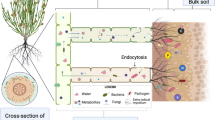
Abstract
We employed an Illumina-based high-throughput metagenomics sequencing approach to unveil the rhizosphere and root endosphere microbial community associated with an organically grown Camellia population located at the Experimental Garden for Plantation Crops, Assam (India). The de novo assembled tea root endosphere metagenome contained 24,231 contigs (total 7,771,089 base pairs with an average length of 321 bps), while tea rhizosphere soil metagenome contained 261,965 sequences (total 230,537,174 base pairs, average length 846). The most prominent rhizobacteria belonged to the genera, viz., Bacillus (10.35%), Candidatus Solibacter (6.36%), Burkholderia (5.19%), Pseudomonas (3.9%), Streptomyces (3.52%), and Bradyrhizobium (2.77%), while the root endosphere was dominated by bacterial genera, viz., Serratia (46.64%), Methylobacterium (8.02%), Yersinia (5.97%), Burkholderia (2.05%), etc. The presence of few agronomically important bacterial genera, Bradyrhizobium, Rhizobium (each 0.93%), Sinorhizobium (0.34%), Azorhizobium, and Flavobacterium (0.17% each), was also detected in the root endosphere. KEGG pathway mapping indicated the presence of microbial metabolic pathway genes related to tyrosine metabolism, tryptophan metabolism, glyoxylate, and dicarboxylate metabolism which play important roles in endosphere activities, including survival, growth promotion, and host adaptation. The root endosphere microbiome also contained few important plant growth promoting traits related to phytohormone production, abiotic stress alleviation, mineral solubilization, and plant disease suppression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tea [Camellia sinensis (L.) O. Kuntze] is a perennial tree and source of the popular beverage produced from processed young leaves of the plant (Mukhopadhyay et al. 2016). Cultivated tea plants generally belong to two major varieties, namely, Camellia sinensis (China type tea) and Camellia assamica (Assam type tea). The Assam type tea, one of the two cultivated tea plants, is processed from the indigenous Camellia sinensis var. assamica (Masters) (Meegahakumbura et al. 2018). The Assam tea is a distinctive black tea known for its malty sweetness and earthy flavor with several reported potential health benefits (Baruah 2017).
Due to the monocropping nature of cultivation covering large areas, the tea plantations are vulnerable to attacks from a large number of pests and diseases. The recurrent episodes of pest attacks necessitate their management through the application of agrochemicals, many of which find their way into tea brew (Zhang et al. 2017). Tea is a dynamic “bioactive” beverage perceived as health value and a majority of consumers prefer organically grown tea over conventionally cultivated or processed teas (de Godoy et al. 2013). Organic tea is usually considered as a value-added product and the recent trend in the global market shows that such teas command a premium of 30–40% over conventionally grown teas (Singh et al. 2016a). In addition, organically grown tea has longer shelf-life and better keeping quality than conventionally grown teas (Yugandhar et al. 2018). Exclusion of chemical inputs, including fungicides, herbicides, and pesticides, is an important aspect of organic tea cultivation which inevitably exposes the plantation to various soil- and air-borne tea diseases, such as blister blight, black rot (leaf diseases), dieback or seed decay (stem diseases), charcoal stump rot, and brown rot (root diseases) (Doanh et al. 2018). Climate change-induced erratic precipitation patterns coupled with nutrient leaching are other contributing factors leading to significant losses in crop productivity and quality of processed tea (Jayasinghe and Kumar 2020).
A comprehensive knowledge of microbial community structure associated with the Camellia sinensis var. assamica root system, including the root endosphere and the rhizosphere regions, was expected not only to provide a better understanding of their influence on the plants’ health and productivity but also to help in harnessing beneficial microorganisms with plant growth promoting traits (Fernández-González et al. 2019). Plant-associated beneficial microbes can help implant ecologically compatible and sustainable organic tea cultivation practices for improving productivity and stimulating production of enhanced bioactive compounds (Hu et al. 2006; Nath et al. 2013). Among the plethora of beneficial microorganisms, the endosphere and rhizosphere components (particularly, fungi, and bacteria) draw attention due to their plant growth promoting traits, such as secretion of indole acetic acid, gibberellin, cytokinin, siderophores, phosphate-solubilizing enzymes, and 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase (Xie et al. 2020). The ability of the plant microbiome to colonize and endure plant endosphere aids in the plant’s ability to withstand biotic and abiotic stress conditions and enhance the fitness of the host plant (Meena et al. 2017; Hassani et al. 2018; Xie et al. 2020). We have previously reported culture-dependent studies that revealed the microbial diversity and their functional aspects prevalent in tea garden soils of Assam (Nath et al. 2013, 2015; Goswami et al. 2017). However, our current understanding of the Camellia sinensis var. assamica-associated microbiota still remains sketchy. Therefore, the present high-throughput sequencing-based study was undertaken to unravel the microbial community structure of belowground rhizosphere and endosphere, and their relevant functional attributes were associated with an organically grown Camellia population of Assam, India. The following objectives were pursued: (a) to elucidate the overall rhizosphere as well as belowground endosphere microbial community structure of Camellia sinensis var. assamica; (b) to unveil distinctive functional signatures between the rhizosphere and endosphere communities.
Methods and materials
Sites description and sample collection
This study was conducted at the organically cultivated area of the Experimental Garden for Plantation Crops (26° 72′ 12.83″ N, 94° 19′ 72.30″ E, elevation 88 m), Assam Agricultural University, Jorhat district of Assam, India (sample collection date: 25-07-2020). The total area of the organically maintained plot is 0.26 ha cultivating an indigenous large leaf cultivar, Betjan, planted in the period of 1951–1957. The tea plantation management system has a biofertilization regime of vermicompost and cow dung manure applied at the rate of 2 tons/ha/year and 8 tons/ha/year, respectively. Additionally, the mature tea bushes receive a foliar spray of Azolla water and vermiwash (10% v/v) six times a month. The tea garden has a history of high infestations of tea mosquito bugs (mainly, Helopeltis theivora), and low occurrences of red spider mite (Oligonychus coffeae) and bunch caterpillar (Andraca Bipunctata). To counteract such pest infestations, an aqueous mixture of leaf extracts from Azadirachta indica (neem) and Pteridium aquilinum L. (vernacular name: Bih-dhekia) at 5% (w/v) concentration is applied three times a week during incidences. A pruning cycle of 4 years along with intermittent skiffing (light trimming) is maintained to achieve a uniform plant height.
For the collection of the rhizosphere and endosphere samples, three disease-free healthy mature tea plants were selected, and their lateral roots were partially uprooted. These lateral roots were vigorously shaken to eliminate loose soils and tertiary fine roots. Finally the lateral roots were washed with 100 ml of 10 mM NaCl solution. Wash solutions from the three lateral root samples were mixed proportionately and collected into 50-ml tubes to constitute the rhizosphere sample. For root endosphere sampling, samples of lateral roots were thoroughly rinsed with distilled water and then immediately transported to the laboratory for surface sterilization. In the laboratory, the surface sterilization of the roots started with washing the samples with double-distilled H2O for three times followed by serially immersing in H2O2 (3%) and absolute ethanol for 30 s each, NaOCl 6.15% (in the presence of 2–3 drops of Tween 20 per 100 ml) for 3 min followed by a wiping with H2O2 (3%) again for 30 s. The roots were given a final washing with sterile dd. H2O and stored at − 80 °C for future use. Characterization of different soil edaphic factors was performed at the Laboratory of Microbial Biotechnology, Assam Agricultural University. The characteristics of the Camellia trees (diameter, height, etc.) and soil parameters (texture, percent C, percent N, etc.) are presented in Table 1. All the observations and experiments related the characteristics of the Camellia trees (diameter, height, etc.) and soil parameters (texture, percent C, percent N, etc.) were performed in triplicate and the variations (analysis of variance) in the data were evaluated using the XLSTAT package program.
Metagenomic DNA extraction
In case of the rhizosphere soil sample, 1 g of the soil slurry centrifuged at 2500g for 2 min was considered for DNA extraction following the manufacturer’s protocol mentioned in the Environmental gDNA isolation kit (Xcelgen, India). While dealing with DNA extraction from the endosphere sample, the same gDNA isolation kit was used with powdered surface-sterilized roots (using liquid nitrogen). The lysis buffer was amended with 50 µl of 10% cetyltrimethylammonium bromide to enhance the lysis process. The extracted gDNA samples were quantified using Qubit® 2 fluorometer as per manufacturer’s protocol.
Library preparation and sequencing
The quality pair-end sequencing library was prepared using Illumina TruSeq DNA Library Preparation Kit. For library preparation, 1.0 µg gDNA was fragmented followed by paired-end adapter ligation. The ligated product was purified using 1X ampure beads. The purified ligated product was subjected to size selection on E-gel at ~ 400–650 bp. The size-selected product was PCR amplified as described in the kit protocol. The amplified library was analyzed in Bioanalyzer 2100 (Agilent Technologies) using High Sensitivity (HS) DNA chip as per manufacturer's instructions. After obtaining the Qubit concentration for the library and the mean peak size from Bioanalyzer profile, 10 pM of the library was loaded into the reagent cartridge of Miseq V2 Reagent Kit 500 cycles PE. The library fragments were captured on a lawn of oligos (complementary to the adapters linked to the fragments) coated on the surface of flow cells. Later, each immobilized fragment was amplified through bridge amplification to develop into a clonal cluster. These clonal clusters were finally subjected to a reversible terminator-based sequencing method. Paired-end sequencing approach allowed the bidirectional (forward and reverse) sequencing of template fragments.
Metagenome assembly and ORF prediction
Prior to the assembly, the quality of the sequencing data, overall GC content, repeat abundance, or the proportion of duplicated reads were assessed using FastQC that provides summary statistics. The high-quality metagenome reads were then screened for contamination with the human genome using the CLC Genomics Workbench 6.0. After removal of host plant-related sequences and adapter sequences from both the endosphere and rhizosphere samples, the resulting clean reads were assembled using the same workbench with default parameters (minimum contig length: 200, mismatch cost: 2, insertion cost: 3, deletion cost: 3, length fraction: 0.5, similarity fraction: 0.8). Total rRNAs were predicted for mining residue samples by aligning the assembled scaffolds against in-house RNA databases using BlastN. The Open Reading Frames were predicted for the mining residue sample using Prodigal (v2.6.1) (Hyatt et al. 2010). The predicted ORFs along with their rRNAs containing scaffolds were then taken for Taxonomic and Functional Annotation. The 16S regions were identified using webMGA (Wu et al. 2011).
Selection and assembly of sequencing reads
The raw reads were first refined by trimming adapter as well as low-quality score sequences (minimum-quality score: 20; minimum read length: 50 bp). The SOAPdenovo was used to assemble the quality reads with Kmer range of 39–47. Scaffolds with minimum 500 bp length were then broken into continuous contigs which were later used for functional and taxonomic predictions. For quality assurance of SOAPdenovo-based assembly, all the sequencing reads were remapped to the contigs using the Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net/) BWA tool (Li and Durbin 2009).
Gene prediction, taxonomy, and functional assignment
The publically available MetaGene Annotator was used to mine the contigs for ORFs and annotations (Ma et al. 2013). Only ORFs with a minimum 100 bp of length were considered for translations through NCBI ORF finder. The translated sequences were tested for BLASTP (BLAST Version 2.2.28+) against the NCBI-nr database (E value threshold: 1e−5). For taxonomic classification, the ORFs containing the small subunit (SSU) rRNA gene tags were aligned with the SILVA database and Ribosomal database project (E value threshold: 1e−5, alignment length: 100 bp, and cutoff: 80%) (Cole et al. 2014). The difference in microbial abundance between the two metagenomes was examined by the two-tailed Wilcoxon rank-sum test, performed using the base R package “stats” (v. 3.4.1) wilcox.test function. Alpha (within sample) diversity indices (Species richness, Shannon diversity, Simpson’s index, Inverse Simpson and Shannon evenness index) were estimated for each of the samples using the R package “vegan.”
For functional analysis, the eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) database version 4.5 was used to align the ORFs and produce corresponding Cluster of Orthologous groups of protein (COG) (Huerta-Cepas et al. 2016). This annotation was accomplished through BLASTP (hosted under the latest BLAST + package version 2.2.28+) against the NCBI-nr database with optimized E value threshold of 1e−5. The web server-based KOBAS version 3.0 (KEGG Orthology Based Annotation System) was used for KEGG GENES (Kyoto Encyclopedia of Genes and Genomes database)-based functional annotation (default E value threshold: 1e−5) (Xie et al. 2011). The KEGG database was used to investigate a few microbial genes linked to plant growth-promoting traits (PGPTs), viz., production of siderophore, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, indole acetic acid (IAA), mineral (phosphate) solubilization, N2 fixation, abiotic stress alleviation (betaine utilization), soil health improvement (lipopolysaccharide production), as well as plant disease suppression (chitinase and endoglucanase). The frequency of the genes was calculated as follows: frequency = (number of sequence reads for a particular gene/total length of the gene) × (100,000/Total number of sequence reads). Finally, the number of the particular gene was normalized by the gene length (kbp) (Tsurumaru et al. 2015).
Results
The average diameter of the three disease-free trees at their breast height was 73 ± 4.58 cm, while crown height and crown diameter were 86.67 ± 1.15 cm and 417 ± 11.36 cm, respectively. Rhizosphere soil samples from these trees were analyzed for various physical and biochemical parameters, including total carbon and nitrogen, soil organic matter, and available ammonium and total nitrate ions. All of the soil characteristics did not vary among the three samples (Table 1). The soil samples were moderately acidic in nature with pH values less than 4.50 (range 4.3–4.5). The average soil organic matter (SOM) ranged from 4.85 to 5.46%, while TC and TN ranged from 1.66 to 1.92 and 0.183–0.223, respectively.
Overview of metagenomic sequencing
The assembled tea root endosphere metagenome contained 24,231 contigs (total 7,771,089 base pairs with an average length of 321 bps), while the tea rhizosphere soil metagenome contained 261,965 contigs (total number of bases: 230,537,174 bp, average length: 846 bps) which corresponded to ~ 2.8–3.1 GB data. The metagenomes were deposited in the Sequence Read Archive hosted at the NCBI (accession number: PRJNA698063). For this study, the metagenomes were further trimmed and filtered by using WebMGA, a web server in order to remove the adaptors and low-quality reads. After this quality control step, the root endosphere metagenome finally contained 21,721 and the rhizosphere metagenome contained 241,280 high-quality reads, respectively. The summary of metagenome sequencing and annotations, and the rarefaction curves are presented in the additional files (Table S1, Figs. S1 and S2).
Taxonomic category hits distribution
Abundance and functional groups estimations are critical in metagenomic studies. A two-tailed Wilcoxon rank-sum test was performed between the microbial abundances of rhizosphere and root endosphere and it was found that there was a significant difference (p value < 0.00001) between the two metagenomes. The rhizosphere metagenome contained a total of 1592 (bacteria: 1344, fungi: 84, Archaea: 85) microbial species, while the endosphere metagenome harbored 223 species (bacteria: 176, fungi: 26, Archaea: 5). The Shannon diversity index (H) and Simpson's Index of Diversity (1-D) for the rhizosphere metagenome were calculated to be 5.648 and 0.986, respectively; these values were higher than that of the endosphere metagenome (H: 4.053; 1-D: 0.926) which confirmed the presence of higher microbial diversity and abundance in the rhizosphere as compared to the endosphere. A higher Shannon evenness index of the rhizosphere metagenome (0.766) than that of the endosphere one (0.750) implied that the endosphere was over-represented by dominant microbial taxa. Information on various alpha diversity indices is available in the additional file (Table S2).
Class-level taxonomic hits distribution showed that the rhizosphere was highly populated by Proteobacteria (relative abundance 40.98%), Acidobacteria (19.74%), Firmicutes (14.43%), Actinobacteria (10.95%), Bacteroidetes (4.70%), Verrucomicrobia (3.05%), and Cyanobacteria (1.9%), while the root endosphere was inhabited by Proteobacteria (89.17%), Actinobacteria (5.08%), Bacteroidetes (1.52%), Firmicutes, and Synergistetes (both with 0.85%). Order-level taxonomic hits distribution revealed the dominance of Bacillales (12.57%), Rhizobiales (11.03%), Actinomycetales (10.22%), Solibacterales (6.35%), Burkholderiales (7.73%), Enterobacteriales (6.55%), Acidobacteriales (5.37%), and Pseudomonadales (4.14%), in the rhizosphere with approximately 10.25% remained as undefined. Bacterial orders, such as Enterobacteriales (57.36%), dominated the root endosphere microflora followed by Rhizobiales (14.04%), Burkholderiales (5.41%), and Actinomycetales (4.57%). The majority of genus-level hits belonged to Bacillus (10.35%), Candidatus Solibacter (6.36%), Burkholderia (5.19%), Pseudomonas (3.9%), Streptomyces (3.52%), and Bradyrhizobium (2.77%) in the rhizosphere, while in case of the endosphere, most of the hits were occupied by bacterial genera, viz., Serratia (46.64%), Methylobacterium (8.02%), Yersinia (5.97%), Burkholderia (2.05%), etc. The presence of few agronomically important bacterial genera, such as Bradyrhizobium, Rhizobium (0.93% each), Sinorhizobium (0.37%), Azorhizobium, and Flavobacterium (0.17% each), was also detected in the root endosphere (Fig. 1).
In case of the fungal population, Sordariomycetes (49.06%), Eurotiomycetes (26.19%), Saccharomycetes (14.76%), Agaricomycetes (12.79%), and Leotiomycetes (2.47%) were the major taxa in the rhizosphere, while the endosphere was dominated by class Eurotiomycetes (20.75%), followed by Sordariomycetes (13.81%), Agaricomycetes, and Leotiomycetes (each with 5.66%). Order-level taxonomic distribution hits demonstrated that fungal orders, such as Eurotiales (endosphere: 9.43%; rhizosphere: 21.15%), Saccharomycetales (endosphere: 1.89%; rhizosphere: 14.76%), Agaricales (endosphere: 5.66%; rhizosphere: 11.57%), Sordariales (endosphere: 15.09%; rhizosphere: 5.40%), and Hypocreales (endosphere: 33.96%; rhizosphere: 5.51%), were prevalent in both the environments. Fungi belonging to the genera, viz., Ustilago (8.36%), Schizosaccharomyces (8.25%), Neosartorya (8.07%), Filobasidiella (6.27%), Aspergillus (5.92%), Gibberella (4.41%), Penicillium (4.26%), Laccaria (3.95%), Coprinopsis (3.8%), Neurospora (3.35%), Yarrowia (2.86%), and Schizophyllum (2.81%), were abundant in the rhizosphere, and genera, such as Gibberella (18.18%), Phaeosphaeria (14.55%), Podospora, Hypocrea (both 10.91%), Aspergillus (5.45%), Coprinopsis, Neurospora, Coccidioides, and Botryotinia (each with 3.64% relative abundance), were prevalent in the root endosphere. Fungal populations, viz., Chaetomium, Coniothyrium, Cladosporium, Gliocladium, Trichoderma, Penicillium, Fusarium, Verticillium, Lecanicillium, Phoma, Alternaria, Aspergillus, Metarhizium, Phomopsis, Curvularia, Colletotrichum, Xylaria, Nigrospora, Humicola, Rhizoctonia, and Tilletiopsis, were prevalent in both the environment (i.e., rhizosphere and endosphere) (Fig. 2). The fungi could be broadly categorized into three groups: (A) ubiquitous soil inhabitant with no- or latent pathogenicity; (B) phytopathogens responsible for various plant diseases; and (C) ecologically important fungi with potential implications as plant growth promoters. Species of the genera Tilletiopsis, Humicola, Chaetomium, Alternaria, Curvularia, and Xylaria are ubiquitous in various environmental matrices, such as dead or living plant materials and decomposing soils. The second group consisted of members belonging to genera Nigrospora, Cladosporium, Fusarium, Lecanicillium, Phoma, Phomopsis, Colletotrichum, and Rhizoctonia which are essentially phytopathogens responsible for plant diseases, such as leaf spots, scab, rot, and wilt. The third group consisting of ecologically important fungi exhibits diverse functionality, such as biocontrol activities (mycoparasitic activity: Coniothyrium, Gliocladium, Trichoderma, and Penicillium; entomopathogenic activity: Beauveria, Metarhizium, and Lecanicillium) and plant growth-promoting activities (Aspergillus and Trichoderma).
COG annotation and analysis
The functional analysis through COG analysis showed that the associated genes could be deciphered into four categories, viz., (1) metabolism, (2) cellular processes and signaling, (3) information storage and processing, and (4) poorly characterized. The category for “metabolism” was most prevalent in both the metagenomes (rhizosphere: 43.5%; endosphere 33.9%) followed by “Cellular processing” (rhizosphere: 21.3%; endosphere: 19.93%). In case of “Information storage and processing,” the endosphere metagenome harbored more genes (26.5%) than its rhizosphere counterpart (18.0%). A significant number of genes (rhizosphere: 17.23%; endosphere: 19.6%) could not be assigned to any function and remained in the “poorly characterized” category. This COG-based functional analysis indicated that the endosphere environment promoted information storage and processing of the microorganisms which is integral to any microbial colonization (War Nongkhlaw and Joshi 2017). Functional categories, viz., “general function prediction only,” “amino acid transport and metabolism,” “energy production and conversion,” “carbohydrate transport and metabolism,” “replication, recombination and repair,” “translation,” “ribosomal structure and biogenesis,” “cell wall/membrane/envelope biogenesis,” and “inorganic ion transport and metabolism,” dominated the COG functional annotations in both the metagenomes. The relative abundance of reads assigned to cytoskeleton, cell motility, RNA processing and modification, chromatin structure and dynamics, etc., were low (≤ 1.0%) in both the environments (Fig. 3).
KEGG function annotation and analysis
KEGG-based functional annotations showed that the rhizosphere metagenome contained more genes, pathways, and enzymes as compared to the endosphere metagenome. We defined dominant pathways as those with a relative abundance greater than 0.5% in any of the metagenomes and as such 55 various pathways were mined with this criterion (Table S3). The most prominent pathways in this group linked to cysteine and methionine metabolism, RNA degradation, plant–microbe interaction, RNA transport, RNA polymerase, cell cycle, oxidative phosphorylation, spliceosome, peroxisome, pyrimidine metabolism, protein processing in endoplasmic reticulum, pyruvate metabolism, phenylpropanoid biosynthesis, etc. The pathway ko02010 assigned to “ABC transporter” was most abundant in the two environments (endosphere: 4.8%; rhizosphere: 6.4%) followed by the pathway “Two-component system-ko02020” (3.7% in both). Among the 55 dominant pathways, the relative abundance of 22 pathways was more in the endosphere metagenome than in the rhizosphere one. Few pathways associated with plant–microbe interaction, such as bacterial chemotaxis (ko02030), vitamin B6 metabolism (ko00750), folate biosynthesis (ko00790), steroid biosynthesis (ko00100), flagellar assembly (ko02040), calcium signaling pathway (ko04020), and glutathione metabolism (ko00480), were significantly more abundant in the endosphere than in the rhizosphere (Fig. 4). A total of 233 functional pathways determined in the three metagenomes are presented in the additional file.
The KEGG-based classification data were further mined for the frequency detection of few functional genes liked to plant growth promoting traits (PGPTs) (Table 2). Among the categories of PGPTs, Cytochrome o ubiquinol oxidase subunit I (cyoB) and Glycine betaine/proline transport system (proV) linked to betaine utilization were most frequently detected (12.46 and 11.37 per 105 reads, respectively), followed by ACC deaminase, Betaine-aldehyde dehydrogenase (betB, gbsA), Glycine betaine/proline transport system (proW), D-glycero-D-manno-heptose 1,7-bisphosphate phosphatase (gmhB), and non-ribosomal peptide synthetase (dhbF) (11.14, 10.42, 9.20, 8.96, and 7.45 per 105 reads, respectively) (Table 2). Apart from this, other important PGPTs frequently detected in the rhizosphere were lipopolysaccharide production, N2 fixation, plant disease suppression, phosphate solubilization, ACC deaminase production and IAA production (20.52, 13.28, 12.00, 11.97, 11.14, and 9.15 per 105 reads, respectively). In the case of the endosphere metagenome, the most frequently assigned PGPTs were Chitinase, β-1,3-glucanase, ACC deaminase, and YUCCA family monooxygenase (3.14, 2.64, 2.15, and 2.14 per 105 reads, respectively). Meanwhile, siderophore production-related sequences (vibF, dhbF, entB, entA, entF) were detected at a low frequency (0.36, 1.14, 0.14, 0.11, 1.17, respectively, per 105 reads). Overall, the endosphere metagenome harbored genes for all the PGPTs except for lipopolysaccharide production.
Discussion
Based on alpha diversity indices, it was evident that the microbial richness and diversity was greater in the rhizosphere than the endosphere which could definitely be attributed to the rhizosphere being the primary and dynamic interface of plant–microbe interactions (Mohanram and Kumar 2019). These interacting microbes produce an array of hormones and metabolites which are known to signal and promote plant growth, antagonize pathogens, and increase availability of essential micro- and macronutrients for plant uptake (Singh et al. 2019). In this study, we observed a high abundance of Proteobacteria both in the rhizosphere (relative abundance 40.98%) as well as the endosphere (89.17%). Our previous study using culture-dependent approach had revealed the dominance of Firmicutes (more than 70%) in the tea garden bulk soils (Goswami et al. 2017) which indicated substantial differences in the two interfaces, i.e., bulk–rhizosphere and rhizosphere–endosphere. Environmental conditions, such as pH, water, oxygen, and nutrient content, function as drivers for such differential microbial pattern between these dynamic interfaces (Vieira et al. 2020).
The tea rhizosphere harbored a diverse array of the ubiquitous soil microbiota, while the endosphere harbored functionally and strategically important subset of the rhizosphere microbiota, viz., symbiotic Bradyrhizobium, Mesorhizobium, and Rhizobium, and non-symbiotic rhizobacteria, viz., Azotobacter, Azospirillum, Azomonas, Bacillus, Pseudomonas, etc. Microorganisms, such as rhizobia, mycorrhizal fungi, actinomycetes, and diazotrophic, are known to be aggressive colonizers which mobilize and allocate nutrients through symbiotic and non-symbiotic associations with plant roots (Barea et al. 2005). Healthy stature of the tea bushes indicated the rhizosphere–microbial taxa playing a key role in disease suppression, nutrient cycling, and healthy status of the soil. Most of the endosphere bacteria mined through this study have been reported to carry promising plant growth promoting traits, such as solubilization of minerals (zinc and phosphate), production of siderophore and indole acetic acid, and exhibition of biocontrol activities (Aeron et al. 2011; Fernández-González et al. 2017; Verma et al. 2019). These functionally important traits have been proven to increase tea productivity right from tea seed germination to seedling growth in nursery and subsequent field establishment (Nath et al. 2013; Bhattacharyya and Sarmah 2018). The potentials of free-living N2-fixing bacteria, such as Azotobacter (aerobic) and Azospirillum (anaerobic, microaerobic), that play an important role in nitrogen geocycle have been realized in the form of organic amendments in tea soils (Gebrewold 2018). Both Azotobacter and Azospirillum are suitable candidates for biofertilizer development targeted for better tea productivity, while Azotobacter is suited for aerated, light-textured soil and Azospirillum is indicated for waterlogged hard-textured soils (Aquilanti et al. 2004). Microbial pesticides intended against a range of phytopathogens, including tea pests, are an important component of integrated pest and disease management systems in tea cultivation (Idris et al. 2020). Several microbial species revealed in this metagenomics-based study, viz., Bacillus spp., Pseudomonas spp., Aspergillus sp., Beauveria bassiana, Gliocladium sp., Metarhizium anisopliae, Paecilomyces sp., Penicillium sp., Trichoderma spp., Lecanicillium, and Streptomyces sp., have been reported to show biopesticidal activities and as such their introduction as biocontrol agents has the potential to elevate tea productivity (Krauss et al. 2004; Scheepmaker and Butt 2010; Sandhu et al. 2012). Coniothyrium minitans (a coelomycete) was detected in both the rhizosphere and endosphere regions. Coniothyrium is a potential biocontrol candidate with mycoparasitic activity against Sclerotinia sclerotiorum (Zhao et al. 2020). Many studies have reported that C. minitans can inhibit mycelial growth and sclerotia formation of Sclerotinia ascospores, thereby suppressing leaf blight infection (Smolińska and Kowalska 2018). Gliocladium virens is another important mycoparasitic fungus which exhibits broad-spectrum biocontrol activity (van Tilburg and Thomas 1993). This common fungus exhibited strong hyperparasitism against plant–pathogens, viz., Rhizoctonia solani, Sclerotium rolfsii, and Pythium aphanidermatum (Sreenivasaprasad and Manibhushanrao 1990). The presence of enzymes, viz., 1-glucanase, N-acetylglucosaminidase, lipase, and proteinase, makes it a successful wide-spectrum biocontrol agent (van Tilburg and Thomas 1993). In terms of plant growth-promoting endofungal agents, it has been generally observed that fungus belonging to Trichoderma genus interact with plants by inducing the plant’s defense system and promotes the plant growth (Zhang et al. 2016). The endosphere ability of Trichoderma has been reported in many plants, like maize, cucumber, cotton, and tomato (Yuan et al. 2017; Contreras-Cornejo et al. 2018; Harman et al. 2019). Trichoderma colonizes in the intracellular spaces, as well as the spaces between the plasma membrane and the cell wall of the root tissue (Ramírez-Valdespino et al. 2019). Once in the endosphere, the fungus can promote seed germination, development of root, shoot, and biomass, and increase tiller number and overall crop yield (Hajieghrari and Mohammadi 2016). In addition, Trichoderma can exhibit antagonistic activities against its adversaries through competition for space and nutrients, antibiosis and mycoparasitism (Chen et al. 2016).
The relative abundance of microbial metabolic genes, pathways, and orthologies were more abundant in rhizosphere metagenome compared to the endosphere counterpart. An increase in the relative abundance of few agriculturally important traits (genes and metabolic pathways) in the endosphere suggested that the tea plant system can selectively permit the penetration and establishment of an elite group of symbionts which help the plant in overcoming various stress conditions. For instance, KEGG-based analogies showed that abundance of several previously reported genes related to vitamin B6 and B9 metabolism (ko00750, ko00790), steroid biosynthesis (ko00100), glutathione metabolism (ko00480), etc., was relatively higher in the endosphere than in the rhizosphere. These pathways and metabolisms improve plant biomass production and empower the plant defense mechanism against various biotic and abiotic stress conditions (Zhu et al. 1999; del Barrio-Duque et al. 2019). In the presence of phytopathogens in soils, the plant has to modulate or reprogram its defense signaling processes and activates suitable yet costly pathogenesis-related pathways. To subsidize this metabolic cost, the resilient nature of the plant defense mechanism partly engineers (active recruitment) the beneficial rhizosphere soil microbiome and depends on it in a semi-intrinsic manner (Singh et al. 2016b). Therefore, it was assumed that the presence of potential phytopathogenic agents, such as Gibberella, Aspergillus, and Ustilago, induced the plant to actively recruit a few beneficial endospheres as a step of disease priming. Metabolic pathway genes linked to “ABC transporter” was most abundant in the two environments (endosphere: 4.8%; rhizosphere: 6.4%) followed by “Two-component system” (3.7% in both). In bacteria, the ATP binding cassette (ABC) superfamily comprises mainly ATP-dependent pump proteins dedicated toward active mobilization of metabolites (Lewis et al. 2012). These superfamily proteins mobilize hormones, lipids, peptides, and primary as well as secondary metabolites, etc., and therefore, have immense significance in plant growth and adaptations (Vasiliou et al. 2009). A few members also have intrinsic channel properties with functions related to drug resistance, heavy metal detoxification, etc. (Lewis et al. 2012). In addition, these proteins also have defense-related implications as evidenced by the presence of an early defense gene in rice which is induced in the presence of salicylic acid and phytopathogenic Magnaporthe grisea (Lee et al. 2005). The two-component system consists of a first component protein with transmembrane domain and a second component protein with phosphotransfer histidine kinase property (Zschiedrich et al. 2016). These systems play an important role in plant–microbe interaction by recognizing the active metabolites present in the root exudates and thereby facilitate chemotaxic movement of the microbial population toward the root surface. The GacS/GacA present in pseudomonads and enteric bacteria exemplifies such systems that are responsible for microbial root colonization (Heeb and Haas 2001). Further, we analyzed potential functional genes linked with plant growth promotion activities (ACC deaminase production, IAA production, N2 fixation, phosphate solubilization, siderophore production), abiotic stress alleviation (betaine utilization), plant defense mechanism (lipopolysaccharide production), and plant disease suppression (chitinase and β-1,3-glucanase) prevalent in the belowground tea microbial community. The rhizosphere microbiota exhibited a significant prevalence of PGPTs related to betaine utilization; this important trait boosts up osmolyte concentration and antioxidant activity in plants under various abiotic stresses (Malekzadeh 2015). The second most frequently detected PGPT in the rhizosphere was related to siderophore production. Siderophores sequester iron through chelation, thereby playing a crucial role in increasing bioavailability and internalization of the ions to the plant (Hider and Kong 2010). The rhizosphere metagenome harbored important PGPTs related to lipopolysaccharide production (toward soil health improvement), N2 fixation, plant disease suppression, phosphate solubilization, ACC deaminase production, and IAA production. Among these many traits, production of lipopolysaccharide, a ubiquitous component of Gram-negative bacteria holds special ground for the induction of systemic resistance in plants against various phytopathogens (Erbs and Newman 2003). However, the endosphere metagenome did not harbor any detectable frequency of lipopolysaccharide genes probably due to high abundance of Proteobacteria (89.17%) and Actinobacteria (5.08%) and low levels of Gram-negative Bacteroidetes (1.52%) in the metagenome. Nonetheless, the endosphere microbiota contained some important PGPTs related to plant disease suppression (Chitinase, β-1,3-glucanase), N2 fixation, betaine utilization, IAA production, siderophore production, phosphate, and ACC deaminase production (5.78, 5.15, 4.81, 3.65, 2.92, 2.68, 2.15 per 105 reads, respectively). Tea plants require a large amount of N fertilizer (90 kg N/10 acres/year) for theanine accumulation in the first flush shoots; therefore, the amount of applied biofertilizer may have modulated the diversity and functionality of the tea microbiome especially in the endosphere.
Conclusion
Our study using culture-independent approach provided a comparative insight into the microbial diversity and function associated with an organically grown tea ecosystem of Assam. EggNOG-based COG analysis of the metagenomes revealed that microbial metabolism was most prevalent in both the rhizosphere and root endosphere environments. The root endosphere metagenome carried a higher relative abundance of genes linked to “information storage and processing” which is integral to any kind of plant–microbe interaction (mutualism or antagonism). KOBAS-mediated KEGG Orthology analysis of the metagenomes showed that the “ABC transporter pathway” genes were highly abundant followed by the “Two-component system pathway”-related genes in both the environments. As compared to the rhizosphere, the root endosphere contained higher abundances of PMI-important genes linked to bacterial chemotaxis, vitamin B6 and folate metabolism, glutathione metabolism, etc. The Shannon diversity indices confirmed higher bacterial diversity in the rhizosphere compared to the endosphere. Bacterial species, such as Serratia, Methylobacterium, Yersinia, and Burkholderia, populated the root endosphere and also had minor representations of few agriculturally important symbiotic and non-symbiotic taxa, such as Bradyrhizobium, Mesorhizobium, Azotobacter, Azospirillum, Azomonas, Bacillus, and Pseudomonas. Overall, a vast array of agriculturally important microbes linked to phytohormone secretion, nitrogen fixation, phosphate and potash solubilization, biotic–abiotic stress tolerance and biocontrol (entomopathogenesis) activity were detected. The exposition of microbial diversity, function, and indicators revealed in this study was supported by our earlier reports of the PGP activities of some of these isolates. This study will pave a way for including natural enemies prevalent in tea plantations for protecting the plants from various pests and diseases. Further studies on the cross-talks of a tripartite interaction encompassing tea plants, bioinoculum, and natural enemies can be considered for the promotion of a sustainable and healthy tea ecosystem.
Availability of data and materials
The metagenomes are publically available in the NCBI-SRA database (PRJNA698063).
References
Aeron A, Kumar S, Pandey P, Maheshwari DK (2011) Emerging role of plant growth promoting rhizobacteria in agrobiology. In: Maheshwari DK (ed) Bacteria in agrobiology: crop ecosystems. Springer, Berlin, pp 1–36
Aquilanti L, Favilli F, Clementi F (2004) Comparison of different strategies for isolation and preliminary identification of Azotobacter from soil samples. Soil Biol Biochem 36:1475–1483. https://doi.org/10.1016/j.soilbio.2004.04.024
Barea J-M, Pozo MJ, Azcón R, Azcón-Aguilar C (2005) Microbial co-operation in the rhizosphere. J Exp Bot 56:1761–1778. https://doi.org/10.1093/jxb/eri197
Baruah P (2017) Wild teas of Assam and North East India. J Tea Sci Res. https://doi.org/10.5376/jtsr.2017.07.0007
Bhattacharyya PN, Sarmah SR (2018) The role of microbes in tea cultivation. In: Sharma VS, Gunasekare MTK (eds) Global tea science: current status and future needs, first. Burleigh Dodds science publishing, Cambridge, pp 135–168
Chen JL, Sun SZ, Miao CP et al (2016) Endophytic Trichoderma gamsii YIM PH30019: a promising biocontrol agent with hyperosmolar, mycoparasitism, and antagonistic activities of induced volatile organic compounds on root-rot pathogenic fungi of Panax notoginseng. J Ginseng Res 40:315–324. https://doi.org/10.1016/j.jgr.2015.09.006
Cole JR, Wang Q, Fish JA et al (2014) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633. https://doi.org/10.1093/nar/gkt1244
Contreras-Cornejo HA, Macías-Rodríguez L, del-Val E, Larsen J (2018) The root endophytic fungus Trichoderma atroviride induces foliar herbivory resistance in maize plants. Appl Soil Ecol 124:45–53. https://doi.org/10.1016/j.apsoil.2017.10.004
de Godoy RCB, Deliza R, Gheno LB et al (2013) Consumer perceptions, attitudes and acceptance of new and traditional mate tea products. Food Res Int 53:801–807. https://doi.org/10.1016/j.foodres.2013.02.054
del Barrio-Duque A, Ley J, Samad A et al (2019) Beneficial endophytic bacteria-serendipita indica interaction for crop enhancement and resistance to phytopathogens. Front Microbiol. https://doi.org/10.3389/fmicb.2019.02888
Doanh N, Thuong N, Heo Y (2018) Impact of conversion to organic tea cultivation on household income in the mountainous areas of Northern Vietnam. Sustainability 10:4475. https://doi.org/10.3390/su10124475
Erbs G, Newman M-A (2003) The role of lipopolysaccharides in induction of plant defence responses. Mol Plant Pathol 4:421–425. https://doi.org/10.1046/j.1364-3703.2003.00179.x
Fernández-González AJ, Martínez-Hidalgo P, Cobo-Díaz JF et al (2017) The rhizosphere microbiome of burned holm-oak: potential role of the genus Arthrobacter in the recovery of burned soils. Sci Rep 7:6008. https://doi.org/10.1038/s41598-017-06112-3
Fernández-González AJ, Villadas PJ, Gómez-Lama Cabanás C et al (2019) Defining the root endosphere and rhizosphere microbiomes from the World Olive Germplasm Collection. Sci Rep 9:20423. https://doi.org/10.1038/s41598-019-56977-9
Gebrewold AZ (2018) Review on integrated nutrient management of tea (Camellia sinensis L.). Cogent Food Agric 4:1543536. https://doi.org/10.1080/23311932.2018.1543536
Goswami G, Deka P, Das P et al (2017a) Diversity and functional properties of acid-tolerant bacteria isolated from tea plantation soil of Assam. 3 Biotech 7:229. https://doi.org/10.1007/s13205-017-0864-9
Hajieghrari B, Mohammadi M (2016) Growth-promoting activity of indigenous trichoderma isolates on wheat seed germination, seedling growth and yield. Aust J Crop Sci 10:1339–1347. https://doi.org/10.21475/ajcs.2016.10.09.p7857
Harman GE, Doni F, Khadka RB, Uphoff N (2019) Endophytic strains of Trichoderma increase plants’ photosynthetic capability. J Appl Microbiol. https://doi.org/10.1111/jam.14368
Hassani MA, Durán P, Hacquard S (2018) Microbial interactions within the plant holobiont. Microbiome 6:58
Heeb S, Haas D (2001) Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol Plant Microbe Interact 14:1351–1363
Hider RC, Kong X (2010) Chemistry and biology of siderophores. Nat Prod Rep 27:637. https://doi.org/10.1039/b906679a
Hu X, Chen J, Guo J (2006) Two Phosphate- and Potassium-solubilizing Bacteria Isolated from Tianmu Mountain, Zhejiang, China. World J Microbiol Biotechnol 22:983–990. https://doi.org/10.1007/s11274-006-9144-2
Huerta-Cepas J, Szklarczyk D, Forslund K et al (2016) EGGNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. https://doi.org/10.1093/nar/gkv1248
Hyatt D, Chen GL, LoCascio PF et al (2010) Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11:119. https://doi.org/10.1186/1471-2105-11-119
Idris AL, Fan X, Muhammad MH et al (2020) Ecologically controlling insect and mite pests of tea plants with microbial pesticides: a review. Arch Microbiol 202:1275–1284. https://doi.org/10.1007/s00203-020-01862-7
Jayasinghe SL, Kumar L (2020) Climate change may imperil tea production in the four major tea producers according to climate prediction models. Agronomy 10:1536. https://doi.org/10.3390/agronomy10101536
Krauss U, Hidalgo E, Arroyo C, Piper SR (2004) Interaction between the Entomopathogens Beauveria bassiana, Metarhizium anisopliae and Paecilomyces fumosoroseus and the Mycoparasites Clonostachys spp., Trichoderma harzianum and Lecanicillium lecanii. Biocontrol Sci Technol 14:331–346. https://doi.org/10.1080/09583150410001665196
Lee Y-J, Yamamoto K, Hamamoto H et al (2005) A novel ABC transporter gene ABC2 involved in multidrug susceptibility but not pathogenicity in rice blast fungus, Magnaporthe grisea. Pestic Biochem Physiol 81:13–23. https://doi.org/10.1016/j.pestbp.2004.07.007
Lewis VG, Ween MP, McDevitt CA (2012) The role of ATP-binding cassette transporters in bacterial pathogenicity. Protoplasma 249:919–942. https://doi.org/10.1007/s00709-011-0360-8
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Ma L, Cao YH, Cheng MH et al (2013) Phylogenetic diversity of bacterial endophytes of Panax notoginseng with antagonistic characteristics towards pathogens of root-rot disease complex. Antonie Van Leeuwenhoek 103:299–312. https://doi.org/10.1007/s10482-012-9810-3
Malekzadeh P (2015) Influence of exogenous application of glycinebetaine on antioxidative system and growth of salt-stressed soybean seedlings (Glycine max L.). Physiol Mol Biol Plants 21:225–232. https://doi.org/10.1007/s12298-015-0292-4
Meegahakumbura MK, Wambulwa MC, Li MM et al (2018) Domestication origin and breeding history of the tea plant (Camellia sinensis) in China and India based on nuclear microsatellites and cpDNA sequence data. Front Plant Sci. https://doi.org/10.3389/fpls.2017.02270
Meena KK, Sorty AM, Bitla UM et al (2017) Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front Plant Sci. https://doi.org/10.3389/fpls.2017.00172
Mohanram S, Kumar P (2019) Rhizosphere microbiome: revisiting the synergy of plant–microbe interactions. Ann Microbiol 69:307–320
Mukhopadhyay M, Mondal TK, Chand PK (2016) Biotechnological advances in tea (Camellia sinensis [L.] O. Kuntze): a review. Plant Cell Rep 35:255–287. https://doi.org/10.1007/s00299-015-1884-8
Nath R, Sharma GD, Barooah M (2013) Screening of endophytic bacterial isolates of tea (Camellia sinensis L.) roots for their multiple plant growth promoting activities. Int J Agric Environ Biotechnol 6:371. https://doi.org/10.5958/j.2230-732X.6.3.005
Nath R, Sharma GD, Barooah M (2015) Plant growth promoting endophytic fungi isolated from tea (Camellia sinensis) shrubs of Assam, India. Appl Ecol Environ Res. https://doi.org/10.15666/aeer/1303_877891
Ramírez-Valdespino CA, Casas-Flores S, Olmedo-Monfil V (2019) Trichoderma as a model to study effector-like molecules. Front Microbiol 10:1030. https://doi.org/10.3389/fmicb.2019.01030
Sandhu SS, Sharma AK, Beniwal V et al (2012) Myco-biocontrol of insect pests: factors involved, mechanism, and regulation. J Pathog 2012:1–10. https://doi.org/10.1155/2012/126819
Scheepmaker JWA, Butt TM (2010) Natural and released inoculum levels of entomopathogenic fungal biocontrol agents in soil in relation to risk assessment and in accordance with EU regulations. Biocontrol Sci Technol. https://doi.org/10.1080/09583150903545035
Singh AK, Bisen JS, Chauhan RK et al (2016a) Tea research for Darjeeling tea industry: various aspects. In: Bag N, Bag A, Palni LMS (eds) Tea: technological initiatives, first. New India Publishing Agency, New Delhi, pp 195–239
Singh UB, Malviya D, Wasiullah D et al (2016b) Bio-protective microbial agents from rhizosphere eco-systems trigger plant defense responses provide protection against sheath blight disease in rice (Oryza sativa L.). Microbiol Res 192:300–312. https://doi.org/10.1016/j.micres.2016.08.007
Singh HB, Keswani C, Reddy MS et al (2019) Secondary metabolites of plant growth promoting rhizomicroorganisms: discovery and applications. Springer, Singapore
Smolińska U, Kowalska B (2018) Biological control of the soil-borne fungal pathogen Sclerotinia sclerotiorum—a review. J Plant Pathol 100:1–12
Sreenivasaprasad S, Manibhushanrao K (1990) Antagonistic potential of Gliocladium virens and Trichoderma longibrachiatum to phytopathogenic fungi. Mycopathologia 109:19–26. https://doi.org/10.1007/BF00437002
Tsurumaru H, Okubo T, Okazaki K et al (2015) Metagenomic analysis of the bacterial community associated with the taproot of sugar beet. Microbes Environ 30:63–69. https://doi.org/10.1264/jsme2.ME14109
van Tilburg AU, Thomas MD (1993) Production of extracellular proteins by the biocontrol fungus Gliocladium virens. Appl Environ Microbiol 59:236–242. https://doi.org/10.1128/AEM.59.1.236-242.1993
Vasiliou V, Vasiliou K, Nebert DW (2009) Human ATP-binding cassette (ABC) transporter family. Hum Genom 3:281–290. https://doi.org/10.1186/1479-7364-3-3-281
Verma M, Mishra J, Arora NK (2019) Plant growth-promoting rhizobacteria: diversity and applications. In: Sobti RC, Arora NK, Kothari R (eds) Environmental biotechnology: for sustainable future. Springer, Singapore, pp 129–173
Vieira S, Sikorski J, Dietz S et al (2020) Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J 14:463–475. https://doi.org/10.1038/s41396-019-0543-4
War Nongkhlaw F, Joshi S (2017) Microscopic study on colonization and antimicrobial property of endophytic bacteria associated with ethnomedicinal plants of Meghalaya. J Microsc Ultrastruct 5:132. https://doi.org/10.1016/j.jmau.2016.09.002
Wu S, Zhu Z, Fu L et al (2011) WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genom 12:444. https://doi.org/10.1186/1471-2164-12-444
Xie C, Mao X, Huang J et al (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316. https://doi.org/10.1093/nar/gkr483
Xie H, Feng X, Wang M et al (2020) Implications of endophytic microbiota in Camellia sinensis: a review on current understanding and future insights. Bioengineered 11:1001–1015. https://doi.org/10.1080/21655979.2020.1816788
Yuan Y, Feng H, Wang L et al (2017) Potential of endophytic fungi isolated from cotton roots for biological control against verticillium wilt disease. PLoS One 12:e0170557. https://doi.org/10.1371/journal.pone.0170557
Yugandhar V, Chandrashekhar G, Bhattacharjee H (2018) Economics of growing tea organically in the initial years in terai zone of West Bengal condition. Int J Curr Microbiol Appl Sci 7:387–391. https://doi.org/10.20546/ijcmas.2018.705.050
Zhang S, Gan Y, Xu B (2016) Application of plant-growth-promoting fungi Trichoderma longibrachiatum T6 enhances tolerance of wheat to salt stress through improvement of antioxidative defense system and gene expression. Front Plant Sci 07:1405. https://doi.org/10.3389/fpls.2016.01405
Zhang Z, Zhou C, Xu Y et al (2017) Effects of intercropping tea with aromatic plants on population dynamics of arthropods in Chinese tea plantations. J Pest Sci (2004) 90:227–237. https://doi.org/10.1007/s10340-016-0783-2
Zhao H, Zhou T, Xie J et al (2020) Mycoparasitism illuminated by genome and transcriptome sequencing of Coniothyrium minitans, an important biocontrol fungus of the plant pathogen Sclerotinia sclerotiorum. Microb Genom. https://doi.org/10.1099/mgen.0.000345
Zhu YL, Pilon-Smits EAH, Jouanin L, Terry N (1999) Overexpression of glutathione synthetase in Indian mustard enhances cadmium accumulation and tolerance. Plant Physiol 119:73–79. https://doi.org/10.1104/pp.119.1.73
Zschiedrich CP, Keidel V, Szurmant H (2016) Molecular mechanisms of two-component signal transduction. J Mol Biol 428:3752–3775. https://doi.org/10.1016/j.jmb.2016.08.003
Acknowledgements
The authors are grateful to the Department of Biotechnology (DBT), Ministry of Science and Technology, Govt. of India for funding the research. The authors also kindly acknowledge the timely suggestions, encouragement, and reviews received from Professor B. K. Sarmah (Director, DBT-NECAB, AAU, Jorhat) throughout the research work.
Funding
This study was funded by the Department of Biotechnology, Govt. of India (DBT, GoI), through the DBT-North East Centre for Agricultural Biotechnology, Programme-III entitled “Gene prospecting from acid soil microbes.”
Author information
Authors and Affiliations
Contributions
MB had conceived and designed the study. SSB and MBorah had carried out the experiments and collected the data. SSB, KKD, MG, MKD, and MR had analyzed the data and prepared the charts. The manuscript was jointly written by SSB and MB.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
This study involved metagenomics of environmental samples only.
Consent to participate
NA.
Consent for publication
All the authors have given consent for publication.
Additional information
Communicated by Govarthanan Muthusamy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bora, S.S., Dey, K.K., Borah, M. et al. Niche differentiation of belowground microorganisms and their functional signatures in Assam type tea (Camellia sinensis var. assamica). Arch Microbiol 203, 5661–5674 (2021). https://doi.org/10.1007/s00203-021-02547-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02547-5