Abstract
A white-coloured, rod-shaped, motile, aerobic, and Gram-stain-positive bacterial strain S3N08T was isolated from agricultural soil. The strain grew at temperature 10–40 °C, at 0–1.0% (w/v) NaCl concentration, and at pH 6.5–8.0. Catalase was negative and oxidase was positive. The phylogenetic analysis inferred that the strain S3N08T belonged to the genus Paenibacillus, with the closest relative being Paenibacillus periandrae PM10T (95.6% 16S rRNA gene sequence similarity). The only menaquinone was MK-7 and the major polar lipids were phosphatidylmonomethylethanolamine, phosphatidylglycerol, and phosphatidylethanolamine. The predominant fatty acids were antiso-C15:0, C16:0, and iso-C15:0. The DNA G + C content was 45.1%. The average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values between strain S3N08T and with closest members were < 72.0% and < 19.0%, respectively. Altogether, the phylogenetic, genomics, phenotypic, and chemotaxonomic evidence illustrated in this study suggested that strain S3N08T represents a novel species of the genus Paenibacillus, for which the name Paenibacillus agricola sp. nov. is proposed. The type strain is S3N08T (= KACC 19666 T = NBRC 113430 T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Paenibacillus, belonging to the phylum Bacillota, was described by Ash et al. in 1993, with the type species Paenibacillus polymyxa (Ash et al. 1993; Tindall 2000). At the time of preparing the manuscript, this genus accommodates a total of 293 species with a validly published and correct names (https://lpsn.dsmz.de/genus/paenibacillus). The members of Paenibacillus have been reported from diverse sources including rhizosphere, phyllosphere, hot spring, root nodules, freshwater wetland, necrotic wound, soil, water, food, faeces, and insects (Baik et al. 2011; Glaeser et al. 2013; Menendez et al. 2016; Kämpfer et al. 2022; Wang et al. 2022). Recently, Paenibacillus rhizolycopersici (Thin et al. 2023) and Paenibacillus sabuli (Gao et al. 2022) have been retrieved from tomato plant and sea environment. Based on polyphasic approach, this study aimed to characterize and assign the taxonomic position of strain S3N08T, which was isolated from agricultural soil and proposed the name as Paenibacillus agricola sp. nov.
Materials and methods
Isolation of strain
Strain S3N08T was isolated from an agricultural soil (rice paddy field) sample collected from Seongju-gun, Republic of Korea (GPS coordinates: 35°53′56.8′′N 128°14′04.9′′E) using the standard dilution plating technique on R2A medium (MB Cell, South Korea). After plating, all the plates were incubated at 28 °C for 7 days. A white-pigmented colonies of bacterial strain, S3N08T was obtained after repeated streaking in R2A agar. The pure colonies of strain S3N08T were then stored at -80 °C as a suspension in R2A broth with 20% (w/v) glycerol for long-term preservation. The strain S3N08T was deposited in the Korean Agricultural Culture Collection and NITE Biological Resource Center.
16S rRNA gene sequence and phylogenetic analysis
Genomic DNA from strain S3N08T was extracted using commercial DNA extraction kit (InstaGene Matrix, Bio-Rad, USA). The 16S rRNA gene was amplified using forward (27F) and reverse (1492R) primers (Frank et al. 2008). The sequencing and analysis of amplified 16S rRNA gene was performed as described previously (Chaudhary et al. 2017). The nearest phylogenetic members were concluded by evaluating 16S rRNA gene sequence in the the EzBioCloud server (Yoon et al. 2017b). The 16S rRNA gene sequences of all the phylogenetically affiliated species were downloaded from EzBioCloud database and aligned with SINA (v1.2.11) according to the SILVA seed alignment (https://www.arb-silva.de) (Pruesse et al. 2012). The generation of phylogenetic trees was accomplished by maximum–likelihood (ML) (Felsenstein 1981) and neighbour–joining (NJ) (Saitou and Nei 1987) algorithms using MEGA X software (Kumar et al. 2018). The topology of the phylogenetic trees was evaluated by the bootstrap resampling method with 1000 replicates (Felsenstein, 1985). Evolutionary distances were determined by Kimura’s two-parameter model (Kimura 1980).
Genome analysis
The genome of strain S3N08T was sequenced by Illumina MiSeq platform and assembled by SPAdes ver. 3.14.1 (Macrogen, South Korea). The quality control and contamination of the sequenced genome were investigated using ContEst16S algorithm (Lee et al. 2017). The annotation of the assembled genome sequence was performed by Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al. 2016) and Rapid Annotation Subsystem technology (RAST server) (Aziz et al. 2008). The putative secondary metabolites were detected with the program antiSMASH 5.0 (Blin et al. 2019). The whole genome similarities between strain S3N08T and the closest members (Paenibacillus periandrae PM10T, Paenibacillus vulneris CCUG 53270T, Paenibacillus rigui JCM 16352T, Paenibacillus phytorum LMG 31458T, and Paenibacillus alginolyticus DSM 5050T) were assessed by average nucleotide identity (ANI) tool using OrthoANIu algorithm (Yoon et al. 2017a) and Genome-to-Genome Distance Calculator (Meier-Kolthoff et al. 2013). The phylogenomic tree was generated on the Type (Strain) Genome Server (Meier-Kolthoff and Göker 2019) using FastME 2.1.6.1 tools (Lefort et al. 2015).
Morphological, physiological, and biochemical analysis
For morphological studies, colony properties were determined by observing the colonies of strain S3N08T grown on R2A agar for 7 days at 28 °C. Cellular structure and flagella was visualized using transmission electron microscopy (TEM; Talos L120C; FEI). Gram reaction was performed using Color Gram 2 kit (bioMérieux). Endospore formation was observed by phase-contrast microscopy using BX53-DIC microscope (Olympus) after staining with 0.5% (w/v) malachite green (Oktari et al. 2017). Motility was evaluated in sulphide indole motility medium (SIM; Oxoid). Catalase and oxidase tests were conducted using ID Color Catalase and Oxidase Reagents (bioMérieux), respectively. The anaerobic growth was analysed by cultivating strain S3N08T on R2A agar for 15 days at 28 °C in an anaerobic jar with an anaerobe atmosphere generation bag. The growth at various temperatures (4, 10, 15, 20, 25, 28, 35, 37, 40, and 45 °C) was assessed on R2A agar after 10 days incubation. NaCl tolerance was investigated in R2A broth formulated with different NaCl content (0–1.5%, w/v, at 0.5% intervals). Growth at various pH 0.5–10.0 (at intervals of 0.5 pH unit) was studied in R2A broth. The pH of the medium was adjusted prior autoclaving using suitable buffers (Breznak and Costilow 2007). Casein, DNA, and starch hydrolysis tests were conducted as described (Smibert 1994). Other various biochemical studies were executed using commercial kits API ZYM, API 20NE, and API 20E (bioMérieux).
Chemotaxonomic characterization
The cellular fatty acids profile was determined after cultivating target and reference strains on R2A agar at 28 °C. Fatty acids were extracted from biomass of all strains harvested at late log phase. The extraction, analysis, and identification of fatty acids were accomplished by MIDI protocol (Sasser 1990). The peptidoglycan was analyzed as described previously (Staneck and Roberts 1974). Both polar lipids and quinones were analysed using freeze-dried cells (Collins and Jones 1981; Minnikin et al. 1984).
Results and discussion
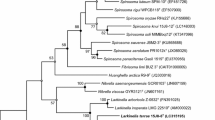
The nearly full-length of 16S rRNA gene of strain S3N08T was 1481 bp (NCBI nucleotide accession number MH159222). The 16S rRNA gene sequence similarity data showed that the strain S3N08T was affiliated to the genus Paenibacillus, with phylogenetically closest members being P. periandrae PM10T (95.6%), P. vulneris CCUG 53270T (94.5%), P. rigui JCM 16352T (94.1%), P. phytorum LMG 31458T (93.1%), and P. alginolyticus DSM 5050T (92.3%). The 16S rRNA gene sequence similarities of strain S3N08T with all closest species were below the cut-off values of < 98.7% used for species delineation (Stackebrandt 2006; Yarza et al. 2008). Furthermore, the phylogenetic trees (ML and NJ) found that strain S3N08T formed a clade with P. periandrae PM10T with high bootstrap values (Figs. 1 and S1). Overall, the 16S rRNA gene sequence and phylogenetic results supported the assignment of strain S3N08T as a novel species in the genus Paenibacillus.
Maximum likelihood tree constructed using 16S rRNA gene sequences of strain S3N08T and closely affiliated taxa. Filled circles represent nodes recovered by both phylogenetic trees (maximum-likelihood and neighbor-joining). The numbers at the nodes indicate the percentage of 1,000 bootstrap replicates (values > 70% are only illustrated). NCBI nucleotide accession numbers are illustrated in parentheses. Geobacillus stearothermophilus IFO 12550T was used as an out-group. The scale bar represents 0.05 substitutions per nucleotide position
The quality control and contamination test of genome sequence data assured that the generated genome sequence was valid to the strain S3N08T. The genome size of strain S3N08T was 8,375,108 bp. The entire genome sequence was assembled in 76 contigs with N50 value of 319,977 bp and genome coverage of 122.6x (Table S1). The annotation of genome executed in the RAST program revealed 316 subsystem features (Fig. S2). The annotated genome data showed various functional genes and proteins related to synthesis of plant growth promoting factors (Table S2). The genome of S3N08T comprised tryptophan synthase enzymes for auxin biosynthesis (GenBank accession numbers: NHN28712.1 and NHN28713.1) (Kriechbaumer and Glawischnig 2005), ammonium transporter for ammonia assimilation (GenBank accession numbers: NHN29230.1 and NHN31246.1), and iron-siderophore protein for iron acquisition (GenBank accession numbers: NHN30782.1 and NHN31881.1). These functional proteins of strain S3N08T help to enhance the plant growth activities (Grady et al. 2016). The biosynthetic gene clusters (BGCs) assessment revealed several genes in the genome of strain S3N08T that are responsible for putative secondary metabolites (terpene, cyclic-lactone-autoinducer, and type III polyketide synthase) (Table S3).
The DNA G + C content analyzed from genome data was 45.1%. The ANI and digital DNA–DNA hybridization (dDDH) values between strain S3N08T and reference strains were in the ranges of 69.9–76.1% and 19.1–26.9%, respectively (Table S4). These genome relatedness data were below than the threshold values of ANI (95.0%) and dDDH (70.0%), indicating that the strain S3N08T differs genomically from closest species of the genus Paenibacillus (Wayne et al. 1987; Richter and Rosselló-Móra 2009). The phylogenomic tree generated with genome sequence data assured the affiliation of strain S3N08T with the genus Paenibacillus, forming clade with P. periandrae PM10T, P. phytorum LMG 31458T, and P. alginolyticus DSM 5050T, but generating separate lineage (Fig. S3).
Cells of strain S3N08T were Gram-stain-positive, and rod shaped with peritrichous flagella (Fig. S4). Catalase test was negative for S3N08T and P. periandrae LMG 28691T and positive for P. vulneris DSM 27954T and P. rigui KCTC 13282T. Oxidase test was positive for strain S3N08T and all reference strains except P. alginolyticus KACC 11445T. Strain S3N08T was able to grow at 10 °C and can tolerate 1.0% NaCl. Nitrate reduction was negative for S3N08T, P. periandrae LMG 28691T, P. vulneris DSM 27954T and P. alginolyticus KACC 11445T, but positive for P. rigui KCTC 13282T and P. phytorum LMG 31458T. Alkaline phosphatase and acid phosphatase were positive for strain S3N08T, whereas most of the other enzymatic and assimilation tests were negative. Leucine arylamidase, D-glucose, L-arabinose, and malic acid were negative for strain S3N08T, but were positive for all reference strains. Several other distinguishing phenotypic features of strain S3N08T are presented in the species description and illustrated along with closest reference strains in Table 1. The detail enzymatic and assimilation properties obtained from API commercial test kits are provided in Table S5.
The only menaquinone detected in strain S3N08T was menaquinone MK-7, which was consistent with other species of the genus Paenibacillus (Baik et al. 2011; Kämpfer et al. 2022). Phosphatidylmonomethylethanolamine (PME), phosphatidylglycerol (PG), and phosphatidylethanolamine (PE) were major polar lipids found in strain S3N08T. Additionally one unidentified phospholipid (PL1) was observed as minor polar lipid (Fig. S5). The polar lipids profiles were identical with other species of the genus Paenibacillus (Glaeser et al. 2013; Menendez et al. 2016). The predominant fatty acids of strain S3N08T were antiso-C15:0 (54.9%), C16:0 (13.2%), and iso-C15:0 (11.2%). The major fatty acids contents of strain S3N08T were similar with closely related reference strains. However, some proportional differences were observed with minor fatty acids composition between strain S3N08T and reference strains. Strain S3N08T reported C17:0 cyclo (1.2%) which was absent from reference strains (Table 2).
Taxonomic conclusion
In overall, the polyphasic taxonomic data provided in this study confirmed that the strain S3N08T represents a novel species in the genus Paenibacillus for which the name Paenibacillus agricola sp. nov. is proposed.
Description of Paenibacillus agricola sp. nov.
Paenibacillus agricola sp. nov. [a.gri′co.la. L. masc. n. ager field, L. suff. cola (from L. n. incola) a dweller, inhabitant, L. masc. n. agricola field dwelling].
Cells (3.3–4.5 × 0.7–0.9 µm) are Gram-stain-positive, aerobic, rod-shaped, and motile with flagella. Colonies on R2A agar are white, convex, circular, smooth, and translucent with 1.0–1.5 mm in diameter. Cells grow at temperature 10–40 °C (optimum, 25–28 °C), at pH 6.5–8.0 (optimum, 6.5), and at 0–1.0% NaCl concentration (optimum without NaCl). Endospores are formed in a sub-terminal position. Negative for catalase and positive for oxidase tests. Aesculin is hydrolysed, but gelatine, urea, casein, DNA, and starch are not hydrolysed. Positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, naphthol-AS-BI-phosphohydrolase, and β-galactosidase. The cells assimilate d-maltose and produce acetoin. The only menaquinone is MK-7; principal polar lipids are PME, PG, and PE; and main cellular fatty acids are antiso-C15:0, C16:0, and iso-C15:0. The DNA G + C content of the type strain is 45.1%.
The type strain, S3N08T (= KACC 19666 T = NBRC 113430 T), was isolated from an agricultural soil in Seongju-gun, Republic of Korea.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA sequence and whole genome shotgun sequence of strain S3N08T are MH159222 and JAAOIW000000000, respectively.
Data availability
The GenBank/EMBL/DDBJ accession number for the 16S rRNA sequence of strain S3N08T is MH159222. The whole genome shotgun sequence of strain S3N08T has been deposited at DDBJ/ENA/GenBank under the accession JAAOIW000000000. The version described in this paper is version JAAOIW010000000.
Abbreviations
- KACC:
-
Korean agricultural culture collection
- KCTC:
-
Korean collection for type cultures
- LMG:
-
Laboratorium voor microbiologie
- DSM:
-
Deutsche Sammlung von Mikroorganismen
- NBRC:
-
NITE biological resource center
- ANI:
-
Average nucleotide identity
- dDDH:
-
Digital DNA-DNA hybridization
References
Ash C, Priest FG, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test: proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64:253–260. https://doi.org/10.1007/BF00873085
Aziz RK et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
Baik KS, Lim CH, Choe HN, Kim EM, Seong CN (2011) Paenibacillus rigui sp. nov., isolated from a freshwater wetland. Int J Syst Evol Microbiol 61:529–534. https://doi.org/10.1099/ijs.0.021485-0
Blin K et al (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. https://doi.org/10.1093/nar/gkz310
Breznak JA, Costilow RN (2007) Physicochemical factors in growth. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Synder LR (eds) Methods gen mol microbiol, 3rd edn. American Society of Microbiology, Washinton, DC, pp 309–329
Chaudhary DK, Dahal RH, Altankhuu K, Kim J (2017) Ravibacter arvi gen. nov., sp. nov., isolated from farmland soil during development of new culture techniques. Int J Syst Evol Microbiol 67:5252–5260. https://doi.org/10.1099/ijsem.0.002456
Collins MD, Jones D (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45:316–354. https://doi.org/10.1128/mr.45.2.316-354.1981
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ (2008) Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74:2461–2470. https://doi.org/10.1128/AEM.02272-07
Gao Q-j, Mo K-l, Hu Y-h, Liu Z-y, Huang H-q (2022) Paenibacillus sabuli sp. Nov., isolated from the South China Sea. Int J Syst Evol Microbiol. 72:005568. https://doi.org/10.1099/ijsem.0.005568
Glaeser SP, Falsen E, Busse H-J, Kämpfer P (2013) Paenibacillus vulneris sp. nov., isolated from a necrotic wound. Int J Syst Evol Microbiol 63:777–782. https://doi.org/10.1099/ijs.0.041210-0
Grady EN, MacDonald J, Liu L, Richman A, Yuan Z-C (2016) Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Factories 15:1–18. https://doi.org/10.1186/s12934-016-0603-7
Kämpfer P et al (2022) Paenibacillus allorhizoplanae sp. nov. from the rhizoplane of a Zea mays root. Arch Microbiol 204:630. https://doi.org/10.1007/s00203-022-03225-w
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kriechbaumer V, Glawischnig E (2005) Auxin biosynthesis within the network of tryptophan metabolism. J Nano Bio Tech 2:53–58
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547. https://doi.org/10.1093/molbev/msy096
Lee I, Chalita M, Ha S-M, Na S-I, Yoon S-H, Chun J (2017) ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol 67:2053–2057. https://doi.org/10.1099/ijsem.0.001872
Lefort V, Desper R, Gascuel O (2015) FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol 32:2798–2800. https://doi.org/10.1093/molbev/msv150
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Menendez E et al (2016) Paenibacillus periandrae sp. nov., isolated from nodules of Periandra mediterranea. Int J Syst Evol Microbiol 66:1838–1843. https://doi.org/10.1099/ijsem.0.000953
Minnikin DE et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Oktari A, Supriatin Y, Kamal M, Syafrullah H (2017) The bacterial endospore stain on schaeffer fulton using variation of methylene blue solution. In: Journal of Physics: Conference Series. IOP Publishing. p 012066
Pruesse E, Peplies J, Glöckner FO (2012) SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. https://doi.org/10.1093/bioinformatics/bts252
Qi SS, Cnockaert M, Carlier A, Vandamme PA (2021) Paenibacillus foliorum sp. nov., Paenibacillus phytohabitans sp. nov., Paenibacillus plantarum sp. nov., Paenibacillus planticolens sp. nov., Paenibacillus phytorum sp. nov., and Paenibacillus germinis sp. nov., isolated from the Arabidopsis thaliana phyllosphere. Int J Syst Evol Microbiol 71:004781. https://doi.org/10.1099/ijsem.0.004781
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sasser M (1990) Bacterial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME). MIDI technical note 101. Midi Inc, Newark
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997) Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Evol Microbiol 47:289–298. https://doi.org/10.1099/00207713-47-2-289
Smibert R (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Stackebrandt E (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231. https://doi.org/10.1128/am.28.2.226-231.1974
Tatusova T et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1093/nar/gkw569
Thin KK, He S-W, Ma R, Wang X, Han J-G, Zhang X-X (2023) Paenibacillus rhizolycopersici sp. nov., an oligotrophic bacterium isolated from a tomato plant in China. Int J Syst Evol Microbiol 73:005698. https://doi.org/10.1099/ijsem.0.005698
Tindall B (2000) What is the type species of the genus Paenibacillus? Request for an opinion. Int J Syst Evol Microbiol 50:939–940. https://doi.org/10.1099/00207713-50-2-939
Wang J et al (2022) Paenibacillus hamazuiensis sp. nov., a bacterium isolated from Hamazui hot spring in Yunnan province, south-west China. Arch Microbiol 204:1–5. https://doi.org/10.1007/s00203-022-03282-1
Wayne LG et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464. https://doi.org/10.1099/00207713-37-4-463
Yarza P et al (2008) The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol 31:241–250. https://doi.org/10.1016/j.syapm.2008.07.001
Yoon S-H, Ha S-m, Lim J, Kwon S, Chun J (2017a) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Yoon S-H et al (2017b) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Acknowledgements
This work was supported by Rural Development Administration, South Korea (Project No. PJ014897032023).
Author information
Authors and Affiliations
Contributions
HL and DK designed the study. HL, DKC and OBL contributed the experimental work, data analysis, and original draft preparation. HL and DKC reviewed and finalized the manuscript. DK supervised the project.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest regarding the publication of this manuscript.
Ethical approval
This study does not describe any experimental work related to human and animal.
Consent to publication
All the authors have given their consent for submission and publication of this manuscript to ‘Archives of Microbiology’.
Additional information
Communicated by Yusuf Akhter.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA sequence and whole genome shotgun sequence of strain S3N08T are MH159222 and JAAOIW000000000, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, H., Chaudhary, D.K., Lim, O.B. et al. Paenibacillus agricola sp. nov., isolated from agricultural soil. Arch Microbiol 205, 248 (2023). https://doi.org/10.1007/s00203-023-03578-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-023-03578-w