Abstract
A Gram-stain-negative, aerobic, rod-shaped and motile bacterium, named LAMW06T, was isolated from greenhouse soil in Beijing, China. In the 16S rRNA gene sequence comparison, strain LAMW06T had the highest similarity with Pseudomonas cuatrocienegasensis 1NT. Phylogenetic analysis based on the 16S rRNA and three housekeeping gene sequences (gyrB, rpoB and rpoD) indicated that strain represented a member of the genus Pseudomonas. The genome sequence size of the isolate was 5.5 Mb, with a DNA G + C content of 63.5 mol%. The average nucleotide identity and DNA–DNA hybridization values between strain LAMW06T and closely related members of Pseudomonas borbori R-20821T, Pseudomonas taeanensis MS-3T and P. cuatrocienegasensis 1NT were 90.9%, 82.4%, 81.5% and 43.0%, 25.9%, 24.6% respectively. The major fatty acids contained summed feature 3 (C16:1 ω6c and/or C16:1 ω7c), C18:1 ω7c and C16:0. The primary respiratory quinone was ubiquinone-9. The main polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, six aminophospholipids, six phospholipids, one aminolipid and one glycolipid. According to the genotypic, phylogenetic and chemotaxonomic data, strain LAMW06T represents a novel species within the genus Pseudomonas, for which the name Pseudomonas tumuqii sp. nov. is proposed. The type strain is LAMW06T (= GDMCC 1.2003T = KCTC 72829T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The first species of genus Pseudomonas was described by Migula in 1894 (1900). At the time of writing, there are 256 validly named species (https://lpsn.dsmz.de/genus/pseudomonas) with validly published names on the List of Prokaryotic names with Standing in Nomenclature (Parte et al. 2020). Members of the genus Pseudomonas are described to be Gram-negative, rod-shaped, motile and catalase- and oxidase-positive (Anwar et al. 2017; Wang et al. 2020). The DNA G + C contents calculated from available genomes ranges from 58 to 69 mol% and the major respiratory quinone is ubiquinone-9 (Sun et al. 2018). Members of the genus Pseudomonas have been isolated from a wide variety of environments comprising soils (Zou et al. 2019), waters (Romanenko et al. 2008), plants (Timilsina et al. 2018) and animals (Lick et al. 2020). We isolated a novel strain designated as LAMW06T from soil sample collected from a greenhouse. Based on the genotypic, phylogenetic, phenotypic and chemotaxonomic characterization, the isolate is considered to represent a new species of the genus Pseudomonas.
Materials and methods
Isolation and culture conditions
Strain LAMW06T was isolated from soil sample collected from a greenhouse in Beijing (39°96.73' N, 116°33.53' E), using an isolation medium of trypticase soy agar (TSA; Difco). The soil samples were diluted and cultured on the TSA medium after at 30 °C for 3 days, separated colonies were picked and serially streaked onto TSA plates incubating at 30 °C to obtain single colony. Strain LAMW06T was obtained after several re-streaking and transfer onto TSA plates. The pure culture of strain LAMW06T was preserved at − 80 °C in tryptic soy broth (TSB; Difco) medium with 25% (v/v) glycerol. The strains Pseudomonas cuatrocienegasensis DSM 23418T (= 1NT) and Pseudomonas borbori DSM 17834T (= R-20821T) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM); Pseudomonas taeanensis JCM 16046T (= MS-3T) was obtained from the Japan Collection of Microorganisms (JCM). All the strains were chosen as reference strains in this study.
Bacterial strain and culture condition
Cells morphology were examined using light microscope (Nikon 80i, Tokyo, Japan) and transmission electron microscope (Hitachi 7500, Tokyo, Japan) after incubation on TSA agar plates for 3 days at 30 °C (Ruan et al. 2014). The temperature range for growth was determined by incubating strain LAMW06T at 10, 15, 20, 25, 30, 35, 37, 40 and 45 °C. The pH value range was determined in tryptic soy broth (TSB; Difco) with pH values of 4.0–11.0 at intervals of 0.5 pH units. The optimal concentration of NaCl for growth was investigated using NaCl-free TSB medium (prepared according to the TSB formula without NaCl) with different NaCl concentrations (0, 0.5 and 1–12% at 1.0% increments, w/v). The pH of the media was adjusted using the following buffer systems: MES (pH 4.0–6.0), MOPS (pH 7.0), Tricine (pH 8.0), TAPS (pH 9.0), CAPS (pH 10.0) and Na2CO3/ NaHCO3 (pH 11.0) (Wang et al. 2017). Catalase and oxidase activities were detected by observing bubble production in 3% (v/v) H2O2 solution and color variance of 1% (w/v) tetramethyl-p-phenylenediamine, respectively. Cell motility was examined using the hanging-drop technique. Anaerobic growth was tested on TSA plates at 30 °C for 10 days in GasPak EZ anaerobic container system (BD) added with sodium nitrite (10 mM) or sodium nitrate (20 mM) as potential electron acceptors (Skerman 1967). Gram-stain reaction, H2S production and methyl red test, hydrolysis of starch, gelatin and Tween 20, 40, 60 and 80 were detected as described by Smibert and Krieg (1994). Additional physiological and biochemical features were determined using API ZYM, 20NE and 50CH strips (bioMérieux) according to the manufacturers’ instructions. Antibiotic susceptibility test was performed on TSA plates using antibiotic disks (Hangzhou Microbial Reagent) containing the following concentrations (μg per disk unless otherwise stated): tetracycline (30 μg), gentamicin (10 μg), erythromycin (15 μg), ampicillin (10 μg), sulfaisoxazole (300 μg), vancomycin (30 μg), clindamycin (2 μg) and amikacin (30 μg).
Phylogenetic analysis based on 16S rRNA gene sequences
The 16S rRNA gene of strain LAMW06T was amplified by PCR using the bacterial universal primers 27F and 1492R (Weisburg et al. 1991) and compared with available sequences using the EzBioCloud identify service (Yoon et al. 2017a). The genomic DNA was prepared using the method described by Sun et al. (2016). The purified PCR product was inserted into pGEM-T vector and sequenced by Shanghai Life Technologies Company. Multiple sequences were aligned using clustal x program (Thompson et al. 1997). Phylogenetic relationships were analysed with neighbour-joining (NJ) (Saitou and Nei 1987), maximum-likelihood (ML) (Felsenstein 1981) and maximum-parsimony (MP) methods (Fitch 1971) using mega 7.0 software (Kumar et al. 2016). According to the algorithm of the Kimura’s two-parameter model, evolutionary distances were calculated using the NJ, ML and MP trees. The topologies of phylogenetic trees were evaluated by bootstrap analysis based on 1000 replicates (Kimura 1980). Multilocus sequence analysis (MLSA) was performed based on the three housekeeping genes gyrB, rpoB and rpoD that gathered from the genome assembly (Table S1), following the method described by Anurat et al. (2019) based on the same taxonomic sampling.
Genomic analysis
The genomic DNA G + C content was calculated according to the draft genome sequence of strain LAMW06T, which was done on the Illumina MiSeq platform by Guangzhou Magigene Company. SOAPdenovo assembler software was applied to assemble the reads (Yoon et al. 2017b). Gene annotation was performed using NCBI Prokaryotic Genome Annotation Pipeline and Swiss-Prot database. The genes involved in metabolic pathways were analysed using the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database. Other genome sequences of closest phylogenetic relatives were obtained from the GenBank sequence database. The average nucleotide identity (ANI) and in silico DNA–DNA hybridization (DDH) values were calculated according to the minimal standards proposed by Chun et al. (2018). The ANI values between strain LAMW06T and the relative reference species of strains P. cuatrocienegasensis 1NT (accession number FOFP00000000 of the ncbi GenBank database), P. borbori R-20821T (FOWX00000000), P. neuropathica P155T (JACOPX000000000) and P. taeanensis MS-3T (AWSQ00000000), and P. peli DSM 17833T (JAAQXQ000000000) were calculated using the OrthoANIu algorithm (https://www.ezbiocloud.net/tools/ani). The in silico DDH value between two strains was calculated by Genome-to-Genome Distance Calculator 2.1 (http://ggdc.dsmz.de). Antibiotic resistance genes (ARGs) were predicted with Comprehensive Antibiotic Resistance Database (CARD) software based on genome, protein or metagenomics data (Jia et al. 2017).
Chemotaxonomic analyses
For fatty acids analyses, strain LAMW06T, P. cuatrocienegasensis DSM 23418T, P. taeanensis JCM 16046T and P. borbori DSM 17834T were incubated on TSA medium after 2 days at 30 °C. According to the manufacturers’ instructions, cellular fatty acids were analysed by the Sherlock Microbial Identification System with the standard MIS Library Generation Software (VERSION 6.0 and Date 4, Microbial ID) and a 6890 N gas chromatograph (Agilent) (Sakamoto et al. 2002). Respiratory quinones of strain LAMW06T were extracted from freeze-dried cells and analysed using LC–MS (Minnikin et al. 1984). The polar lipid extracts were isolated by two-dimensional TLC using silica gel 60 F 254 aluminium-backed thin-layer plates (Merck) and identified by the method described by Xu et al. (2011). Molybdophosphoric acid was used for the detection of all lipids, ninhydrin reagent for aminolipids, molybdenum blue reagent for phospholipids and p-anisaldehyde reagent for glycolipids (Kates 1986).
Results
Morphological and physiological characteristics
Cells of strain LAMW06T were 1.2–2.0 μm length and 0.6–1.0 μm width rod-shaped in Fig. S1. Positive for oxidase and catalase activities, negative for hydrolysis of starch, casein, chitin, Tween 20, 40 and 60 as same as the related strains P. cuatrocienegasensis DSM 23418T, P. borbori DSM 17834T and P. taeanensis JCM 16046T. Negative for gelatin hydrolysis unlike P. cuatrocienegasensis DSM 23418T. In the API ZYM strip test, esterase lipase (C8) and leucine arylamidase were positive for strain LAMW06T and the related strains P. cuatrocienegasensis DSM 23418T, P. borbori DSM 17834T and P. taeanensis JCM 16046T. The lists of differing physiological and biochemical characteristics of strain LAMW06T and their closely related species are shown in Table 1. Strain LAMW06T was resistant to ampicillin, sulfaisoxazole and clindamycin, intermediately susceptible to gentamicin, and susceptible to tetracycline, erythromycin, vancomycin and amikacin. According to the antibiotic susceptibility tests.
Phylogenetic analysis
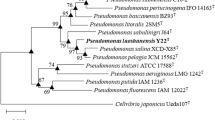
Based on 16S rRNA gene sequence similarity, strain LAMW06T (1418 bp) was closely related to P. cuatrocienegasensis 1NT (97.4%), P. borbori R-20821T (97.3%), P. neuropathica P155T (97.0%), P. taeanensis MS-3T (97.0%) and lower than 97.0% to other species. Phylogenetic analysis based on the NJ, ML and MP method revealed that strain LAMW06T did not cluster with other type species (Fig. 1, S2 and S3). The phylogenetic trees analysis indicated that strain LAMW06T belonged to the genus Pseudomonas. Alignment and comparison of the gyrB, rpoB and rpoD genes indicated that strain LAMW06T and the type strain Pseudomonas borbori R-20821T formed a clade by its own (Fig. 2).
Neighbor-joining tree based on 16S rRNA gene sequences showing the phylogenetic relationships between strain LAMW06T and its related taxa. The sequence of Azotobacter chroococcum LMG 8756T was used as outgroup. Filled circles indicate that the corresponding nodes were also recovered in the trees generated with the maximum-likelihood method and maximum-parsimony method. Bootstrap values (those above 50%) are shown as percentages of 1000 replicates. Bar, 0.005 substitutions per nucleotide position
Phylogenetic tree based on MLSA using the three housekeeping genes gyrB, rpoB and rpoD of type strains of species of the genus Pseudomonas, reconstructed using the neighbour-joining method. Bootstrap values were expressed based on 1000 replications; only values 50% or above are shown at the nodes. Bar, 0.02 nucleotide substitutions per 100 nucleotides
Genomic analysis
The genome of strain LAMW06T was 5.51 Mbp including 65 contigs with N50 as 351,006 coding sequences. The DNA G + C content of strain LAMW06T was 63.5 mol%, which was consistent with the range of 58–69 mol% reported for the genus Pseudomonas. The ANI value between strain LAMW06T and the relative reference species of genus Pseudomonas were less than 91.0%, for example, with P. cuatrocienegasensis 1NT, P. borbori R-20821T, P. neuropathica P155T and P. taeanensis MS-3T, and P. peli DSM 17833T were 81.5%, 90.9%, 76.8% and 82.4%, respectively, which was significantly less than the threshold used for species recognition (94–96%) (Meier-Kolthoff et al. 2013). The in silico DDH values between strain LAMW06T and the relative reference species of genus Pseudomonas were less than 43.0%, for example, with P. cuatrocienegasensis 1NT, P. borbori R-20821T, P. neuropathica P155T and P. taeanensis MS-3T were 24.6%, 43.0%, 21.4% and 25.9% (Table S2), respectively, which was also lower than the threshold value recommended for the assignment of strains to the same genomic species (70%) (Goris et al. 2007). Strain LAMW06T and the relative reference species of P. cuatrocienegasensis 1NT, P. borbori R-20821T, P. neuropathica P155T and P. taeanensis MS-3T had the similar antibiotic resistance genes adeF and rsmA from genomes that involved in the resistance-nodulation-cell division (RND) antibiotic efflux pump (Table S3).
Chemotaxonomic characteristics
The predominant fatty acids of strain LAMW06T (> 10%) were summed feature 3 (C16:1 ω6c and/or C16:1 ω7c) (38.2%), C18:1 ω7c (18.1%) and C16:0 (14.9%) consistent with strains P. cuatrocienegasensis DSM 23418T, P. borbori DSM 17834T and P. taeanensis JCM 16046T. Strain LAMW06T contained iso-C12:0 (6.3%) was lower than that of strain P. cuatrocienegasensis DSM 23418T shown in Table S4. The predominant menaquinone of strain LAMW06T was ubiquinone-9 (Q-9) like other species of the genus Pseudomonas. Major polar lipids were diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), six aminophospholipids (APL1-6), six phospholipids (PL1-6), one aminolipid (AL) and one glycolipid (GL) (Fig S4).
Therefore, based on the above phenotypic, phylogenetic analysis and genotypic data, it is proposed that strain LAMW06T should be classified as representative of a novel species of the genus Pseudomonas with the name Pseudomonas tumuqii sp. nov.
Description of Pseudomonas tumuqii sp. nov.
Pseudomonas tumuqii (tu.mu'qi.i. N.L. gen. n. tumuqii, of the Tumuqi biotechnological company, China, where taxonomic studies on this species were performed).
Cells are Gram-stain-negative, rod-shaped (0.6–1.0 × 1.2–2.0 μm), and motile by means of a single flagellum. Colonies are circular, smooth, light brown and with approximately 1.0–2.5 mm in diameter incubation on TSA after 3 days at 30 °C. Growth occurs at 15–37 °C (optimum, 30 °C), pH 5.0–10.0 (optimum, 8.0) and 0–4% (w/v) NaCl (optimum, 1%). Positive for catalase and oxidase, and negative for hydrolysis of starch, gelatin, casein, chitin, Tween 20, 40, 60 and 80. In the API ZYM and 20NE strip tests, positive for esterase (C4), esterase lipase (C8), leucine arylamidase and naphthol-AS-BI-phosphohydrolase, nitrate reduction, and assimilation of glucose, maltose, gluconate, capric acid, malic acid and citrate. In the API 50CH strip test, all results are negative. The major cellular fatty acids (> 10%) are summed feature 8 (C18:1 ω7c and/or C18:1 ω6c), summed feature 3 (C16:1 ω6c and/or C16:1 ω7c) and C16:0. The major menaquinone and polar lipids are ubiquinone-9 and diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, six aminophospholipids, one aminolipid, six phospholipids and one glycolipid, respectively. The genomic DNA G + C content is 63.5 mol%.
The type strain LAMW06T (= GDMCC 1.2003T = KCTC 72829T), was isolated from greenhouse soil in Beijing, China.
Data availability
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain LAMW06T is MN503522. The GenBank accession number for the draft genome sequence of strain LAMW06T is JAAOJA000000000.
References
Anurat P, Duangmal K, Srisuk N (2019) Pseudomonas mangiferae sp. nov., isolated from bark of mango tree in Thailand. Int J Syst Evol Microbiol 69:3537–3543. https://doi.org/10.1099/ijsem.0.003657
Anwar N, Rozahon M, Zayadan B, Mamtimin H, Abdurahman M, Kurban M, Abdurusul M, Mamtimin T, Abdukerim M, Rahman E (2017) Pseudomonas tarimensis sp. nov., an endophytic bacteria isolated from Populus euphratica. Int J Syst Evol Microbiol 67:4372–4378. https://doi.org/10.1099/ijsem.0.002295
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, Da CM, Costa MS, Rooney AP, Yi H, Xu XW, Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Escalante AE, Caballero-Mellado J, Martínez-Aguilar L, Rodríguez-Verdugo A, González-González A, Toribio-Jiménez J, Souza V (2009) Pseudomonas cuatrocienegasensis sp. nov. isolated from an evaporating lagoon in the Cuatro Cienegas valley in Coahuila Mexico. Int J Syst Evol Microbiol 59:1416–1420. https://doi.org/10.1099/ijs.0.006189-0
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. https://doi.org/10.1007/BF01734359
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416. https://doi.org/10.1093/sysbio/20.4.406
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo PY, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(D1):D566–D573. https://doi.org/10.1093/nar/gkw1004
Kates M (1986) Techniques of Lipidology: Isolation, Analysis and Identification of lipids, 2nd edn. rev, pp 106–107, 241–246. Elsevier, Amsterdam
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Kumar S, Stecher G, Tamura K (2016) mega 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lee DH, Moon SR, Park YH, Kim JH, Kim H, Parales RE, Kahng HY (2010) Pseudomonas taeanensis sp. nov. isolated from a crude oil-contaminated seashore. Int J Syst Evol Microbiol 60:2719–2723. https://doi.org/10.1099/ijs.0.018093-0
Lick S, Krockel L, Wibberg D, Winkler A, Blom J, Goesmann A, Kalinowski J (2020) Pseudomonas bubulae sp. nov., isolated from beef. Int J Syst Evol Microbiol 70:292–301. https://doi.org/10.1099/ijsem.0.003751
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. https://doi.org/10.1186/1471-2105-14-60
Migula W (1900) System der Bakterien, vol 2. Gustav Fischer, Jena
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M (2020) List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612. https://doi.org/10.1099/ijsem.0.004332
Romanenko LA, Uchino M, Tebo BM, Tanaka N, Frolova GM, Mikhailov VV (2008) Pseudomonas marincola sp. nov., isolated from marine environments. Int J Syst Evol Microbiol 58:706–710. https://doi.org/10.1099/ijs.0.65406-0
Ruan Z, Wang Y, Song J, Jiang S, Wang H, Li Y, Zhao B, Jiang R, Zhao B (2014) Kurthia huakuii sp. nov., isolated from biogas slurry, and emended description of the genus Kurthia. Int J Syst Evol Microbiol 64:518–521. https://doi.org/10.1099/ijs.0.056044-0
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sakamoto M, Suzuki M, Umeda M, Ishikawa I, Benno Y (2002) Reclassification of Bacteroides forsythus (Tanner et al. 1986) as Tannerella forsythensis corrig., gen. nov., comb. nov. Int J Syst Evol Microbiol 52:841–849. https://doi.org/10.1099/00207713-52-3-841
Skerman VBD (1967) A guide to the identification of the genera of bacteria, 2nd edn. Williams & Wilkins, Baltimore
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–654
Sun C, Fu GY, Zhang CY, Hu J, Xu L, Wang RJ, Su Y, Han SB, Yu XY, Cheng H, Zhang XQ, Huo YY, Xu XW, Wu M (2016) Isolation and complete genome sequence of Algibacter alginolytica sp. nov., a novel seaweed-degrading bacteroidetes bacterium with diverse putative polysaccharide utilization loci. Appl Environ Microbiol 82:2975–2987. https://doi.org/10.1128/AEM.00204-16
Sun J, Wang W, Ying Y, Zhu X, Liu J, Hao J (2018) Pseudomonas profundi sp. nov., isolated from deep-sea water. Int J Syst Evol Microbiol 68:1776–1780. https://doi.org/10.1099/ijsem.0.002748
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustal x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Timilsina S, Minsavage GV, Preston J, Newberry EA, Paret ML, Goss EM, Jones JB, Vallad GE (2018) Pseudomonas floridensis sp. nov., a bacterial pathogen isolated from tomato. Int J Syst Evol Microbiol 68:64–70. https://doi.org/10.1099/ijsem.0.002445
Vanparys B, Heylen K, Lebbe L, De Vos P (2006) Pseudomonas peli sp. nov. and Pseudomonas borbori sp. nov., isolated from a nitrifying inoculum. Int J Syst Evol Microbiol 56:1875–1881. https://doi.org/10.1099/ijs.0.64224-0
Wang XL, Wang YN, Yang XT, Sun H, Li B, Zhang XH (2017) Photobacterium alginatilyticum sp. nov., a marine bacterium isolated from bottom seawater. Int J Syst Evol Microbiol 67:1912–1917. https://doi.org/10.1099/ijsem.0.001886
Wang X, He SW, Guo HB, Thin KK, Gao JS, Wang Y, Zhang XX (2020) Pseudomonas rhizoryzae sp. nov., isolated from rice. Int J Syst Evol Microbiol 70:944–950. https://doi.org/10.1099/ijsem.0.003852
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Xu XW, Huo YY, Wang CS, Oren A, Cui HL, Vedler E, Wu M (2011) Pelagibacterium halotolerans gen. nov., sp. nov. and Pelagibacterium luteolum sp. nov., novel members of the family Hyphomicrobiaceae. Int J Syst Evol Microbiol 61:1817–1822. https://doi.org/10.1099/ijs.0.023325-0
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017a) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017b) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Zou Y, He S, Sun Y, Zhang X, Liu Y, Cheng Q (2019) Pseudomonas urumqiensis sp. nov., isolated from rhizosphere soil of Alhagi sparsifolia. Int J Syst Evol Microbiol 69:1760–1766. https://doi.org/10.1099/ijsem.0.003390
Funding
This research was partially supported by the National Natural Science Foundation of China (32070004, 31670006). This study was also supported by the Fundamental Research Funds for Central Non-profit Scientific Institution (1610132020009), Central Public-interest Scientific Institution Basal Research Fund (Y2021GH18) and Key Laboratory of Microbial Resources Exploitation and Application of Gansu Province (GK2019-01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kong, D., Li, Q., Zhou, Y. et al. Pseudomonas tumuqii sp. nov., isolated from greenhouse soil. Arch Microbiol 204, 249 (2022). https://doi.org/10.1007/s00203-022-02869-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02869-y