Abstract
A novel sulfate-reducing bacterium, strain PPLLT, was isolated from marsh soil. Cells of strain PPLLT were rod-shaped with length of 1.5 μm and width of 0.7 μm. Growth was observed at 22–37 °C (optimum 35 °C) and pH 6.8–8.4 (optimum 7.3). Lactate, succinate, fumarate, formate and malate were utilized as electron donors for sulfate reduction. Fermentative growth was not observed on tested organic acids. Besides sulfate, sulfite, thiosulfate and elemental sulfur were utilized as electron acceptors. Hydrogen is used only in the presence acetate or yeast extract. The major fatty acid was C16:0. The complete genome of strain PPLLT was composed of a circular chromosome with length of 4.2 Mbp and G + C content of 57.7 mol%. Sequence analysis of the 16S rRNA gene showed that strain PPLLT was affiliated with the genus Desulfofustis in the family Desulfocapsaceae. On the basis of differences in the phylogenetic and phenotypic properties between the strain and the type strain of the genus Desulfofustis, strain PPLLT (DSM 110475T = JCM 39161T) is proposed as the type strain of a new species, with name of Desulfofustis limnaeus sp. nov.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Desulfofustis contains the sole species Desulfofustis glycolicus, which can degrade glycolate and glyoxylate with sulfate respiration (Friedrich et al. 1996). The glycolate is mainly produced as an intermediate of photosynthesis, and, therefore, Desulfofustis species may occur in cyanobacterial mats in marine brackish ecosystem (Galushko and Kuever 2019). The strain PerGlyST, the type strain of D. glycolicus, is a mesophilic and neutrophilic sulfate-reducing bacterium isolated with glycolate from anoxic marine mud in Italy. Based on the genome-based phylogeny (Parks et al. 2018), the genus Desulfofustis was proposed to be reclassified in the family Desulfocapsaceae within the order Desulfobulbales in the class Desulfobulbia (Waite et al. 2020). In this study, a novel freshwater sulfate-reducing bacterium designated as the strain PPLLT was isolated and characterized, as a representative of a new species within the genus Desulfofustis.
Materials and methods
Enrichment and isolation
Strain PPLLT was isolated from soil of a marsh in Hokkaido, Japan (43° 04′ 08′′ N 141° 31′ 11′′ E). From the same site, a spore-forming sulfate-reducing bacterium was isolated in a previous study (Watanabe et al. 2017). Approximately, 1 ml of the soil slurry was inoculated into 40 ml bicarbonate-buffered sulfide-reduced defined basal medium containing sulfate (Widdel and Bak 1992) to establish the first enrichment culture. One milliliter of 2% (v/v) p-xylene solution [in 2, 2, 4, 4, 6, 8-heptamethylnonane, which served as carrier phase (Rabus et al. 1993)] was added to 40 ml of the medium as a sole carbon and energy source. The headspace of the bottle was filled with N2/CO2 (80: 20, v/v), and incubation was carried out in the dark at 28 °C. The grown enrichment culture was subjected to agar shake dilution with 2% (v/v) p-xylene solution as described in a previous study (Higashioka et al. 2009). After several months, a beige colony was picked and inoculated into the fresh medium same as for the first enrichment. Subsequently, the grown culture was transferred and diluted to extinction in the bottle of fresh medium filled with H2 + CO2 gasses (H2/N2/CO2, 50: 40: 10; 2 atm total pressure). After the growth of enrichment, the substrate was changed to lactate (5 mM). After colony isolation from the agar tube dilution using 5 mM lactate, a single colony of strain PPLLT was obtained. The purity of culture was ascertained routinely by microscopy and verified by denaturing gradient gel electrophoresis of the 16S rRNA gene (Muyzer et al. 1993) for cultures used to perform physiological tests.
Phenotypic characterization
In experiments for phenotypic characterizations, strain PPLLT was cultured at 35 °C in the basal medium supplemented with 5 mM lactate and 0.05% (w/v) yeast extract, unless otherwise specified. Cell morphology was confirmed by phase-contrast microscopy (Axioplan 2; Zeiss). The fatty acid profile was carried out by the identification services of Techno Suruga Laboratory with the Sherlock Microbial Identification System (MIDI) version 6.0 (database; MOORE6). For cellular fatty acid analysis, strain PPLLT was grown in the basal medium supplemented with 10 mM lactate and 0.05% (w/v) yeast extract.
Effect of temperature on growth was examined by culturing at 18, 22, 25, 28, 32, 35, 37, 42, 45 and 50 °C. In the test of pH effect on growth, bicarbonate in the basal medium was replaced with MES, MOPS, or TAPS (20 mM). The pH of the modified media was adjusted with NaOH, to pH 5.8, 6.2, 6.4, 6.5 and 6.8 (MES), pH 6.5, 7.0, 7.3, 7.5 and 7.8 (MOPS), pH 7.8, 8.0, 8.2, 8.4, 8.6, 8.8 and 9.0 (TAPS). Effect of salt concentration on growth was examined by culturing with various concentration of NaCl, ranging from 0 to 3.0% (w/v) at 0.5% intervals.
Utilization of growth substrates was tested with the basal medium, supplemented with one of the following substrates (mM; unless otherwise specified); formate (5), acetate (5), propionate (5), lactate (5), glycolate (5), butyrate (2), isobutyrate (2), malate (5), succinate (5), fumarate (5), benzoate (2), citrate (5), methanol (5), ethanol (5), glucose (5) and yeast extract (0.05% w/v). Hydrogen-dependent growth was tested under a gas mixture of H2, N2 and CO2 (50:40:10 v/v/v, 200 kPa total pressure), with or without supplement of acetate (1 mM) or yeast extract (0.05%). Fermentative growth was tested in sulfate-free medium supplemented with lactate (5), succinate (5), malate (5), pyruvate (5) or fumarate (5). As electron accepters, thiosulfate (10), elemental sulfur (0.5% w/v), sulfite (1 and 5), nitrate (10) and poorly crystalline Fe (III) oxide (20) were tested. The production of acetate during sulfate reduction was assessed using acetate colorimetric assay kit (Sigma-Aldrich) and supernatant of well-grown culture supplemented with 5 mM lactate.
Genomic characterization and phylogenetic analysis
Whole-genome sequencing was performed using the platforms of Illumina NextSeq and Nanopore GridION. Short and long reads from the platforms were subjected to hybrid assembly using Unicycler (Ver 0.4.7). The assembled genome sequence was annotated with DFAST web pipeline (Tanizawa et al. 2018). In order to evaluate genomic distance between strain PPLLT and related organisms, values of average amino acid identity (AAI) (Rodriguez R and Konstantinidis 2014) and percentage of conserved proteins (POCP) (Qin et al. 2014) were calculated, using EzAAI version 1.0 (Kim et al. 2021) and modified scripts based on data_file_4.sh (Rodriguez-R and Konstantinidis 2016), respectively. The Genome-to-Genome Distance Calculator (GGDC) calculation was carried out using GGDC 3.0 server (Meier-Kolthoff et al. 2022). For reconstruction of metabolic pathway, eggNOG-mapper version 2.1.6 annotation server (Cantalapiedra et al. 2021), NCBI BLASTP analysis was utilized for amino acid sequences of strain PPLLT genome. The phylogenetic affiliation based on genome-based taxonomy of strain PPLLT was investigated using GTDB-Tk version 1.5.0 (Chaumeil et al. 2020) with the GTDB database release 202. The 16S rRNA gene sequence of strain PPLLT was retrieved from the complete genome and was aligned with reference sequences using the program CLUSTAL X version 2.1 (Larkin et al. 2007). Selection of the best nucleotide substitution models and construction of phylogenetic trees were performed using MEGA version 10.0.5 (Kumar et al. 2018).
Results
Physiological and chemotaxonomic characteristics
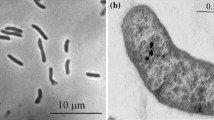
Cells of strain PPLLT were non-motile, rod-shaped, 0.7 μm in width, 1.5 μm in length. In the cellular fatty acid profile, C16: 0 accounting for 32.7% was predominant. Other major components accounting > 10% of total were C18:1ω7c summarized with unknown acids (17.0%), C16: 1ω7c (14.4%) and C16: 1ω5c (13.3%).
Strain PPLLT grew at 22–37 °C with optimum growth at 35 °C, and grew at pH range of 6.8–8.4 with the optimum pH of 7.2. No growth was observed in the presence of 0.5% or higher concentrations of NaCl. In the presence of sulfate, formate, lactate, succinate, fumarate and malate supported heterotrophic growth of PPLLT. Growth on H2 gas was observed only in the presence of acetate or yeast extract. Sulfate, sulfite, thiosulfate and elemental sulfur were used as electron acceptor. Nitrate and Fe (III) oxide did not support growth of strain PPLLT as electron acceptor. Fermentative growth was not observed with the tested substrates. Acetate production was observed under sulfate-reducing conditions with lactate.
The complete genome of strain PPLLT was reconstructed as a 4,232,141 bp of circular chromosome with 57.7 mol% of G + C content. It was predicted to contain 3843 protein-coding genes, 50 tRNA genes, and 9 rRNA genes. Based on the metabolic pathway reconstruction, gene sets for anaerobic alkylbenzene degradation were not identified in the genome of strain PPLLT. The genome possesses all genes for complete Wood–Ljungdahl pathway, although strain PPLLT did not grow autotrophically. Strain PPLLT did not use nitrate as electron acceptor, but its genome has narGHI and nrfHA genes, that are the key components for dissimilatory nitrate reduction to ammonia. In addition, nirK and norB genes, that are essential genetic components for nitrate reduction to nitrous oxide were also identified. The gene of nosZ was not found in the genome. As key enzyme for glycolate oxidation, membrane-associated methylene-blue-dependent glycolate dehydrogenase (E.C. 1.1.99.14) is encoded in the genome of D. glycolicus PerGlyST, but its gene was not found in the genome of strain PPLLT.
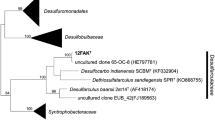
In the genome-based taxonomy according to the GTDB, strain PPLLT was classified into the genus Desulfofustis, but not into any existing species. The POCP value between strain PPLLT and D. glycolicus PerGlyST was 73.1%, within the genus boundary range of 60–80%. The AAI between them was 75.8%, which is clearly higher than proposed threshold for genera delineation, 50%. These genomic distance calculations suggested that the strain PPLLT should be a new species of the genus Desulfofustis. Based on the 16S rRNA gene sequence, the sequence similarity between strain PPLLT and its closest relative D. glycolicus PerGlyST was 97.2% (compared length: 1,406 nucleotide positions). This value is lower than 98.7–99% of the threshold value for species delineation between two bacterial strains (Stackebrandt 2006). In addition, a low ANI value between these strains (78.8%) was shown in the analysis with GTDB-tk. An ANI value of 95% is widely utilized for the species demarcation, and therefore the result indicated that these strains should not be classified to same species. The GGDC value between these species was 20.4% and was lower than 70% of species boundary, which is also supported that these strains can be discriminated at a species level. The phylogenetic tree reconstruction based on the 16S rRNA gene sequences revealed that strain PPLLT could be a representative of a novel species of the genus Desulfofustis in the family Desulfocapsaceae (Fig. 1).
Maximum-likelihood tree based on 16S rRNA gene sequences of strains PPLLT and reference strains of the family Desulfocapsaceae. A total of 1477 positions were used in the final dataset. Desulfobulbus propionicus DSM 2032 T was used as an outgroup. The phylogenetic tree was inferred using the maximum likelihood method and Kimura 2-parameter model + Gamma distributed with Invariant sites. Bootstrap values (percentages of 1000 replications) only 50% or more are shown at nodes
The differential properties between strain PPLLT and D. glycolicus PerGlyST are shown in Table 1. The utilization of electron acceptors and donors indicated that these strains have different metabolic characteristics. In addition, these strains differ in tolerance and requirement of NaCl. The type strain of D. glycolicus does not grow in freshwater medium, whereas strain PPLLT grows only under freshwater conditions. Based on the phenotypic, genomic and phylogenetic analysis, we conclude that the strain PPLLT represents a novel species of the genus Desulfofustis for which the name D. limnaeus sp. nov. is proposed.
Description of Desulfofustis limnaeus sp. nov.
Desulfofustis limnaeus (lim’nae.us. Gr. Masc. adj. limnaîos, living in limnic waters, swamps; N.L. masc. adj. limnaeus, living in freshwaters and swamps).
In addition to the characteristics given in the description of the genus, the following properties are observed. Growth occurs at 22–37 °C (optimum 35 °C), at pH 6.8–8.4 (optimum pH 7.2), and with 0% NaCl. Grows on lactate, succinate, formate, fumarate and malate; acetate, propionate, glycolate, citrate, butyrate, iso-butyrate, glucose and alcohols are not utilized. Uses sulfate, sulfite, thiosulfate and elemental sulfur as an electron acceptor; nitrate and Fe (III) are not utilized. Growth on H2 gas is observed only in the presence of yeast extract or acetate. Fermentative growth is not observed. Predominant fatty acid is C16:0. The G + C content of genomic DNA of the type strain is 57.7 mol%. The type strain PPLLT (= DSM 110475 = JCM 39161) was isolated from marsh soil. The GenBank/EMBL/DDBJ accession number for the complete genome and the 16S rRNA gene sequence of strain PPLLT are AP025516 and LC702440.
Data availability
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene and genome sequence of strain PPLLT are LC702440 and AP025516. The genome sequence of the strain PPLL was published in public database.
References
Cantalapiedra CP, Hernández-Plaza A, Letunic I et al (2021) eggNOG-mapper v2: functional annotation, orthologyassignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38(12):5825–5829
Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH (2020) GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz848
Friedrich M, Springer N, Ludwig W, Schink B (1996) Phylogenetic positions of Desulfofustis glycolicus gen. nov., sp. nov., and Syntrophobotulus glycolicus gen. nov., sp. nov., two new strict anaerobes growing with glycolic acid. Int J Syst Bacteriol 46:1065–1069. https://doi.org/10.1099/00207713-46-4-1065/CITE/REFWORKS
Galushko A, Kuever J (2019) Desulfofustis. In: DeVos P, Dedysh S, Hedlund B, Kämpfer P, Rainey F, Trujillo ME, Bowman J, Brown DR, Glöckner FO, Oren A, Paster BJ, Wade W, Ward N, Busse HJ, Reysenbach AL, Whitman WB (eds) Bergey’s manual of systematic bacteriology. Wiley, Hoboken
Higashioka Y, Kojima H, Fukui M (2009) Isolation and characterization of novel sulfate-reducing bacterium capable of anaerobic degradation of p-xylene. Microbes Environ 27(3):273–277
Kim D, Park S, Chun J (2021) Introducing EzAAI: a pipeline for high throughput calculations of prokaryotic average amino acid identity. J Microbiol 59:476–480
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal w and Clustal x version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M (2022) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Parks DH, Chuvochina M, Waite DW et al (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996. https://doi.org/10.1038/nbt.4229
Qin Q-L, Xie B-B, Zhang X-Y et al (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215
Rabus R, Nordhaus R, Ludwig W, Widdel F (1993) Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol 59:1444–1451
Rodriguez-R LM, Konstantinidis KT (2014) Bypassing cultivation to identify bacterial species. Microbe 9:111–118
Rodriguez-R LM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints, UK
Stackebrandt E (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 6:152–155
Tanizawa Y, Fujisawa T, Nakamura Y (2018) DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34:1037–1039
Waite DW, Chuvochina M, Pelikan C et al (2020) Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol 70(11):5972–6016
Watanabe M, Kojima H, Fukui M (2017) Desulfocucumis palustris gen. nov., sp. nov., a mesophilic sulfate reducer belonging to Desulfotomaculum subcluster Ig. Int J Syst Evol Microbiol 67:2679–2682. https://doi.org/10.1099/ijsem.0.002005
Widdel F, Bak F (1992) Gram-negative mesophilic sulfate-reducing bacteria. The Prokaryotes, vol 4, 2nd edn. Springer-verlag, New York, pp 3352–3378
Acknowledgements
We thank A. Shinohara for technical assistance. Genome analysis was partially performed on the NIG supercomputer at ROIS National Institute of Genetics
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Watanabe, M., Takahashi, A., Kojima, H. et al. Desulfofustis limnaeus sp. nov., a freshwater sulfate-reducing bacterium isolated from marsh soil. Arch Microbiol 204, 647 (2022). https://doi.org/10.1007/s00203-022-03261-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03261-6