Abstract
A novel marine Gram-stain-negative, aerobic, rod-shaped bacterium, designated as strain PS1T, was isolated from the deep-sea sediments of the Mariana Trench and characterized phylogenetically and phenotypically. Bacterial optimal growth occurred at 35 °C (ranging 10–45 °C), pH 6 (ranging pH 5–10) and with 11% (w/v) NaCl (ranging 0–17%). The 16S rRNA gene sequence similarity results revealed that strain PS1T was most closely related to Pseudomonas stutzeri ATCC 17588T, Pseudomonas nitrititolerans GL14T, Pseudomonas zhaodongensis NEAU-ST5-21T, Pseudomonas xanthomarina DSM 18231T and Pseudomonas kunmingensis HL22-2T with 98.3–98.7%. The digital DNA–DNA hybridization values and the average nucleotide identity between strain PS1T and the reference strains were 20.4–40.1% and 78.7–79.4%, respectively. The major respiratory quinone is ubiquinone Q-9. The major polar lipids were phosphatidylethanolamine, diphosphatidyglycerol, phosphatidylglycerol, phosphatidylcholine, aminoglycolipid, two unidentified glycolipids and one unidentified lipid. The predominant cellular fatty acids of strain PS1T were summed feature 8 (C18:1ω7c and/or C18:1ω6c), summed feature 3 (C16:1ω7c and/or C16:1ω6c), C16:0 and cyclo-C19:0 ω8c. The G + C content of the genomic DNA was 63.0%. The combined genotypic and phenotypic data indicated that strain PS1T represents a novel species of the genus Pseudomonas, for which the name Pseudomonas marianensis sp. nov. is proposed, with the type strain PS1T (= DSM 112238T = MCCC 1K05112T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Pseudomonas, first proposed by Migula (1984), represents a group of Gram-stain-negative bacteria that are aerobic, non-spore-forming, catalase-positive, oxidase-positive, motile and rod shaped (Migula 1984; Timmis 2002). Ubiquinone Q-9 is the major respiratory quinone and summed feature 8 (C18:1ω7c and/or C18:1ω6c) is the major fatty acid component in this genus. The DNA G + C content of bacteria of the genus ranges from 58 to 69 mol% (Wei et al. 2018; Garrity et al. 2005). At the time of writing (April 2022), the genus currently comprises more than 299 species (http://www.bacterio.net/pseudomonas.html). They play important roles in the different marine environments, including seawater (Pascual et al. 2012; Wang et al. 2016; Yoshida et al. 2015), sediments (Carrión et al. 2021; Tamegai et al. 1997), and marine animals (Romanenko et al. 2008). Some of the species have also been isolated from the Challenger Deep of the Mariana Trench (Wei et al. 2018; Tamegai et al. 1997; Quigley and Colwell 1968) and the Japan Trench (Yoshida et al. 2015). Here, we report the taxonomic characteristics of a novel Pseudomonas species isolated from deep-sea sediments of the Mariana Trench.
Materials and methods
Isolation and cultivation
Strain PS1T was isolated from a deep-sea sediment from the Mariana Trench (11.33 °N, 142.2 °E) on January 12, 2020. The sample was serially diluted and spread on marine agar 2216. After incubation for 3 days at 28 °C under aerobic conditions, strain PS1T was selected, and pure culture was obtained after three successive transfers to fresh medium. The strain was routinely cultivated on marine broth 2216 (MB; Difco) under aerobic conditions and stored at −80 °C in liquid medium supplemented with 20% (v/v) glycerol. Pseudomonas stutzeri DSM 5190T, Pseudomonas zhaodongensis DSM 27559T and Pseudomonas xanthomarina DSM 18231T were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). Pseudomonas nitrititolerans CGMCC 1.13874T and Pseudomonas kunmingensis CGMCC 1.12273T were obtained from the China General Microbiological Culture Collection Center (CGMCC). All strains were cultured under comparable conditions as experimental control strains.
Phenotypic characteristics
Gram reactions were carried out according to previously established procedures (Wei et al. 2018). Cell morphology, size and the presence of flagella were determined using transmission electron microscopy (TEM-1230, JEOL, Japan). Motility was observed using the hanging-drop method described by Skerman (1967). Anaerobic growth was evaluated in 10% MB in the presence of NaNO3 (10 mM), prepared with a N2 gas phase (200 kPa) in sealed sterile vials and incubated at 35 °C for 14 days. The temperature ranges for growth were determined in MB incubated at > 45 °C for 7 days and at 10–45 °C for 3 days. Tolerance to NaCl was evaluated in an artificial marine broth medium according to the MB formula, except for the modification of NaCl concentrations to be 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 13, 14, 15, 16 or 18% (w/v) (Wei et al. 2018). The pH ranges for growth were evaluated in MB adjusted to pH 2.0–11.0, at 1 pH unit intervals, with citrate/phosphate (pH 2.0–7.0), Tris/HCl (pH 8.0–9.0) or sodium carbonate/sodium bicarbonate (pH 10.0–12.0) buffers. Catalase activity was evaluated by the production of oxygen bubbles in 3% (v/v) H2O2 and oxidase activity was determined using oxidase reagent (bioMérieux). Hydrolysis of starch, casein, DNA, gelatin, and Tween 80 was tested according to the methods described by Smibert and Krieg (1994). Production of fluorescent pigments was tested on King B medium (King et al. 1954). Other phenotypic characteristics, such as the hydrolysis of starch were determined according to the methods described by Tindall et al. (2007). Other biochemical tests were carried out using API 20NE, API ZYM (both from bioMérieux) and GEN III MicroPlate kit (Biolog) according to the manufacturer’s instructions, except for adjusting the NaCl concentration in all tests to 3.0%.
Molecular analysis
Genomic DNA was extracted using the Fast DNA SPIN Kit for Soil (Qbiogene, MP Biomedicals, Illkirch, France) according to the manufacturer’s instructions. The 16S rRNA gene was amplified by PCR using universal primer pair 27F (5′-GAGAGTTTGATCCTGGCTCAG-3′)/1492R (5′-GTCGTAACAAGGTAGCCGT A-3′). Purified PCR product was ligated to pUCm-T Vector (Sangon Biotech) and cloned according to the manufacturer’s instructions. Sequence similarity was determined using the EzTaxon-e server (http://www.ezbiocloud.net/) (Kim et al. 2012). Phylogenetic trees based on 16S rRNA gene sequences were constructed using MEGA version X (Kumar et al. 2018). Distances were calculated using the Kimura two-parameter model and clustering was performed with the neighbor-joining (NJ) (Saitou and Nei 1987) and maximum likelihood (ML) (Felsenstein 1981). Bootstrap analysis based on 1000 replications was used to estimate the confidence level of tree topologies and the complete deletion option was implemented.
DNA–DNA relatedness
The draft genome sequence of strain PS1T was sequenced at Shanghai Majorbio Bio-pharm Technology (Shanghai, China) using Solexa paired-end (500 bp library) sequencing technology. The de novo assembly of the reads was performed using SOAPdenovo v2.04. The genome sequences of P. stutzeri ATCC 17588T (CP002881), P. nitrititolerans GL14T (RFFL01000000), P. zhaodongensis NEAU-ST5-21T (RFFM00000000), P. xanthomarina DSM 18231T (FQXA00000000) and P. kunmingensis HL22-2T (FORS00000000) were obtained from the NCBI database. The DNA–DNA hybridization (DDH) estimate value was analyzed using the genome to genome distance calculator (GGDC2.0) (Goris et al. 2007; Meier-Kolthoff et al. 2013). The average nucleotide identity (ANI) among the three genomes was calculated using JSpecies (V1.2.1) as described by Richter and Rosselló-Móra (2009). The G + C content of the genomic DNA was determined from the draft genome sequence. A whole-genome-based phylogenomic tree was also reconstructed based on the whole-genome nucleotide sequences using the Type (Strain) Genome Server (TYGS) (Meier-Kolthoff and Göker 2019).
Chemotaxonomy
For cellular fatty acid analysis, strains PS1T and the reference strains were cultured in MB for 3 days at 35 °C. Fatty acids were saponified, methylated and extracted using the standard protocol of MIDI (Sherlock Microbial Identification System, version 6.0) and identified by using the RTSBA6.0 database of the Microbial Identification System (Sasser 1990). Polar lipids of strain PS1T were extracted and separated on silica gel 60 F254 aluminium-backed thin-layer plates (10 × 10 cm; Merk 5554) which had dried for 30 min at 55 °C and further analyzed according to Minnikin et al. (1984). The first dimension of the solvent system was chloroform/methanol/water (65:24:4, by vol.) and the second dimension was chloroform/glacial acetic acid/methanol/water (80:15:12:4, by vol.). Then the plates were sprayed with 5% phosphomolybdic acid (w/v, dissolved in alcohol) and heated at 160 °C for 10–15 min to reveal total lipids. Other reagents such as a-naphthol, ninhydrin and molybdenum blue (sigma) were used to detect glycolipids, aminolipids and phospholipids according to Tindall (1990). The respiratory quinones were extracted using the method described by Minnikin et al. (1984) and analyzed by HPLC as described by Tindall (1990).
Results and discussion
Morphological and physiological characteristics
Cells of strain PS1T were Gram-stain-negative, motile by single polar flagellum, rod-shaped, non-pigmented, 0.4–0.5 μm in width and 1.0–1.9 μm in length (Supplementary Fig. S1). Cells grow in the presence of 0–17% (w/v) (11% optimum) NaCl, at 10–45 °C (35 °C optimum) and pH 5–10 (pH 6 optimum). Differentiating phenotypic characteristics of strain PS1T from its closest phylogenetic neighbours are given in Table 1. All negative phenotypic traits of strain PS1T are given in Supplementary Tab. S1.
Molecular analysis
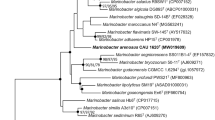
The nearly complete 16S rRNA gene sequence (1483 nt) of strain PS1T was obtained. As shown in Fig. 1, strain PS1T, P. stutzeri ATCC 17588T and P. nitrititolerans GL14T formed an independent monophyletic cluster. High sequence similarities were observed between the isolate and its closest relatives, P. stutzeri ATCC 17588T (98.7%), followed by P. nitrititolerans GL14T (98.6%), P. zhaodongensis NEAU-ST5-21T (98.5%), P. xanthomarina DSM 18231T (98.3%) and P. kunmingensis HL22-2T (98.3%). Sequence similarities between strain PS1T and other type strains were less than 98.0%. The high similarity of the 16S rRNA gene sequence to those of the nearest related type strains confirms that strain PS1T belongs to the genus Pseudomonas. The whole-genome-based phylogenomic tree showed that strain PS1T, P. zhaodongensis NEAU-ST5-21T and P. xanthomarina DSM 18231T formed a tight cluster to its most closely related strains within the genus Pseudomonas (Supplementary Fig. S2).
Neighbour-joining tree based on 16S rRNA gene sequences, showing the phylogenetic position of strain PS1T and related members within the genus Pseudomonas. Solid circles indicate that the corresponding nodes (groupings) are also recovered in maximum-likelihood trees. Cellvibrio ostraviensis LMG 19434T (GenBank accession number: AJ493583) was used as an outgroup. Bootstrap values (expressed as percentages of 1000 replications) of above 50% are shown at the branch points. Bar, 0.01 substitutions per nucleotide position (MEGA X)
The genomic DNA G + C content of strain PS1T was 63.0% according to its draft genome sequence. The genomic G + C content of members of the genus is 58–69% (Wei et al. 2018; Garrity et al. 2005). The DDH estimate values between strain PS1T and its most closely related strains were below 35%, which are far below the cutoff values recommended for bacterial species delineation (Thompson et al. 2013; Wayne et al. 1987). The ANI values between strain PS1T and its most closely related strains were below 80%, which are below the standard criteria for classifying strains as the same species (95–96%) (Richter and Rosselló-Móra 2009). Thus, DDH and ANI both confirm that strain PS1T represents a novel species.
Chemotaxonomic characteristics
The major cellular fatty acids of strain PS1T were summed feature 8 (C18:1ω7c and/or C18:1ω6c) (23.5%), C16:0 (21.8%), summed feature 3 (C16:1ω7c and/or C16:1ω6c) (15.4%) and cyclo-C19:0 ω8c (12.4%) (Supplementary Tab. S2). In strain PS1T, C10:0 3-OH (3.4%), C11:0 3-OH (0.7%) and C12:0 3-OH (3.6%) were present, which are the common characteristics of the genus Pseudomonas (Timmis 2002; Wei et al. 2018). Strain PS1T could be differentiated from its five closest phylogenetic type strains by fatty acid compositions, as detailed in Supplementary Tab. S2, such as the much larger amount of cyclo-C19:0 ω8c (12.4%).
The major polar lipids found in strain PS1T were phosphatidylethanolamine, diphosphatidyglycerol, phosphatidylglycerol, phosphatidylcholine, aminoglycolipid, two unidentified glycolipids and one unidentified lipid (Supplementary Fig. S3). Menaquinone was extracted using the method described by Minnikin et al. (1984) and analyzed by HPLC as described by Tindall (1990). The major respiratory quinone is ubiquinone Q-9, which was consistent with other species of the genus Pseudomonas (Wei et al. 2018).
On the basis of phylogenetic, phenotypic and chemotaxonomic characterization, strain PS1T shares many common characteristics with the closely related species. However, there are some the differences in physiological, biochemical and chemotaxonomic characteristics among them (Table 1), as well as low values of DDH and ANI. It is proposed that strain PS1T is classified as a novel species of the genus Pseudomonas, for which the name Pseudomonas marianensis sp. nov. is proposed.
Description of Pseudomonas marianensis sp. nov.
Pseudomonas marianensis (ma.ri.an.en´sis. N.L. fem. adj. marianensis pertaining to the Mariana Trench, the source of the type strain).
Cells are Gram-stain-negative, rod-shaped, non-pigmented, 0.4–0.5 μm in width and 1.0–1.9 μm in length and are motile by single polar flagellum. Colonies are circular, smooth, non-pigmented, whitish and transparent when incubated on MA. Fluorescent pigment production was not observed on King B media. The cells grow in the presence of 0–17% (w/v) (11% optimum) NaCl and at 10–45 °C, pH 5–10. The genomic DNA G + C content of the type strain is 63.0%. Positive for oxidase and catalase activities; negative for nitrate reduction and hydrolysis of Tweens 40, 80, gelatin and starch; positive for arginine dihydrolase and urease. Assimilates d-glucose, l-arabinose, d-mannitol, d-maltose, potassium gluconate, capric acid, malic acid and trisodium citrate. In the API ZYM strip, positive for alkaline phosphatase, esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, acid phosphatase and naphthol-AS-BI-phosphohydrolase. When assayed with the GEN III MicroPlate kit, positive for utilization of α-cyclodextrin, l-arabinose, d-arabitol, maltose, d-melibiose, xylitol, mono-methyl-succinate, cis-aconitic acid, d-gluconic acid, α-hydroxy butyric acid, β-hydroxy butyric acid, α-keto valeric acid, d,l-lactic acid, propionic acid, d-saccharic acid, sebacic acid, succinic acid, succinamic acid, glucuronamide, d-alanine, l-alanine, l-alanyl-glycine, l-aspartic acid, l-glutamic acid, hydroxy-l-proline, l-ornithine, l-phenylalanine, d-serine, l-serine, d,l-carnitine, γ-amino butyric acid, urocanic acid, thymidine, 2-aminoethanol, 2,3-butanediol, d,l-α-glycerol phosphate and glucose-1-phosphate; weakly positive or negative utilization of the other substrates. The major fatty acids of strain PS1T were summed feature 8 (C18:1ω7c and/or C18:1ω6c), C16:0, summed feature 3 (C16:1ω7c and/or C16:1ω6c) and cyclo-C19:0ω8c. The major polar lipids found in strain PS1T were phosphatidylethanolamine, diphosphatidyglycerol and phosphatidylglycerol.
The type strain is PS1T (= DSM 112238T = MCCC 1K05112T), isolated from deep-sea sediments of the Mariana Trench.
Data availability
All data generated or analysed during this study are included in this published article, its supplementary information files and GenBank/EMBL/DDBJ. The Gen-Bank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain PS1T is MZ670768 and the number for the whole genome sequence is JALGRD000000000. Two supplementary figures are available with the online version of this paper.
Abbreviations
- ANI:
-
Average nucleotide identity
- AAI:
-
Average amino acid identity
- DDH:
-
DNA–DNA hybridization
- DPG:
-
Diphosphatidyglycerol
- PG:
-
Phosphatidylglycerol
- PE:
-
Phosphatidylethanolamine
- PC:
-
Phosphatidylcholine
- AGL:
-
Aminoglycolipid
References
Carrión O, Miñana-Galbis D, Montes MJ, Mercadé E (2021) Pseudomonas deceptionensis sp. nov., a psychrotolerant bacterium from the Antarctic. Int J Syst Evol Microbiol 61:2401–2405. https://doi.org/10.1099/ijs.0.024919-0
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/bf01734359
Garrity GM, Bell JA, Lilburn T (2005) Genus I. Pseudomonas Orla-Jensen 1921, 270 AL. In: Garrity GM, Brenner DJ, Krieg NR, Staley JR (eds) Bergey’s manual of systematic bacteriology (the proteobacteria part B the gammaproteobacteria), vol 2, 2nd edn. Springer, New York, pp 323–379
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. https://doi.org/10.1099/ijs.0.038075-0
King EO, Ward MK, Rainey DE (1954) Two simple media for demonstration of pyocyanin and fluorescein. J Lab Clin Med 44:301–307. https://doi.org/10.1371/journal.pone.0057409
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Mandel M (1966) Deoxyribonucleic acid base composition in the genus Pseudomonas. J Gen Microbiol 43:273–292. https://doi.org/10.1099/00221287-43-2-273
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Migula W (1984) Über ein neues system der bakterien. Arb Bakteriol InstKarlsruhe 1:235–238
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Pascual J, Lucena T, Ruvira MA, Giordano A, Gambacorta A, Garay E, Arahal DR, Pujalte MJ, Macián MC (2012) Pseudomonas litoralis sp. nov., isolated from Mediterranean seawater. Int J Syst Evol Microbiol 62:438–444. https://doi.org/10.1099/ijs.0.029447-0
Peng JS, Liu Y, Yan L, Hou TT, Liu HC, Zhou YH, Liu ZP (2019) Pseudomonas nitrititolerans sp. nov. a nitrite-tolerant denitrifying bacterium isolated from a nitrification/denitrification bioreactor. Int J Syst Evol Microbiol 69:2471–2476. https://doi.org/10.1099/ijsem.0.003516
Quigley MM, Colwell RR (1968) Proposal of a new species Pseudomonas bathycetes. Int J Syst Bacteriol 18:241–252. https://doi.org/10.1099/00207713-18-3-241
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:9126–19131. https://doi.org/10.1073/pnas.0906412106
Romanenko LA, Uchino M, Falsen E, Lysenko AM, Zhukova NV, Mikhailov VV (2005) Pseudomonas xanthomarina sp. nov., a novel bacterium isolated from marine ascidian. J Gen Appl Microbiol 51:65–71. https://doi.org/10.2323/jgam.51.65
Romanenko LA, Uchino M, Tebo BM, Tanaka N, Frolova GM, Mikhailov VV (2008) Pseudomonas marincola sp. nov., isolated from marine environments. Int J Syst Evol Microbiol 58:706–710. https://doi.org/10.1099/ijs.0.65406-0
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. MIDI Inc., Newark
Skerman V (1967) A guide to the identification of the genera of bacteria: with methods and digests of generic characteristics, 2nd edn. Williams & Wilkins, Baltimore
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–655
Tamegai H, Li L, Masui N, Kato C (1997) A denitrifying bacterium from the deep sea at 11,000-m depth. Extremophiles 1:207. https://doi.org/10.1007/s007920050035
Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL (2013) Microbial genomic taxonomy. BMC Genom 14:913. https://doi.org/10.1186/1471-2164-14-913
Timmis KN (2002) Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ Microbiol 4:779–781. https://doi.org/10.1046/j.1462-2920.2002.00365.x
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202. https://doi.org/10.1111/j.1574-6968.1990.tb03996.x
Tindall B, Sikorski J, Smibert R, Krieg N (2007) Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR (eds) Methods for general and molecular microbiology, 3rd edn. American Society for Microbiology, Washington, pp 330–393
Wang MQ, Sun L (2016) Pseudomonas oceani sp. nov., isolated from deep seawater. Int J Syst Evol Microbiol 66:4250–4255. https://doi.org/10.1099/ijsem.0.001343
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky M, Moore L, Moore W, Murray R, Stackebrandt E, Starr M, Trüper H (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464. https://doi.org/10.1016/s0176-6724(88)80120-2
Wei Y, Mao H, Xu Y, Zou W, Fang J, Jochen B (2018) Pseudomonas abyssi sp. nov., isolated from the abyssopelagic water of the Mariana Trench. Int J Syst Evol Microbiol 68:2462–2467. https://doi.org/10.1099/ijsem.0.002785
Xie F, Ma H, Quan S, Liu D, Chen G, Chao Y, Qian S (2014) Pseudomonas kunmingensis sp. nov., an exopolysaccharide-producing bacterium isolated from a phosphate mine. Int J Syst Evol Microbiol 64:559–564. https://doi.org/10.1099/ijs.0.055632-0
Yoshida M, Yoshida-Takashima Y, Nunoura T, Takai K (2015) Identification and genomic analysis of temperate Pseudomonas bacteriophage PstS-1 from the Japan Trench at a depth of 7000 m. Res Microbiol 166:668–676. https://doi.org/10.1016/j.resmic.2015.05.001
Zhang L, Pan Y, Wang K, Zhang X, Zhang C, Zhang S, Fu X, Jiang J (2015) Pseudomonas zhaodongensis sp. nov., isolated from saline and alkaline soils. Int J Syst Evol Microbiol 65:1022–1030. https://doi.org/10.1099/ijsem.0.001755
Funding
This work was supported by the National Natural Science Foundation of China (42073077). BLL acknowledges program on the survey, monitoring and assessment of global fishery resources sponsored by the Ministry of Agriculture and Rural Affairs.
Author information
Authors and Affiliations
Contributions
YLW drafted the manuscript. YXY, YL, DW and YXG performed isolation, deposition, identification and genome analysis. YLW, BLL and YPX designed all the experiments and supervised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethics statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank accession number for the 16S rRNA gene sequence and the genome sequence of strain PS1T is MZ670768 and JALGRD000000000, respectively. Five supplementary figures and tables are available with the online Supplementary Materials.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, Y., Gao, Y., Liu, Y. et al. Pseudomonas marianensis sp. nov., a marine bacterium isolated from deep-sea sediments of the Mariana Trench. Arch Microbiol 204, 638 (2022). https://doi.org/10.1007/s00203-022-03250-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03250-9