Abstract
Identification of the emerging multidrug-resistant yeast Candida auris is challenging. Here, we describe the role of the Mexico national reference laboratory Instituto de Diagnóstico y Referencia Epidemiológicos Dr. Manuel Martínez Báez (InDRE) and the Mexican national laboratory network in the identification of C. auris. Reference identification of six suspected isolates was done based on phenotypic and molecular laboratory methods, including growth in special media, evaluation of isolate micromorphology, and species-specific PCR and pan-fungal PCR and sequencing. The four C. auris isolates identified were able to grow on modified Sabouraud agar with 10% NaCl incubated at 42 °C. With one exception, isolates of C. auris were spherical to ovoid yeast-like cells and blastoconidia, with no hyphae or pseudohyphae on cornmeal agar. C. auris isolates were resistant to fluconazole. Species-specific and pan-fungal PCR confirmed isolates as C. auris. Sequence analysis revealed the presence of two different C. auris clades in Mexico, clade I (South Asia) and clade IV (South America).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Candida auris is an emerging multidrug resistance yeast able to cause invasive fungal infections and hospital outbreaks. This species of Candida differs from other species in that it more readily spreads from patient to patient in healthcare settings through contact with contaminated environmental surfaces or equipment (Welsh et al. 2017; Jeffery-Smith et al. 2018). The laboratory identification of C. auris using commercially available methods for yeast identification is challenging due to frequent misidentification (Jeffery-Smith et al. 2018; Caceres et al. 2019; Kathuria et al. 2015; Carvajal-Valencia et al. 2020). Currently, the most reliable methods for identification of C. auris include the VITEK® 2 with the most current software update, MALDI-TOF, molecular methods using specific PCR assays (Kathuria et al. 2015; Kordalewska et al. 2017; Satoh et al. 2009), or by the amplification and sequencing of the ITS1-ITS2 or the D1/D2 region of the ribosomal cistron (Satoh et al. 2009; Lockhart et al. 2017a, b, c).
In Mexico, C. auris was reported for the first time in May 2020 in a woman with severe endometriosis (Ayala-Gaytán et al. 2021). Three months later, the same hospital reported a large outbreak of C. auris associated with high mortality in patients with severe COVID-19 (Villanueva-Lozano et al. 2021). The impact of the current COVID-19 pandemic resulted in overcrowded hospitals, with larger numbers of critically ill patients at increased risk of invasive candidiasis. These overcrowded conditions set the stage for an increase in hospital-associated infections, including those caused by C. auris (Chowdhary and Sharma 2020).
Here, we describe the role of the Mexico national reference laboratory, Instituto de Diagnóstico y Referencia Epidemiológicos Dr. Manuel Martínez Báez (InDRE), and the Mexican national laboratory network in the detection of emerging problems, and the identification of the emerging yeast C. auris. This report presents a summary of the microbiological characteristics, focused on the phenotypic and molecular characteristics of Mexican isolates proven to be C. auris.
Materials and methods
Isolates
Six isolates suspected to be C. auris following CDC recommendations for C. auris identification ((CDC) CfDCaP 2018a) were submitted to InDRE for species identification. One from blood and two from urine came from a private hospital at the state of Nuevo León (labeled MYC-28, MYC-29, and MYC-30). The other three isolates were referred from public hospitals: a sputum isolate from Tabasco state (MYC-43), a blood isolate from Querétaro state (MYC-121), and a urine isolate from Morelos state (MYC-221). Before remission to InDRE, MYC-28, MYC-29, and MYC-30 were identified as C. auris using MALDI-TOF MS Bruker, and MYC-43 as C. auris, MYC-121 as C. krusei, and MYC-221 as C. parapsilosis using the VITEK® 2 system, v8.01 software.
Phenotypic identification
Suspected isolates were grown on Sabouraud dextrose agar (SDA, Becton Dickinson), incubated at 30 °C for 120 h. In parallel, isolates were grown on a selective modified Sabouraud agar (Welsh et al. 2017), supplemented with 10% NaCl (Sab-10% NaCl) and dulcitol instead of dextrose as carbon source. Isolates on this media were incubated at 42 °C for 72 h. In addition, morphological characteristics of isolates were evaluated, including growth on Cornmeal agar (Becton Dickinson).
Isolates were also tested using the VITEK® 2 system. Identification was performed using VITEK® 2 YST cards and results were analyzed using the software version 8.01 (bioMérieux, Marcy, L'Etoile, France). Additionally, assimilation of the following carbohydrates was evaluated: glucose, galactose, cellobiose, raffinose, melezitose, rhamnose, trehalose, dulcitol, and xylose. Results of carbohydrate assimilation were extracted from the VITEK® 2 system.
Molecular identification
Genomic DNA (gDNA) extraction was performed using the FastDNA™ kit (MP Biomedicals, USA) and the FastPrep® equipment as suggested by the manufacturer. Lysed material was further purified using the QIAamp® DNA Mini Kit (QIAGEN, Hilden, Germany).
Two PCR amplifications were used for molecular identification. The first was a pan-fungal PCR targeting the internal transcribed spacers (ITS1-ITS2) of the ribosomal cistron using primers ITS1F (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4R (5′-TCCTCCGCTTATTGATATGC-3′), with a product size of 380 bp. The second amplification was a species-specific PCR for C. auris using primers ITS1F and CaurisR (5′-CCACCGCGAAGATTGGTG-3), with a product size of 280 bp. The PCR conditions have been previously described (Theill et al. 2018), and PCR products were sequenced by the Sanger method (Sanger et al. 1977). Sequence analysis was done using the nBLAST tool at the NCBI database (http://www.ncbi.nlm.nih.gov/blast). A phylogenetic tree of maximum-likelihood III of the ITS regions of 14 non-redundant sequences extracted from the GenBank database and four sequences obtained here were constructed using MEGA7.0 software and Jukes–Cantor nucleotide substitution model with 1000 bootstraps (Kumar et al. 2016).
Fluconazole susceptibility testing
Fluconazole (FLU) susceptibility testing was performed using the agar dilution method, using Sigma-Aldrich FLU, following InDRE standard operating procedure (SOP) (Pérez et al. 2019). Minimum inhibitory concentration (MIC) was assessed in concentrations ranging from 2 to 256 µg/mL. After 24 h incubation, the MIC was determined as the lowest concentration that inhibits yeast growth (CLSI 2017). For susceptibility testing, Candida parapsilosis (ATCC 22019) was used as a susceptible strain, and Candida krusei (PAHO-ARG13) as a resistant strain (CLSI-M60). Candida auris MIC values for were interpretated using the tentative cut-off values proposed by the CDC (MIC ≥ 32 µg/mL) ((CDC) CfDCaP 2018a).
Results
Four isolates were from men and two from women, and patients were presented with a median age of 59 years (range 54–78 years). All patients were diagnosed with COVID-19 and were hospitalized in the intensive-care unit (ICU). Median ICU stay was 35 days (range 23–71 days). Five of the six patients died, all of them infected with C. auris and C. krusei.
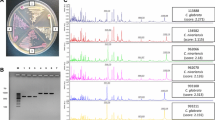
Using phenotypic methods, we found that isolates originally identified as C. auris (MYC-28, MYC-29, MYC-30, and MYC-43) grew on SDA at 37 °C and Sab-10% NaCl at 42 °C. Microscopic examination of these isolates revealed spherical and ovoid yeast-like cells and blastoconidia. Chlamydoconidia and hyphae or pseudohyphae were not observed on cornmeal agar for isolates MYC-28, MYC-29, and MYC-30 (Fig. 1A). In contrast, isolate MYC-43 displayed blastoconidia and hyphae or pseudohyphae on cornmeal agar (Fig. 1B). Isolates originally identified as C. parapsilosis and C. krusei did not grow in Sab-10% NaCl at 42 °C and produced pseudohyphae and chlamydoconidia on cornmeal agar (Fig. 1C and D).
Culture on cornmeal agar at 30 °C, for 3–5 days. 40X. a Candida auris, clade IV, Nuevo León, Mexico. Yeasts alone and some with blastoconidia, the size and spherical shape is a constant characteristic that differentiates it from other species (picture from isolate MYC-28). b Candida auris, clade I, Tabasco, Mexico. Yeasts alone, with blastoconidia and hyphae or pseudohyphae (picture from isolate MYC-43). c Candida parapsilosis. Yeasts alone, with blastoconidia, pseudohyphae alone, or branched. Incipient or immature chlamydoconidia. The cells adopt a characteristic irregular size and shape (picture from isolate MYC-221). d Candida krusei. Septate hyphae with spherical, ovoid, and elongated lateral blastoconidia on the sides. Bilateral symmetry seems to be a differential characteristic in this species (picture from isolate MYC-121). Blastoconidia and chlamydoconidia are indicated with arrow, respectively
Assimilation of carbohydrates testing shows that isolates originally identified as C. auris (MYC-28, MYC-29, MYC-30, and MYC-43) assimilated five of the nine carbohydrates tested: glucose, raffinose, melezitose, trehalose, and dulcitol. Raffinose and dulcitol were only assimilated by C. auris isolates. Final species identification was performed using the species-specific C. auris PCR. An approximately 280 bp band was amplified for MYC-28, MYC-29, MYC-30, and MYC-43 isolates, while no amplification was observed for isolates MYC-121 and MYC-221 (Fig. 2). Using pan-fungal PCR sequencing, C. auris isolates had 99–100% identity with other C. auris sequences within GenBank. Phylogenetic analysis of the sequence of ITS1-ITS2 regions showed that Mexican C. auris isolates were clustered with the C. auris South American clade (clade IV) for isolates MYC-28, MYC-29, and MYC-30, and isolate MYC-43 grouped with C. auris South Asian clade (clade I) (Lockhart et al. 2017c) (Fig. 3). Sequences were deposited at the GenBank database under accession numbers MW358012 (MYC-28), MW358013 (MYC-29), MW358014 (MYC-30), MZ648436 (MYC-43), MZ020643 (MYC-121), and MZ020644 (MYC-221).
Products of the species-specific C. auris PCR. Positive amplification: #1 C. auris (isolate positive control), #3 (MYC-28), #4 (MYC-29), #5 (MYC-30), and #6 (MYC-43). Negative amplification: #2 C. albicans (isolate negative control), #7 (MYC-121), #8 (MYC-228), and #9 (negative control). M: DNA molecular weight marker 1 Kb Plus and its migration is shown on the right side
C. auris phylogenetic analysis. The evolutionary tree was constructed with 18 nucleotide sequences of the Internal Transcribed Spacer (ITS) regions of the 18S rDNA gene using the maximum-likelihood method based on the Jukes–Cantor model with 1000 bootstraps. Analyses were conducted in MEGA v.7 software. The tree includes representative sequences of the four Candida auris clades and four Mexican sequences reported here; they were highlighted with a bold square. C. auris isolates from the state of Nuevo León (MYC-28, MYC-29 and MYC-30) matched with C. auris isolates corresponding to the South American clade (clade IV), and the isolate from Tabasco state (MYC-43) matched with C. auris isolates corresponding to the South Asian clade (clade I). The four clades of C. auris are indicated on the right side of the tree
Fluconazole susceptibility testing of the four C. auris isolates showed high MIC values. Isolates MYC-28, MYC-43, MYC-29, and MYC-30 had MICs of 128 µg/mL; based on proposed breakpoints (MIC ≥ 32), these isolates were considered resistant to fluconazole ((CDC) CfDCaP 2018a).
Discussion
The national laboratory surveillance confirmed four cases of C. auris in Mexico belonging to two different clades; all cases were identified in patients in the ICU with severe COVID-19. With the identification of C. auris in several Mexican states and the increase in the number of patients hospitalized in ICUs as a result of the COVID-19 pandemic, the mycology reference service of the InDRE implemented national laboratory surveillance for C. auris. Several challenges had to be addressed during the implementation of this surveillance, especially the limited availability of reference methods, such as MALDI-TOF, for the confirmation of C. auris.
In combination with several other low-cost tests, we were able to successfully use species-specific PCR for definitive identification of C. auris in a resource-limited setting, obtaining reliable and comparable results as those obtained with the reference methods. Candida auris-specific PCR tests have been used for the identification of colonized patients by the direct detection of C. auris DNA on skin swabs (Walchak et al. 2020). This type of non-culture based methodology has shown relevant utility in the early detection and control of C. auris outbreaks ((CDC) CfDCaP 2018b). Here, we have used this test to definitively identify isolates of C. auris using simple PCR and gel electrophoresis rather than real-time PCR. We have shown that this test could be employed in combination with other simple tests when MALDI-TOF and DNA sequencing are not available.
Without specialized Candida chromogenic media, the use of basic mycology methods, such as the evaluation of growth under different culture conditions, such as high salt and high temperature, were key to identifying potential isolates of C. auris. On cornmeal agar, C. auris generally has homogeneous size and shaped blastoconida and no hyphae and pseudohyphae. However, one of the four isolates (MYC-43) did present pseudohyphae, showing that is not a definitive characteristic of C. auris.
The evaluation of the profile of carbohydrates’ assimilation is not widely used for identification of Candida spp, mainly because it is a complex and laborious laboratory method. We confirmed that dulcitol assimilation is key to identify C. auris to other closely related species of Candida (Welsh et al. 2017; Jeffery-Smith et al. 2018; Caceres et al. 2019). Despite the close genetic relationship between C. auris and the C. haemolunii complex, the biochemical assimilation profile between them is distinct, so that a four carbohydrate assimilation combination (galactose, raffinose, rhamnose, and dulcitol) is decisive for the differentiation of C. auris from the C. haemulonii species complex (Jeffery-Smith et al. 2018).
Using the VITEK® 2 system, we found that all four isolates were correctly identified as C. auris. As reported previously, VITEK® 2 system using the software version 8.01 is able to identify C. auris, especially isolates from the South American clade (clade IV) (Ambaraghassi et al. 2019).
Here, we observed all isolates were resistant to fluconazole, with high MIC values (≥ 128 µg/mL). This high resistance to fluconazole is similar to others reports around the world. Unfortunately, our information about antifungal susceptibly testing (AFST) had the limitation of not being a gold standard methodology, but it is important to highlight that the method used is a viable alternative in settings where access to AFST using broth microdilution or gradient diffusion is not available.
Phylogenetic analysis of sequences showed that the three Mexican C. auris isolates from the north of the country belonged to the South American clade (clade IV). Surprisingly, the Mexican C. auris isolate from the south of the country clustered with the South Asian clade (Clade I), indicating that there have been multiple introductions of C. auris into Mexico and several clades are now circulating through the population.
With limited resources and limited access to reference methods for Candida species identification such as MALDI-TOF, we were able to confirm the species identification of isolates suspected to be C. auris. Here, we describe how conventional mycology laboratory methods, such as microscopy, growth on differential media, and growth at elevated temperatures, were key aspects to tentative identification of C. auris. The use of a simple species-specific PCR test confirmed the tentative identification. These methods can all be easily implemented in laboratories in countries with limited resources, or with logistical difficulties in accessing reference methods for Candida identification.
References
Ambaraghassi G, Dufresne PJ, Dufresne SF, Vallieres E, Munoz JF, Cuomo CA, Berkow EL, Lockhart SR, Luong M (2019) Identification of Candida auris using the updated 801 VITEK(R)2 yeast identification system: a multi-laboratory evaluation study. J Clin Microbiol 57(11):e00884-e919. https://doi.org/10.1128/JCM.00884-19
Ayala-Gaytán JJ, Montoya AM, Martínez-Resendez MF, Guajardo-Lara CE, de Treviño-Rangel RJ, Salazar-Cavazos L, Llaca-Diaz JM, González GM (2021) First case of Candida auris isolated from the bloodstream of a Mexican patient with serious gastrointestinal complications from severe endometriosis. Infection 49(3):523–525. https://doi.org/10.1007/s15010-020-01525-1
Caceres DH, Forsberg K, Welsh RM, Sexton DJ, Lockhart SR, Jackson BR, Chiller T (2019) Candida auris: a review of recommendations for detection and control in healthcare settings. J Fungi 5(4):111. https://doi.org/10.3390/jof5040111
Carvajal-Valencia SK, Lizarazo D, Duarte C, Escandon P (2020) Identificación de aislamientos de Candida auris recuperados a través de la vigilancia por laboratorio en Colombia: un reto para el diagnóstico. Infectio 24:224–228. https://doi.org/10.22354/in.v24i4.880
(CDC) CfDCaP (2018a) Recommendations for Identification of Candida auris. https://www.cdc.gov/fungal/candida-auris/recommendations.html
(CDC) CfDCaP (2018b) Recommendations for Infection Prevention and Control for Candida auris. https://www.cdc.gov/fungal/candida-auris/c-auris-infection-control.html
Chowdhary A, Sharma A (2020) The lurking scourge of multidrug resistant Candida auris in times of COVID-19 pandemic. J Glob Antimicrob Resist 22:175–176. https://doi.org/10.1016/j.jgar.2020.06.003
CLSI (2017) M60 Performance Standards for Antifungal Susceptibility Testing of Yeasts, 1st edn. https://clsi.org/
Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, Candida auris Incident Management Team, Rohini M, Brown CS (2018) Candida auris: a review of the literature. Clin Microbiol Rev 31(1):e00029-e117. https://doi.org/10.1128/CMR.00029-17
Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A (2015) Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J Clin Microbiol 53(6):1823–1830. https://doi.org/10.1128/JCM.00367-15
Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS (2017) Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol 55(8):2445–2452. https://doi.org/10.1128/JCM.00630-17
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Lockhart SR, Jackson BR, Vallabhaneni S, Ostrosky-Zeichner L, Pappas PG, Chiller T (2017a) Thinking beyond the common Candida Species: need for species-level identification of candida due to the emergence of multidrug-resistant Candida auris. J Clin Microbiol 55(12):3324–3327. https://doi.org/10.1128/JCM.01355-17
Lockhart SR, Berkow EL, Chow N, Welsh RM (2017b) Candida auris for the clinical microbiology laboratory: not your grandfather’s Candida species. Clin Microbiol Newsl 39(13):99–103. https://doi.org/10.1016/j.clinmicnews.2017.06.003
Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP et al (2017c) Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64(2):134–140. https://doi.org/10.1093/cid/ciw691
Pérez CC, García PG, Parra LGB, Sánchez SP, Méndez JD (2019) Candida Lusitaniae. Isolation, identification and clinical relevance. WJPR 8(5):606–621. https://doi.org/10.20959/wjpr20195-14720
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74(12):5463–5467. https://doi.org/10.1073/pnas.74.12.5463
Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H (2009) Candida auris sp. Nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53(1):41–44. https://doi.org/10.1111/j.1348-0421.2008.00083.x
Theill L, Dudiuk C, Morales-Lopez S, Berrio I, Rodríguez JY, Marin A, Gamarra S, Garcia-Effron G (2018) Single-tube classical PCR for Candida auris and Candida haemulonii identification. Rev Iberoam Micol 35(2):110–112. https://doi.org/10.1016/j.riam.2018.01.003
Villanueva-Lozano H, Treviño-Rangel RJ, González GM, Ramírez-Elizondo MT, Lara-Medrano R, Aleman-Bocanegra MC et al (2021) Outbreak of Candida auris infection in a COVID-19 hospital in Mexico. Clin Microbiol Infect 27(5):813–816. https://doi.org/10.1016/j.cmi.2020.12.030
Walchak RC, Buckwalter SP, Zinsmaster NM, Henn KM, Johnson KM, Koelsch JM et al (2020) Candida auris direct detection from surveillance swabs, blood, and urine using a laboratory-developed PCR method. J Fungi 6(4):224. https://doi.org/10.3390/jof6040224
Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJ, Litvinseva AP (2017) Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol 55:2996–3005. https://doi.org/10.1128/JCM.00921-17
Acknowledgements
We would like to thank the State Laboratory Network of Public Health (LESP) and the Support Laboratory in Epidemiological Surveillance (LAVE-IMSS) to contribution for the epidemiological surveillance of C. auris and other species.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Instituto de Diagnóstico y Referencia Epidemiológicos Dr. Manuel Martínez Báez (InDRE).
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
González-Durán, E., Contreras-Pérez, C.U., Caceres, D.H. et al. The use of readily available laboratory tests for the identification of the emerging yeast Candida auris in Mexico. Arch Microbiol 204, 592 (2022). https://doi.org/10.1007/s00203-022-03159-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-03159-3