Abstract
A Gram-stain-negative, oval or short rod-shaped, non-motile, aerobic bacterium, designated strain S1109LT, was isolated from a marine sediment in Weihai, PR China. Cells were oxidase positive and catalase positive. Growth of strain S1109LT occurred at 10–40 °C (optimum, 30–33 °C), pH 6.5–10.0 (optimum, 7.0–8.0) and in the presence of 1–21% (optimum, 4–6%) (w/v) NaCl. 16S rRNA gene sequence phylogeny indicated that strain S1109LT was associated with the genus Pontibaca of the family Rhodobacteraceae because it showed the highest sequence similarity to Pontibaca methylaminivorans KCTC 22497T (97.5%). The average nucleotide identity (ANI) and the digital DNA–DNA hybridization (dDDH) scores between strain S1109LT and Pontibaca methylaminivorans KCTC 22497T were 74.6% and 18.7%. The major cellular fatty acids of strain S1109LT were C19:0 cyclo ω8c and C18:1 ω7c. The polar lipids profiles of strain S1109LT were phosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine and two unidentified lipids. Strain S1109LT contained ubiquinone-10 as the major respiratory quinone. The genomic DNA G + C content was 55.9 mol%. On the basis of the evidence presented in this study, strain S1109LT is considered to represent a novel species of the genus Pontibaca, for which the name Pontibaca salina sp. nov. is proposed. The type strain of is S1109LT (= KCTC 82411T = MCCC 1H00441T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Pontibaca, a member of the family Rhodobacteraceae, was described by Kwang Kyu et al. (Kim et al. 2010). Pontibaca methylaminivorans, the only member of the genus Pontibaca, was isolated from coastal sediment of the East Sea, Korea. The aim of the present work was to report on the taxonomic characterization of a Pontibaca-like bacterial strain, S1109LT, which was isolated from marine sediment sample collected from Weihai.
Materials and methods
Isolation of the strain and culture conditions
Strain S1109LT was isolated from marine sediment sample collected from Weihai, PR China (37° 33′ 57.60″ N, 122° 7′ 38.80″ E). The sample, after being appropriately diluted with sterile seawater, was coated on marine sandwich agar plate for cultivation at 28 °C. The marine sandwich agar plate was prepared with marine agar 2216 (MA; Becton Dickinson) as the bottom layer, the growth-promoting bacterium Roseibacterium beibuensis MCCC 1F00103T, which can provide growth promoting effect, as the middle layer and 0.1 × marine agar 2216 (MA; Becton Dickinson) as the top layer (the method has not been published). Strain S1109LT was isolated and purified by sub-culture on MA plates. Strain S1109LT was routinely cultured on MA at 30 °C (certified to be optimum) and preserved in marine broth 2216 (MB) containing glycerol (20%, v/v) at − 80 °C.
16S rRNA gene sequencing and phylogenetic analyses
The 16S rRNA gen was amplified by PCR from cell lysate using the universal oligonucleotide primers 27F (5′-AGAGTTTGATC(A/C)TGGCTCAG-3′) and 1492R (5′-TACGG(C/T)TACCTTGTTACGACTT-3′) (Liu et al. 2014). The amplified production was purified and ligated into the vector pMD18-T (TaKaRa). The ligation product was transformed into Escherichia coli DH5α cells (Trans-Gen Biotech). The clones of transformed Escherichia coli DH5α, which can grow on LB agar containing 0.1% ampicillin, were sequenced by GBI Co., Ltd (Qingdao, PR China). Sequences were compared with the available 16S rRNA gene sequences using BLAST search program (http://www.ncbi.nlm.nih.gov/blast.cgi) and EzBioCloud sever (https://www.ezbiocloud.net/identify). The 16S rRNA gene sequence of strain S1109LT and other related strains were aligned using CLUSTAL_X (Thompson et al. 1997). The phylogenetic trees of 16S rRNA gene were constructed using the neighbour-joining, maximum-parsimony and maximum-likelihood algorithms supported by bootstrap test with 1000 replications in MEGA 7.0 software. (Kimura 1980; Kumar et al. 2016). Evolutionary distance matrices were computed using the Kimura’s two-parameter model (Kimura 1980).
Genome extraction and analysis
Genomic DNA of strain S1109LT was extracted and purified by using a TaKaRa miniBEST universal genomic DNA extraction kit following the instruction’s manual. Whole genome sequencing of strain S1109LT was performed by Beijing Novogene Bioinformatics Technology using the Illumina HiSeq PE150 platform. All good quality paired reads were assembled using SOAP de novo version 2.04 software (Li, 2010). The average nucleotide identity (ANI) values and the digital DNA–DNA hybridization (dDDH) values were analyzed using the average nucleotide identity (ANI) calculator (https://www.ezbiocloud.net/tools/ani) and genome-to-genome distance calculator (GGDC) (http://ggdc.dsmz.de/ggdc.php#). The genes of involved in S1109LT metabolic pathways were annotated using KEGG database (Aziz, 2008) and the RAST Server (Kanehisa et al. 2016).
Physiology and chemotaxonomy
Considering the highest 16S rRNA gene sequence similarities to strain S1109LT from EzBioCloud sever, Pontibaca methylaminivorans KCTC 22497T (type species) and Primorskyibacter marinus KCTC 42952T were chosen as reference strains for taxonomic analysis of strain S1109LT. Pontibaca methylaminivorans KCTC 22497T and Primorskyibacter marinus KCTC 42952T were purchased from Marine Culture Collection of China and Korean Collection for Type Cultures, respectively.
Colony morphology was observed on MA after incubation at 30 °C for 3 days. Cell morphology was observed using light microscopy (E600, Nikon) and scanning electron microscopy (Nova NanoSEM450, FEI). The Gram staining was performed by using the bioMérieux Gram-stain kit as manufacturer’s instructions. Motility was examined described by Gerhardt et al. (PhilippGerhardt et al. 1994) and Bowman et al. (Bowman 2000). The temperature range for growth was tested at 0, 4, 10, 15, 20, 25, 28, 30, 33, 37, 42 and 45 °C on MA. Growth at various concentrations of NaCl was examined using a medium containing 0.1% (w/v) yeast extract, 0.5% (w/v) peptone and 2.0% (w/v) agar and comprising 0.0–30.0% (w/v) NaCl (in increments of 1.0%), prepared with artificial seawater (0.32% MgSO4, 0.12% CaCl2, 0.07% KCl and 0.02% NaHCO3, all w/v). The growth at pH range was measured in MB with pH 5.5 to 9.5 (in increments of 0.5 pH unit), using 20 mM MES (pH 5.5 and 6.0), PIPES (pH 6.5 and 7.0), HEPES (pH 7.5 and 8.0), Tricine (pH 8.5) and CAPSO (pH 9.0 and 9.5) for supplement during pH adjustment. Anaerobic growth was investigated on MA with or without 0.1% (w/v) KNO3 in an anaerobic jar with an atmosphere of 10% H2, 5% CO2 and 85% N2 for 14 days at 30 °C.
Catalase activity was measured by bubble production after dripping 3% (v/v) aqueous hydrogen peroxide solution onto fresh colonies. Oxidase activity was determined using the bioMérieux oxidase reagent kit according to the manufacturer’s protocol. Hydrolysis of starch, casein, alginate, CM cellulose and Tweens (20, 40, 60 and 80) were tested as described by Smibert and Krieg (Smibert and Krieg 1994). Hydrolysis of DNA was tested on DNase test agar (BD Diagnostics), with the modification that artificial seawater was used for the preparation of media. Other physiological and biochemical characteristics were investigated using API 20NE, API 50CHB, API ZYM strips (all from bioMérieux) and the Biolog GEN III MicroPlate kit according to the instruction manual (except salinity of the suspension medium, which was adjusted to 5.0%).
Susceptibility to antibiotics was measured on MA using the diffusion plate technique with the following procedure (Du et al. 2014). The cell suspension (McFarland standard 0.5) was evenly coated on MA; Commercial antibiotic discs (Hangzhou Binhe Microorganism Reagent Co., Ltd) were placed on to the surface; and zones of growth inhibition were registered following CLSI standards after 48 h of incubation at 30 °C.
The quinones and polar lipids were extracted as Minnikin described (Minnikin, 1984). The quinones were separated by a silica-gel thin-layer chromatography (TLC, Merck) plate and tested by HPLC (Kroppenstedt and Reiner 1982; Hiraishi et al. 1996). The polar lipids were separated by two-dimensional TLC on silica gel 60 F254 plates (Merck) using chloroform/methanol/water (65:25:4, by vol.) for the first dimension and chloroform/acetic acid/methanol/water (80:15:12:4, by vol.) for the second dimension. Molybdatophosphoric acid, ninhydrin, a-naphthol and the spray reagent based on the Zinzadze reagent (Vaskovsky and Kostetsky 1968) were used to detect total lipids, aminolipids, glycolipids and phospholipids, respectively (Collins et al. 1980). Major cellular fatty acids were extracted as described (Eder 1995), identified by an Agilent 6890 N gas chromatograph and analyzed using the TSBA40 database of the Microbial Identification System (Sasser 1990).
Results
Phylogenetic analysis
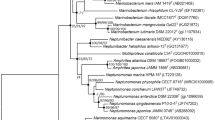
Strain S1109LT shared the highest sequence similarity with Pontibaca methylaminivorans KCTC 22497T (97.5%) and Primorskyibacter marinus KCTC 42952T (95.9%) by analysis of the complete 16S rRNA gene sequence. The neighbour-joining phylogenetic tree is given in Fig. 1. Neighbour-joining, maximum-likelihood and maximum-parsimony trees all showed that strain S1109LT should be classified as a separate species of Pontibaca genus because of clustering with the closely related type strains Pontibaca methylaminivorans KCTC 22497T (Fig. S1).
Neighbour-joining phylogeny reconstructed with 16S rRNA gene sequences showing the position of strain S1109LT among related taxa. The strain characterized in this study is shown in bold type. Only bootstrap values (percentages of 1000 resamplings) above 50% are shown. Antarcticimicrobium sediminis S4J41T (MK120109) is used to root the tree. Solid circles indicate the corresponding nodes are also recovered in the trees generated with the maximum-likelihood and maximum-parsimony algorithms. Bar, 0.005 substitutions per nucleotide position
Only one 16S rRNA gene sequence (1479 bp) was detected at whole genome level and had 99.9% similarity to the cloned sequence (1427 bp). There were two base pairs of differences between the two sequences (12th and 1129th of the cloned sequence). The genome sequence of strain S1109LT, consisting of 41 contigs, was 2,934,809 bp in length (N50 value of 221,932 bp) with 1099.7 × coverage. The genomic DNA G + C content was 55.9 mol%. The genome sequences of Pontibaca methylaminivorans KCTC 22497 T and Primorskyibacter marinus KCTC 42952T were obtained from NCBI with GenBank accession numbers NZ_FTPS00000000.1 and NZ_QJSD00000000.1, respectively. The ANI values between strain S1109LT and Pontibaca methylaminivorans KCTC 22497T and Primorskyibacter marinus KCTC 42952T were 74.6% and 72.3%, respectively. The dDDH values between strain S1109LT and Pontibaca methylaminivorans KCTC 22497T and Primorskyibacter marinus KCTC 42952T were 18.7% and 19.3%, respectively. Both the ANI and dDDH values are below the cutoff values for species differentiation (Richter and Rosselló-Móra 2009; Meier-Kolthoff et al. 2013). These data support that strain S1109LT represents a novel species of the genus Pontibaca.
Sequencing of the draft genome of strain S1109LT allowed genomic analyses. Strain S1109LT contained 168 genes for carbohydrate metabolism and 117 genes for amino acid metabolism. The genome of strain S1109LT contained 2 genes, proA and proB, for carbapenem biosynthesis. Carbapenem antibiotics, relatively resistant to hydrolysis by most β-lactamases, are generally considered to have broad-spectrum antibacterial effect. To test whether strain S1109LT have carbapenem biosynthesis, strain S1109LT was inoculated on MA, with carbapenem-sensitive Escherichia coli KCTC 25922T and carbapenem-resistant Escherichia coli (CR-Eco) on left side and right side, respectively. The results showed that no growth inhibition occurred in Escherichia coli KCTC 25922T around the growth lawn of S1109LT after 48 h of incubation at 30 °C (Fig. S2). These indicated that there was no or inadequate carbapenem biosynthesis in strain S1109LT to inhibit the growth of Escherichia coli KCTC 25922T.
Phenotypic characteristics
Phenotypic characteristics of strain S1109LT are displayed in Table 1 and in the species description.
Strain S1109LT was found to be sensitive to penicillin (10 µg), ampicillin (10 µg), carbenicillin (100 µg), cefotaxime sodium (30 µg), ceftriaxone (30 µg), clarithromycin (15 µg), polymyxin B (300 µg), chloramphenicol (30 µg) and resistant to gentamycin (10 µg), tobramycin (10 µg), kanamycin (30 µg), neomycin (30 µg), tetracycline (30 µg), ofloxacin (5 µg), streptomycin (10 µg), norfloxacin (10 µg) and vancomycin (30 µg).
The major isoprenoid quinone of strain S1109LT was ubiquinone-10 (Q-10), which was the same as that found in Pontibaca methylaminivorans KCTC 22497T and Primorskyibacter marinus KCTC 42952T. The major polar lipids detected in strain S1109LT were phosphatidylglycerol (PG), phosphatidylcholine (PC), phosphatidylethanolamine (PE) and two unidentified lipid (L1-2). The polar lipid profile of strain S1109LT was in line with that detected in Pontibaca methylaminivorans KCTC 22497T and similar to that major lipids in Primorskyibacter marinus KCTC 42952T, but diphosphatidylglycerol (DPG) was only found in Primorskyibacter marinus KCTC 42952T. Further detailed polar lipid images of different specific stains are given in Fig. S3.
The major fatty acids (> 10%) detected in strain S1109LT were C19:0 cyclo ω8c and C18:1 ω7c. The percentages of the fatty acid profile were similar to those in Pontibaca methylaminivorans KCTC 22497T. However, the presence of C16:0 was a distinct characteristic of the fatty acid composition of Pontibaca methylaminivorans KCTC 22497T. Unlike in strain S1109LT and Pontibaca methylaminivorans KCTC 22497T, C18:1 ω7c and C18:1 ω7c 11-methyl were major fatty acids in Primorskyibacter marinus KCTC 42952T. Differences in the fatty acid profiles of strain S1109LT, Pontibaca methylaminivorans KCTC 22497T and Primorskyibacter marinus KCTC 42952T are listed in Table 2.
Based on the phylogenetic, phenotypic and chemotaxonomic results mentioned above, strain S1109LT represents a new species of the genus Pontibaca, for which the name Pontibaca salina sp.nov. is proposed.
Description of Pontibaca salina sp. nov.
Pontibaca salina (sa.li’na. N. L. fem. adj. salina salted, saline)
Cells are Gram-stain-negative, oval or short rod-shaped (0.4–0.5 × 0.5–1.0 µm) and non-motile. Colonies on MA are about 0.5–1 mm in diameter, circular, smooth, slightly convex, translucent and light-brown coloured after incubation for 3 days at 30 °C. Growth occurs at 10–40 °C (optimum, 30–33 °C), pH 6.5–10.0 (optimum, 7.0–8.0) and in the presence of 1–21% (optimum, 4–6%) (w/v) NaCl. Catalase and oxidase are positive. Nitrate is not reduced. Urea and casein are hydrolysed, but starch, aesculin, gelatin, CM cellulose, alginate, DNA and Tweens (20, 40, 60 and 80) are not. Activities of leucine arylamidase and naphthol-AS-BI-phosphohydrolase are present. Acid are produced from d-fructose and potassium 5-keto-gluconate. In carbon source oxidation tests, positive results are obtained for l-alanine, l-arginine, l-glutamic acid, l-histidine, l-serine, glucuronamide, l-lactic acid, α-hydroxy-butyric acid, β-hydroxy-d,l-butyric acid, acetoacetic acid, propionic acid, acetic acid and formic acid. Ubiquinone-10 (Q-10) is the major isoprenoid quinone. The main fatty acids are C19:0 cyclo ω8c and C18:1 ω7c. The major polar lipids are phosphatidylglycerol, phosphatidylcholine, phosphatidylethanolamine and two unidentified lipids. The G + C content of the DNA is 55.9%.
The type strain, S1109LT (= KCTC 82411T = MCCC 1H00441T), was isolated from a marine sediment collected from Weihai, PR China (37° 33′ 57.60″ N, 122° 7′ 38.80″ E).
The GenBank accession number for the 16S rRNA gene sequence of strain S1109LT is MW422814. The Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAEIJD000000000.
Data availability
The GenBank accession number for the 16S rRNA gene sequence and whole-genome shotgun project of strain S1109LT are MW422814 and JAEIJD000000000, respectively.
Abbreviations
- MCCC:
-
Marine culture collection of China
- KCTC:
-
The Korean collection for type cultures
- ANI:
-
Average nucleotide identity
- GGDC:
-
Genome-to-genome distance calculator
- dDDH:
-
Digital DNA–DNA hybridization
- MEGA:
-
Molecular evolutionary genetics analysis
- MIDI:
-
Microbial identification system
- HPLC:
-
High-performance liquid chromatography
- TLC:
-
Thin layer chromatography
- PG:
-
Phosphatidylglycerol
- PE:
-
Phosphatidylethanolamine
- PC:
-
Phosphatidylcholine
- DPG:
-
Diphosphatidylglycerol
- MA:
-
Marine agar 2216
- MB:
-
Marine broth 2216
References
Aziz RK et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Bowman JP (2000) Description of Cellulophaga algicola sp. nov., isolated from the surfaces of Antarctic algae, and reclassification of Cytophaga uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Cellulophaga uliginosa comb nov. Int J Syst Evol Microbiol 50(Pt 5):1861–1868. https://doi.org/10.1099/00207713-50-5-1861
Collins MD, Goodfellow M, Minnikin DE (1980) Fatty acid, isoprenoid quinone and polar lipid composition in the classification of Curtobacterium and related taxa. J Gen Microbiol 118:29–37. https://doi.org/10.1099/00221287-118-1-29
Du ZJ, Wang Y, Dunlap C, Rooney AP, Chen GJ (2014) Draconibacterium orientale gen. nov., sp. nov., isolated from two distinct marine environments, and proposal of Draconibacteriaceae fam. nov. Int J Syst Evol Microbiol 64:1690–1696. https://doi.org/10.1099/ijs.0.056812-0
Eder K (1995) Gas chromatographic analysis of fatty acid methyl esters. J Chromatogr B Biomed Sci Appl 671:113–131. https://doi.org/10.1016/0378-4347(95)00142-6
Gerhardt P, Murray RGE, Wood W, Krieg N (1994) Methods for general and molecular bacteriology. American Society for Microbiology
Hiraishi A, Ueda Y, Ishihara J, Mori T (1996) Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J Gen Appl Microbiol 42:457–469. https://doi.org/10.2323/jgam.42.457
Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. https://doi.org/10.1016/j.jmb.2015.11.006
Kim KK, Lee JS, Lee KC, Oh HM, Kim SG (2010) Pontibaca methylaminivorans gen. nov., sp. nov., a member of the family Rhodobacteraceae. Int J Syst Evol Microbiol 60:2170–2175. https://doi.org/10.1099/ijs.0.020172-0
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/bf01731581
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Liq Chromatogr 5:2359–2367
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Li R et al (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272. https://doi.org/10.1101/gr.097261.109
Liu QQ, Wang Y, Li J, Du ZJ, Chen GJ (2014) Saccharicrinis carchari sp. nov., isolated from a shark, and emended descriptions of the genus Saccharicrinis and Saccharicrinis fermentans. Int J Syst Evol Microbiol 64:2204–2209. https://doi.org/10.1099/ijs.0.061986-0
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Minnikin D et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241. https://doi.org/10.1016/0167-7012(84)90018-6
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:1–6
Smibert RM, Krieg NR (1994) Phenotypic characterization. In methods for general and molecular bacteriology. American Society For Microbiology:611–651
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. https://doi.org/10.1093/nar/25.24.4876
Vaskovsky VE, Kostetsky EY (1968) Modified spray for the detection of phospholipids on thin-layer chromatograms. J Lipid Res 9:396
Wang NN, Sang J, Wang XQ, Li YX, Du ZJ (2018) Primorskyibacter marinus sp. nov., isolated from coastal sediment. Int J Syst Evol Microbiol 68:3169–3174. https://doi.org/10.1099/ijsem.0.002959
Acknowledgements
The scanning electron microscopy was supported by the Physical Chemical Materials Analytical and Testing Center of Shandong University at Weihai.
Funding
None.
Author information
Authors and Affiliations
Contributions
Strain S1109LT was isolated by J-SB. Material preparation, experimental operation, data collection and analysis were performed by J-SB, SW and X-Z Song. The manuscript was written by J-SB. Project guidance and critical revision of manuscripts was performed by Z-JD. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that there is no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bo, JS., Wang, S., Song, XZ. et al. Pontibaca salina sp. nov., isolated from marine sediment. Arch Microbiol 203, 4493–4498 (2021). https://doi.org/10.1007/s00203-021-02434-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02434-z