followed by Tukey–Kramer multiple comparison test or Student’s t test was carried out to assess which treatment group(s) showed a significant difference from the control group. The differences were considered significant when p < 0.05.
Results
Effect of mid-chain HETEs on RL-14 cells viability
To determine the maximum nontoxic concentrations of mid-chain HETEs to be utilized in the current study, RL-14, human ventricular cardiomyocytes, cells were exposed for 24 h to wide range concentrations of 5-, 12- and 15-HETEs (0, 0.5, 1, 2.5, 5, 10, 20 and 40 μM). Thereafter, the cell viability was determined using MTT assay. Our results showed that all mid-chain HETEs concentrations ranging from 0.5 to 20 μM did not significantly affect cell viability (Fig. 1). However, mid-chain HETEs 40 µM significantly decreased the cell viability by approximately 40 %. Based on these findings, mid-chain HETEs concentrations 2.5, 5, 10 and 20 µM were selected to be utilized in all subsequent in vitro experiments in RL-14 cells (Fig. 1).
Effect of mid-chain HETEs on cells viability. RL-14 cells were treated for 24 h with various concentrations of 5-, 12- and 15-HETEs (0, 0.5, 1, 2.5, 5, 10, 20 and 40 μM) using MTT assay. Triplicate reactions were performed for each experiment, and the values represent mean of expression levels ±SEM (n = 6)
Concentration- and time-dependent effects of mid-chain HETEs on α-MHC, β-MHC, ANP and BNP mRNA levels
To investigate the capacity of mid-chain HETEs to induce cellular hypertrophy in RL-14 cells, the mRNA expression of hypertrophy markers, α-MHC, β-MHC, ANP and BNP was determined in RL-14 cells treated for 6 h with increasing concentrations of 5-, 12- and 15-HETE (0, 1, 2.5, 5, 10 and 20 μM) using real-time PCR. Figure 2 shows that mid-chain HETEs increased the mRNA expression of α-MHC, β-MHC and BNP in a concentration-dependent manner. The maximum induction of α-MHC was observed at the highest concentration tested, 20 μM, by approximately 3.5-, 1.5- and 3-folds for 5-, 12- and 15-HETE, respectively (Fig. 2a). 5-, 12- and 15-HETE significantly induced β-MHC by approximately 6.5-, 2.5- and 4.5-folds at 20 μM (Fig. 2b). In contrast to 5-HETE, 12- and 15-HETE significantly induced ANP by about 2.5- and 1.5-folds, respectively (Fig. 2c). 5-, 12- and 15-HETE caused a maximum induction of BNP by approximately 2-, 2.5- and 4-folds at 20 μM concentrations, respectively (Fig. 2d).
Concentration-dependent effects of mid-chain HETEs on α-MHC, β-MHC, ANP and BNP mRNA levels. RL-14 cells were treated for 6 h with various concentrations of 5-, 12- and 15-HETEs (0, 0.5, 1, 2.5, 5, 10 and 20 μM). Thereafter, total RNA was isolated using TRIzol reagent, and the mRNA levels of a α-MHC, b β-MHC, c ANP and d BNP were quantified using real-time PCR and normalized to β-actin as a housekeeping gene. Triplicate reactions were performed for each experiment, and the values represent mean of fold change ± SEM (n = 6). + p < 0.05 compared to control (concentration = 0 µM)
To better understand the kinetics of the induction of hypertrophy markers in response to mid-chain HETEs, β-MHC/α-MHC mRNA was measured at various time points (0, 2, 6, 12, 18 and 24 h) following the incubation of RL-14 cells with a single concentration of mid-chain HETEs (20 µM). The concentration of 20 µM mid-chain HETEs was chosen based on their ability to cause maximal induction of the hypertrophy markers (Fig. 3). Thereafter, the mRNA expressions of hypertrophy genes were determined by real-time PCR. The onset of β-MHC/α-MHC induction was detectable as early as 6 h and reached the maximum induction at least 24 h after treatment (Fig. 3).
Time-dependent effects of mid-chain HETEs on β-MHC/α-MHC mRNA levels. RL-14 cells were treated with 20 µM 5-, 12- and 15-HETEs for different time intervals (0, 2, 6, 12, 18 and 24 h). Thereafter, total RNA was isolated using TRIzol reagent, and the mRNA level of β-MHC/α-MHC was quantified using real-time PCR and normalized to β-actin housekeeping gene. Triplicate reactions were performed for each experiment, and the values represent mean of fold change ±SEM (n = 6). + p < 0.05 compared to control (concentration = 0 µM) or (time = 0 h)
Hypertrophy and increase in RL-14 cell surface area by mid-chain HETEs
To determine whether the mid-chain HETE-induced hypertrophy markers at the mRNA (Figs. 2 and 3) were associated with cellular hypertrophy and increase the cell surface area, RL-14 cells were treated for 24 h with mid-chain HETEs 20 µM; thereafter, cell surface area was determined by phase contrast imaging, which was taken with Zeiss Axio Observer Z1 inverted microscope using 20× objective lens. Figure 4 shows that treatment of RL-14 cells for 24 h with mid-chain HETEs significantly increased the percentage of cell surface area by about 50 % as compared with control.
Effect of mid-chain HETEs on cell surface area. RL-14 cells were treated with 20 µM 5-, 12- and 15-HETEs for 24 h. Thereafter, cell surface area was determined by phase contrast images, which were taken with Zeiss Axio Observer Z1 inverted microscope using 20× objective lens. Triplicate measurements were performed for each experiment, and the values represent mean of fold change ±SEM (n = 6). + p < 0.05 compared to control (concentration = 0 µM)
Effect of mid-chain HETEs on NF-κB signaling pathway
To investigate whether mid-chain HETEs trigger NF-κB activation, we tested the capacity of 5-, 12- and 15-HETE to increase NF-κB binding activity using a commercially available kit. Our results showed that positive control, LPS, significantly increased DNA binding activity by 6- and 2.5-fold of induction for P50 and P65, respectively, compared to controls (Fig. 5). Furthermore, all mid-chain HETEs were able to induce the binding activity of NF-κB to its responsive element in a HETE-dependent manner. Figure 5 showed that 12-HETE was the most potent inducer of P50 NF-κB binding activity by approximately 13-fold of induction, whereas 15-HETE caused the highest induction of P65 NF-κB binding activity by about 7.5-fold. 5-HETE significantly induced P50 NF-κB and P65 NF-κB by approximately 3- and 6-fold, respectively.
Effect of mid-chain HETEs on NF-κB signaling pathway. a RL-14 cells were treated with 20 µM 5-, 12- and 15-HETEs for 2 h. Thereafter, NF-κB binding activity was determined using commercially available kit. b RL-14 cells were treated with 10 µM PDTC for 2 h before the addition of mid-chain HETEs for 6 h. Thereafter, total RNA was isolated using TRIzol reagent, and the mRNA level of β-MHC/α-MHC was quantified using real-time PCR and normalized to β-actin as a housekeeping gene. Triplicate reactions were performed for each experiment, and the values represent mean of fold change ±SEM (n = 6). + p < 0.05 compared to control (concentration = 0 µM)
To further confirm the NF-κB-dependent induction of the cellular hypertrophy and cardiac hypertrophy markers in RL-14 cells by mid-chain HETEs, we tested the effect of specific NF-κB inhibitor, pyrrolidinedithiocarbamate (PDTC), on mid-chain HETE-induced β-MHC/α-MHC mRNA. For this purpose, RL-14 cells were treated for 2 h with PDTC (10 µM) before the addition of mid-chain HETEs for 6 h. Thereafter, the mRNA expression of the hypertrophy markers, β-MHC/α-MHC, were measured by real-time PCR. Figure 5b shows that mid-chain HETEs alone caused a significant induction of β-MHC/α-MHC genes. Importantly, pretreatment with PDTC significantly blocked the induction of β-MHC/α-MHC mRNA in response to mid-chain HETEs, suggesting that NF-κB is essential for mid-chain HETE-mediated induction of cellular hypertrophy.
Effect of mid-chain HETEs on MAPK signaling pathway
To assess the role of MAPK signaling pathway on the mid-chain HETE-induced cellular hypertrophy, RL-14 cells were treated for 2 h with 20 µM 5-, 12- and 15-HETE. Thereafter, phosphorylated MAPK levels were determined using a commercially available kit. Figure 6 shows that incubation of the cells with 20 µM of 5-, 12- and 15-HETE significantly induced the phosphorylated ERK1/2 by approximately 2.2-, 2.5- and 2-fold of induction, respectively, whereas no significant changes were observed on phosphorylated P38 or JNK (Fig. 6).
Effect of mid-chain HETEs on MAPKs signaling pathway. a RL-14 cells were treated with 20 µM 5-, 12- and 15-HETEs for 2 h. Thereafter, MAPKs protein phosphorylation was determined in cytoplasmic protein extracts using the PhosphoTracer ERK1/2 (pT202/Y204) + p38 MAPK (pT180/Y182) + JNK1/2/3 (pT183/Y185) ELISA Kit (Abcam, Cambridge, UK). b RL-14 cells were treated with 10 µM U0126 for 2 h before the addition of mid-chain HETEs for 6 h. Thereafter, total RNA was isolated using TRIzol reagent, and the mRNA level of β-MHC/α-MHC was quantified using real-time PCR and normalized to β-actin as a housekeeping gene. Triplicate reactions were performed for each experiment, and the values represent mean of fold change ±SEM (n = 6). + p < 0.05 compared to control (concentration = 0 µM)
To examine whether the induction of the cellular hypertrophy and cardiac hypertrophy markers in RL-14 cells by mid-chain HETEs is ERK1/2-dependent mechanism, we tested the effect of specific ERK1/2 inhibitor, U0126, on mid-chain HETE-induced β-MHC/α-MHC mRNA. For this purpose, RL-14 cells were treated for 2 h with U0126 (10 µM) before the addition of mid-chain HETEs for 6 h. Thereafter, the mRNA expression of the hypertrophy markers, β-MHC/α-MHC, were measured by real-time PCR. Figure 6b shows that mid-chain HETEs alone caused a significant induction of β-MHC/α-MHC genes. Importantly, pretreatment with U0126 significantly blocked the induction of β-MHC/α-MHC mRNA in response to mid-chain HETEs, suggesting that ERK1/2 is essential for mid-chain HETE-mediated induction of cellular hypertrophy.
Effect of mid-chain HETEs on the expression of CYP epoxygenases and ω-hydroxylases mRNA and protein levels in RL-14 Cells
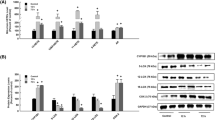
To examine the effect of mid-chain HETEs on the expression of CYP epoxygenases, CYP2B6, CYP2C8, and CYP2J2 and CYP ω-hydroxylases, CYP4F2 and CYP4F11, RL-14 cells were treated with 20 µM 5-, 12- and 15-HETE for 6 h. Thereafter, CYP epoxygenases and ω-hydroxylases genes were measured using real-time PCR. Figures 7a and 8a show that all mid-chain HETEs did not significantly alter the expression of CYP epoxygenases and ω-hydroxylases mRNAs levels.
Effect of mid-chain HETEs on CYP epoxygenases in RL-14 cells. a RL-14 cells were treated with 20 µM 5-, 12- and 15-HETEs for 6 h. Thereafter, total RNA was isolated using TRIzol reagent, and the mRNA levels of CYP epoxygenases were quantified using real-time PCR and normalized to β-actin housekeeping gene. b RL-14 cells were treated for 24 h with 20 µM 5-, 12- and 15-HETEs; thereafter, CYP epoxygenases protein levels were determined by Western blot analysis and detected using the enhanced chemiluminescence method. The intensity of CYP epoxygenases protein bands was normalized to the signals obtained for GAPDH protein and quantified using ImageJ®. Triplicate reactions were performed for each experiment, and the values represent mean of fold change ±SEM (n = 6). + p < 0.05 compared to control (concentration = 0 µM)
Effect of mid-chain HETEs on CYP ω-hydroxylases in RL-14 cells. a RL-14 cells were treated with 20 µM 5-, 12- and 15-HETEs for 6 h. Thereafter, total RNA was isolated using TRIzol reagent, and the mRNA levels of CYP ω-hydroxylases were quantified using real–time PCR and normalized to β-actin housekeeping gene. b RL-14 cells were treated for 24 h with 5-, 12- and 15-HETEs 20 µM; thereafter, CYP ω-hydroxylases protein levels were determined by Western blot analysis and detected using the enhanced chemiluminescence method. The intensity of CYP ω-hydroxylases protein bands were normalized to the signals obtained for GAPDH protein and quantified using ImageJ®. Triplicate reactions were performed for each experiment, and the values represent mean of fold change ±SEM (n = 6). + p < 0.05 compared to control (concentration = 0 µM)
To further examine whether CYP epoxygenases and ω-hydroxylases would be induced in response to mid-chain HETEs at translational level, RL-14 cells were treated for 24 h with 20 µM 5-, 12- and 15-HETE; thereafter, CYP protein expression was determined by Western blot analysis. Figures 7b and 8b show that, in a pattern similar to what was observed with mRNA, all mid-chain HETEs did not significantly alter the expression of CYP epoxygenases and ω-hydroxylases protein levels.
Effect of mid-chain HETEs on CYP-mediated AA metabolism
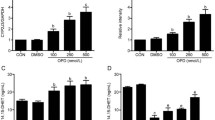
To examine the effect of mid-chain HETEs on CYP-mediated AA metabolism, RL-14 cells were treated with 20 µM 5-, 12- and 15-HETE for 24 h, and then, the cells were incubated with 50 μM AA for 3 h. Thereafter, AA metabolites were measured using LC–ESI–MS. Figure 9A shows that 12-HETE was only able to significantly inhibit the formation of 14,15-EET and 11,12-EET by approximately 60 and 40 %, respectively, compared to control level. On the other hand, 5, 12 and 15-HETE significantly induced the formation of 14,15-DHET by 1.8-, 1.79- and 1.6-fold of induction, respectively, whereas 5- and 12-HETE caused a significant induction of 11,12-DHET by approximately 1.8- and 2-fold of induction, respectively (Fig. 9b).
Effect of mid-chain HETEs on CYP-mediated AA metabolism. RL-14 cells were treated with 20 µM 5-, 12- and 15-HETE for 24 h, and then, the cells were incubated with 50 μM AA for 3 h. Thereafter, a EETs, b DHETs, c 20-HETE were extracted from the media by ethyl acetate and dried using speed vacuum. Reconstituted metabolites were injected into LC–ESI–MS for metabolite determination. Triplicate reactions were performed for each experiment, and the values represent mean ±SEM (n = 6). + p < 0.05 compared to control
To determine the effect of 5-, 12- and 15-HETE on CYP ω-hydroxylases activity, we measured the level of 20-HETE formations. Our results showed that RL-14 all mid-chain HETEs did not significantly change the formation of 20-HETE compared to control level (Fig. 9c).
Role of sEH in mid-chain HETEs mediated effect
To investigate the mechanism responsible for high levels of DHETs in RL-14 cells in response to mid-chain HETEs, the activity level of sEH was determined. Our results showed that incubation of the RL-14 cells with 20 µM of 5-, 12- and 15-HETE caused a significant induction of sEH catalytic activity in RL-14 cells by 2.7-, 4.3- and 4-fold of induction, respectively (Fig. 10).
Role of sEH in mid-chain HETEs mediated effect. a RL-14 cells were treated with 20 µM 5-, 12- and 15-HETE for 24 h, and then, the cells were incubated with 5 μM EETs for 30 min. Thereafter, EETs and DHETs were extracted from the media by ethyl acetate and dried using speed vacuum. Reconstituted metabolites were injected into LC–ESI–MS for metabolite determination. sEH activity was calculated as the ratio of total DHETs/total EETs. b RL-14 cells were treated for 24 h with tAUCB before the addition of mid-chain HETEs for 6 h. Thereafter, total RNA was isolated using TRIzol reagent, and the mRNA level of β-MHC/α-MHC was quantified using real-time PCR and normalized to β-actin as a housekeeping gene. Triplicate reactions were performed for each experiment, and the values represent mean of fold change ± SEM (n = 6). + p < 0.05 compared to control (concentration = 0 µM)
In an attempt to explore whether it is directly involved in the induction of cellular hypertrophy and the cardiac hypertrophy markers, RL-14 cells were treated for 24 h with sEH inhibitor, tAUCB, before the addition of mid-chain HETEs for 6 h. Thereafter, the mRNA expression of the hypertrophy markers, β-MHC/α-MHC, was measured by real-time PCR. Figure 10b shows that mid-chain HETEs alone caused a significant induction of β-MHC/α-MHC genes. Importantly, pretreatment with tAUCB significantly blocked the induction of β-MHC/α-MHC mRNA in response to mid-chain HETEs, suggesting that sEH is essential for mid-chain HETE-mediated induction of cellular hypertrophy.
Discussion
The present work provides the first evidence that mid-chain HETEs typified by 5-, 12- and 15-HETE induced cellular hypertrophy in the human ventricular cardiomyocyte, RL-14 cell line, through MAPK- and NF-κB-dependent mechanism.
Mid-chain HETEs are a biologically active eicosanoids result from the metabolism of AA by both LOX- and CYP-catalyzed bis-allylic oxidation reactions (LOX-like reaction). Recent studies have established the role of mid-chain HETEs in the development of cardiovascular disease (Jenkins et al. 2009). For example, 5-HETE has been reported to have vasoconstrictive and pro-inflammatory actions (Burhop et al. 1988). In addition, 12-HETE has been reported to act as vasoconstrictors in small renal arteries and induce cellular hypertrophy and fibrosis, whereas 15-HETE has been proposed to be implicated in heart failure by induction of cardiac fibrosis. However, most of these previous studies were conducted on vascular smooth muscle, and none of them have utilized in vitro human cardiomyocytes to study the cardiac hypertrophic effect of mid-chain HETEs. In addition, the involvement of NF-κB and MAPKs pathway has never been investigated before in the human cardiomyocytes. Therefore, the current study was conducted to determine the potential cellular hypertrophy effect of mid-chain HETEs in the human ventricular cardiomyocyte, RL-14 cell line, and to explore the role of MAPKs and NF-κB signaling pathway.
One of the hallmarks of cardiac hypertrophy and heart failure in patients is the increase in β-MHC/α-MHC, ANP and BNP expression levels (Barry et al. 2008). Thus, their expressions have been considered as a good predictor of ventricular dysfunction and decompensated heart failure. The ability of mid-chain HETEs to induce cellular hypertrophy, in the current study, is evidenced first by the induction of the cardiac hypertrophy markers β-MHC/α-MHC, ANP and BNP in time- and concentration-dependent manners. Our results are supported by the previous observations that ischemic and dilated cardiomyopathies are associated with the increased level of β-MHC/α-MHC ratio (Reiser et al. 2001). Furthermore, increased ANP and BNP expression levels are associated with heart failure and ventricular hypertrophy induced by isoproterenol (Zordoky et al. 2008). The second evidence for the induction of cellular hypertrophy is the increase in cell surface area. The increase in cell surface area was also reported with hypertrophic agonists treatment, including endothelin and angiotensin II (Gu et al. 2014; Vanezis et al. 2014).
Recent studies have indicated that NF-κB has a wide range of pathophysiological functions of cardiac hypertrophy (Hirotani et al. 2002). In that, NF-κB has been shown to be activated in the failing human heart (Grabellus et al. 2002). Furthermore, it has been demonstrated that blockade of NF-κB ameliorates myocardial hypertrophy in response to aortic banding and chronic infusion of angiotensin II suggesting an important role of NF-κB as a signaling pathway in the regulation of cardiac hypertrophy (Kawano et al. 2005). In this regard, we demonstrated that mid-chain HETEs were able to induce the binding activity of NF-κB to their responsive elements in a HETE-dependent manner. The direct evidence for the involvement of NF-κB in the mid-chain HETE-mediated induction of cellular hypertrophy was supported by the observation that blocking of NF-κB using PDTC significantly resulted in restoration of the mRNA expression of the hypertrophy markers to their normal levels implying that the activation of NF-κB is required for the induction of cardiac hypertrophy. In agreement with our results, it has been shown that activation of NF-κB is required for hypertrophic growth of cardiomyocytes in response to hypertrophic agonist, including phenylephrine, endothelin-1 and angiotensin II (Hirotani et al. 2002). In addition, cardioprotective metabolites, EETs, are reported to have cardioprotective effects through several mechanisms most notably by inhibiting the activation of NF-κB, whereas doxorubicin has been shown to induce myocardial apoptosis through activation of NF-κB (Li et al. 2008; Xu et al. 2006).
MAPKs are intracellular signal transduction factors that are critically involved in the regulation of signaling pathways, ultimately leading to cardiac hypertrophy and heart failure (Zhang et al. 2003). The three best-characterized MAPKs include p38, JNK and ERK1/2. Recent reports demonstrated that persistent activation of p38 and JNK can promote apoptosis, resulting in cardiac dilation and dysfunction (Pearson et al. 2001). ERK1/2 has been proposed to regulate smooth muscle contraction and to promote cellular hypertrophy (Pearson et al. 2001). Importantly, the exact role of MAPK in mid-chain HETE-induced cardiac hypertrophy is still unclear. Thus, the second objective of the current study was to explore the role of MAPK in mid-chain HETE-induced cardiac hypertrophy. Our results showed that mid-chain HETE significantly induced the phosphorylated ERK1/2, whereas no significant changes were observed on phosphorylated p38 or JNK. The direct involvement of ERK1/2 was assessed by determining the effects of the ERK1/2 inhibitor, U0126. Our results clearly demonstrated that the activation of ERK1/2 signaling pathway positively regulates the induction of the cardiac hypertrophy markers in response to mid-chain HETEs. The premise of this observation emerges from the fact that the activation of phosphorylated ERK1/2 is crucial for cellular hypertrophy. This is supported by the finding that angiotensin II induced cellular hypertrophy in H9c2 cells through ERK1/2 but not p38 or JNK (Zong et al. 2013). Of particular interest in this study, baicalin, LOX inhibitor, blocked the cellular hypertrophy effect of angiotensin II through ERK1/2 signaling pathway (Zong et al. 2013).
CYP enzymes have been detected in the cardiovascular tissue, and their specific isoforms have been detected in rat heart (Imaoka et al. 2005) and in different regions of human heart (Delozier et al. 2007; Roman 2002). Several lines of evidence support the role of CYP metabolites in the maintenance of cardiovascular health, including the regulation of vascular tone, extracellular fluid volume, heart contractility and cardiac hypertrophy (Roman 2002). Therefore, we investigated the effect of mid-chain HETEs on the expression of different CYP genes involved in cardiac hypertrophy. Our results demonstrated that all mid-chain HETEs did not significantly alter the expression of CYP ω-hydroxylases and epoxygenases at mRNA and protein levels. However, we found a significant decrease in 14, 15- and 11, 12-EETs formation in RL-14 cells treated with 12-HETE. This decrease in EETs formation was accompanied by a significant increase in the formation of their corresponding DHETs. Therefore, it was necessary to investigate the effect of mid-chain HETEs on the activity of sEH, which promotes the conversion of EETs to their corresponding inactive diols form, DHETs (Imig et al. 2002; Yu et al. 2000). Interestingly, our results clearly demonstrated that mid-chain HETEs caused a significant induction of sEH catalytic activity in RL-14 cells, which explain the high level of DHETs. Importantly, the mid-chain HETE-induced cellular hypertrophy was significantly attenuated by pretreatment of RL-14 cells with selective sEH inhibitor, tAUCB, suggesting that sEH is essential for mid-chain HETE-mediated induction of cellular hypertrophy. The induction of sEH and DHETs has been reported to increase in animal models of angiotensin II- and isoproterenol-induced cardiac hypertrophy (Ai et al. 2009; Zordoky et al. 2008). Moreover, sEH inhibitors have been shown to prevent and/or reverse the development of cardiac hypertrophy in several models (Xu et al. 2006).
In conclusion, our study provides the first evidence that mid-chain HETEs induce cellular hypertrophy in the human ventricular cardiomyocytes RL-14 cells through MAPK- and NF-κB-dependent mechanism and could be used as a novel targets in the treatment of cardiac hypertrophy and heart failure.
References
Ai D, Pang W, Li N et al (2009) Soluble epoxide hydrolase plays an essential role in angiotensin II-induced cardiac hypertrophy. Proc Natl Acad Sci USA 106(2):564–569. doi:10.1073/pnas.0811022106
Akazawa H, Komuro I (2003) Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res 92(10):1079–1088. doi:10.1161/01.RES.0000072977.86706.23
Althurwi HN, Tse MM, Abdelhamid G, Zordoky BN, Hammock BD, El-Kadi AO (2013) Soluble epoxide hydrolase inhibitor, TUPS, protects against isoprenaline-induced cardiac hypertrophy. Br J Pharmacol 168(8):1794–1807. doi:10.1111/bph.12066
Andrews NC, Faller DV (1991) A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res 19(9):2499
Barry SP, Davidson SM, Townsend PA (2008) Molecular regulation of cardiac hypertrophy. Int J Biochem Cell Biol 40(10):2023–2039. doi:10.1016/j.biocel.2008.02.020
Bhattacharya N, Sarno A, Idler IS et al (2010) High-throughput detection of nuclear factor-kappaB activity using a sensitive oligo-based chemiluminescent enzyme-linked immunosorbent assay. Int J Cancer J Int du Cancer 127(2):404–411. doi:10.1002/ijc.25054
Burhop KE, Selig WM, Malik AB (1988) Monohydroxyeicosatetraenoic acids (5-HETE and 15-HETE) induce pulmonary vasoconstriction and edema. Circ Res 62(4):687–698
Capdevila J, Chacos N, Werringloer J, Prough RA, Estabrook RW (1981) Liver microsomal cytochrome P-450 and the oxidative metabolism of arachidonic acid. Proc Natl Acad Sci USA 78(9):5362–5366
Cyrus T, Witztum JL, Rader DJ et al (1999) Disruption of the 12/15-lipoxygenase gene diminishes atherosclerosis in apo E-deficient mice. J Clin Investig 103(11):1597–1604. doi:10.1172/JCI5897
Delozier TC, Kissling GE, Coulter SJ et al (2007) Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos 35(4):682–688. doi:10.1124/dmd.106.012823
Elshenawy OH, Anwar-Mohamed A, El-Kadi AO (2013) 20-Hydroxyeicosatetraenoic acid is a potential therapeutic target in cardiovascular diseases. Curr Drug Metab 14(6):706–719
Esposito G, Rapacciuolo A, Naga Prasad SV et al (2002) Genetic alterations that inhibit in vivo pressure-overload hypertrophy prevent cardiac dysfunction despite increased wall stress. Circulation 105(1):85–92
Fava C, Ricci M, Melander O, Minuz P (2012) Hypertension, cardiovascular risk and polymorphisms in genes controlling the cytochrome P450 pathway of arachidonic acid: a sex-specific relation? Prostaglandins Other Lipid Mediat 98(3–4):75–85. doi:10.1016/j.prostaglandins.2011.11.007
Gonzalez-Nunez D, Claria J, Rivera F, Poch E (2001) Increased levels of 12(S)-HETE in patients with essential hypertension. Hypertension 37(2):334–338
Grabellus F, Levkau B, Sokoll A et al (2002) Reversible activation of nuclear factor-kappaB in human end-stage heart failure after left ventricular mechanical support. Cardiovasc Res 53(1):124–130
Gross GJ, Falck JR, Gross ER, Isbell M, Moore J, Nithipatikom K (2005) Cytochrome P450 and arachidonic acid metabolites: role in myocardial ischemia/reperfusion injury revisited. Cardiovasc Res 68(1):18–25. doi:10.1016/j.cardiores.2005.06.007
Gu S, Zhang W, Chen J et al (2014) EPC-derived microvesicles protect cardiomyocytes from Ang II-induced hypertrophy and apoptosis. PLOS One 9(1):e85396. doi:10.1371/journal.pone.0085396
Hirotani S, Otsu K, Nishida K et al (2002) Involvement of nuclear factor-kappaB and apoptosis signal-regulating kinase 1 in G-protein-coupled receptor agonist-induced cardiomyocyte hypertrophy. Circulation 105(4):509–515
Imaoka S, Hashizume T, Funae Y (2005) Localization of rat cytochrome P450 in various tissues and comparison of arachidonic acid metabolism by rat P450 with that by human P450 orthologs. Drug Metab Pharmacokinet 20(6):478–484
Imig JD, Zhao X, Capdevila JH, Morisseau C, Hammock BD (2002) Soluble epoxide hydrolase inhibition lowers arterial blood pressure in angiotensin II hypertension. Hypertension 39(2 Pt 2):690–694
Jenkins CM, Cedars A, Gross RW (2009) Eicosanoid signalling pathways in the heart. Cardiovasc Res 82(2):240–249. doi:10.1093/cvr/cvn346
Kawano S, Kubota T, Monden Y et al (2005) Blockade of NF-kappaB ameliorates myocardial hypertrophy in response to chronic infusion of angiotensin II. Cardiovasc Res 67(4):689–698. doi:10.1016/j.cardiores.2005.04.030
Kayama Y, Minamino T, Toko H et al (2009) Cardiac 12/15 lipoxygenase-induced inflammation is involved in heart failure. J Exp Med 206(7):1565–1574. doi:10.1084/jem.20082596
Korashy HM, El-Kadi AO (2004) Differential effects of mercury, lead and copper on the constitutive and inducible expression of aryl hydrocarbon receptor (AHR)-regulated genes in cultured hepatoma Hepa 1c1c7 cells. Toxicology 201(1–3):153–172. doi:10.1016/j.tox.2004.04.011
Leychenko A, Konorev E, Jijiwa M, Matter ML (2011) Stretch-induced hypertrophy activates NFkB-mediated VEGF secretion in adult cardiomyocytes. PLoS One 6(12):e29055. doi:10.1371/journal.pone.0029055
Li S, Mingyan E, Yu B (2008) Adriamycin induces myocardium apoptosis through activation of nuclear factor kappaB in rat. Mol Biol Rep 35(4):489–494. doi:10.1007/s11033-007-9112-4
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. doi:10.1006/meth.2001.1262
Maayah ZH, El Gendy MA, El-Kadi AO, Korashy HM (2013) Sunitinib, a tyrosine kinase inhibitor, induces cytochrome P450 1A1 gene in human breast cancer MCF7 cells through ligand-independent aryl hydrocarbon receptor activation. Arch Toxicol 87(5):847–856. doi:10.1007/s00204-012-0996-y
Maayah ZH, Ansari MA, El Gendy MA, Al-Arifi MN, Korashy HM (2014) Development of cardiac hypertrophy by sunitinib in vivo and in vitro rat cardiomyocytes is influenced by the aryl hydrocarbon receptor signaling pathway. Arch Toxicol 88(3):725–738. doi:10.1007/s00204-013-1159-5
Modesti PA, Serneri GG, Gamberi T et al (2008) Impaired angiotensin II-extracellular signal-regulated kinase signaling in failing human ventricular myocytes. J Hypertens 26(10):2030–2039. doi:10.1097/HJH.0b013e328308de68
Nozawa K, Tuck ML, Golub M, Eggena P, Nadler JL, Stern N (1990) Inhibition of lipoxygenase pathway reduces blood pressure in renovascular hypertensive rats. Am J Physiol 259(6 Pt 2):H1774–H1780
Pearson G, Robinson F, Beers Gibson T et al (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22(2):153–183. doi:10.1210/edrv.22.2.0428
Reiser PJ, Portman MA, Ning XH, Schomisch Moravec C (2001) Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am J Physiol Heart Circ Physiol 280(4):H1814–H1820
Roman RJ (2002) P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev 82(1):131–185. doi:10.1152/physrev.00021.2001
Sasaki M, Hori MT, Hino T, Golub MS, Tuck ML (1997) Elevated 12-lipoxygenase activity in the spontaneously hypertensive rat. Am J Hypertens 10(4 Pt 1):371–378
Schwartzman ML, da Silva JL, Lin F, Nishimura M, Abraham NG (1996) Cytochrome P450 4A expression and arachidonic acid omega-hydroxylation in the kidney of the spontaneously hypertensive rat. Nephron 73(4):652–663
Sopontammarak S, Aliharoob A, Ocampo C, Arcilla RA, Gupta MP, Gupta M (2005) Mitogen-activated protein kinases (p38 and c-Jun NH2-terminal kinase) are differentially regulated during cardiac volume and pressure overload hypertrophy. Cell Biochem Biophys 43(1):61–76. doi:10.1385/CBB:43:1:061
Thorburn J, McMahon M, Thorburn A (1994) Raf-1 kinase activity is necessary and sufficient for gene expression changes but not sufficient for cellular morphology changes associated with cardiac myocyte hypertrophy. J Biol Chem 269(48):30580–30586
Tse MM, Aboutabl ME, Althurwi HN, Elshenawy OH, Abdelhamid G, El-Kadi AO (2013) Cytochrome P450 epoxygenase metabolite, 14,15-EET, protects against isoproterenol-induced cellular hypertrophy in H9c2 rat cell line. Vascul Pharmacol 58(5–6):363–373. doi:10.1016/j.vph.2013.02.004
Vanezis A, Thaitirarot C, Butt M, Squire I, Samani N, Rodrigo G (2014) 208 The PKC epsilon/AMPK ALPHA/ENOS pathway is implicated as a mechanism by which remote ischaemic conditioning attenuates endothelin-1 mediated cardiomyocyte hypertrophy. Heart 100(Suppl 3):A114. doi:10.1136/heartjnl-2014-306118.208
Wallukat G, Morwinski R, Kuhn H (1994) Modulation of the beta-adrenergic response of cardiomyocytes by specific lipoxygenase products involves their incorporation into phosphatidylinositol and activation of protein kinase C. J Biol Chem 269(46):29055–29060
Wu CC, Schwartzman ML (2011) The role of 20-HETE in androgen-mediated hypertension. Prostaglandins Other Lipid Mediat 96(1–4):45–53. doi:10.1016/j.prostaglandins.2011.06.006
Xu D, Li N, He Y et al (2006) Prevention and reversal of cardiac hypertrophy by soluble epoxide hydrolase inhibitors. Proc Natl Acad Sci USA 103(49):18733–18738. doi:10.1073/pnas.0609158103
Yousif MH, Benter IF, Roman RJ (2009) Cytochrome P450 metabolites of arachidonic acid play a role in the enhanced cardiac dysfunction in diabetic rats following ischaemic reperfusion injury. Auton Autacoid Pharmacol 29(1–2):33–41. doi:10.1111/j.1474-8673.2009.00429.x
Yu Z, Xu F, Huse LM et al (2000) Soluble epoxide hydrolase regulates hydrolysis of vasoactive epoxyeicosatrienoic acids. Circ Res 87(11):992–998
Zhang W, Elimban V, Nijjar MS, Gupta SK, Dhalla NS (2003) Role of mitogen-activated protein kinase in cardiac hypertrophy and heart failure. Exp Clin Cardiol 8(4):173–183
Zong J, Zhang DP, Zhou H et al (2013) Baicalein protects against cardiac hypertrophy through blocking MEK-ERK1/2 signaling. J Cell Biochem 114(5):1058–1065. doi:10.1002/jcb.24445
Zordoky BN, El-Kadi AO (2010) Effect of cytochrome P450 polymorphism on arachidonic acid metabolism and their impact on cardiovascular diseases. Pharmacol Ther 125(3):446–463. doi:10.1016/j.pharmthera.2009.12.002
Zordoky BN, Aboutabl ME, El-Kadi AO (2008) Modulation of cytochrome P450 gene expression and arachidonic acid metabolism during isoproterenol-induced cardiac hypertrophy in rats. Drug Metab Dispos 36(11):2277–2286. doi:10.1124/dmd.108.023077
Zordoky BN, Anwar-Mohamed A, Aboutabl ME, El-Kadi AO (2010) Acute doxorubicin cardiotoxicity alters cardiac cytochrome P450 expression and arachidonic acid metabolism in rats. Toxicol Appl Pharmacol 242(1):38–46. doi:10.1016/j.taap.2009.09.012
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research [Grant 106665] to A.O.S.E. Z.H.M. is the recipient of University of Alberta Doctoral Recruitment Scholarship. We are grateful to Dr. Vishwa Somayaji for technical assistance with LCMS.
Conflict of interest
There is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Maayah, Z.H., El-Kadi, A.O.S. 5-, 12- and 15-Hydroxyeicosatetraenoic acids induce cellular hypertrophy in the human ventricular cardiomyocyte, RL-14 cell line, through MAPK- and NF-κB-dependent mechanism. Arch Toxicol 90, 359–373 (2016). https://doi.org/10.1007/s00204-014-1419-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00204-014-1419-z