Abstract
A cultivated form of bacteria (strain 2202) was isolated from the hemal fluid of the bivalve mollusk Modiolus kurilensis. Based on the set of data collected by genetic and physiological/biochemical analyses, the strain was identified as the species Pseudoalteromonas piscicida. Strain 2202 exhibits antimicrobial activity against Staphylococcus aureus, Candida albicans, and Bacillus subtilis but not against Escherichia coli and Pseudomonas aeruginosa. These activities characterize the behavior of strain 2202 as predator-like and classify it as a facultative predator. Being part of the normal microflora in the hemolymph of M. kurilensis, when external conditions change, strain 2202 shows features of opportunistic microflora. The strain 2202 exhibits selective toxicity towards larvae of various invertebrates: it impairs the early development of Mytilus edulis, but not of Strongylocentrotus nudus. Thus, the selective manner in which P. piscicida strains interact with various species of microorganisms and eukaryotes should be taken into consideration when using their biotechnological potential as a probiotic in aquaculture, source of antimicrobial substances, and factors that prevent fouling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The new family Pseudoalteromonadaceae, comprising marine Alteromonas-like bacteria, was established relatively recently (Ivanova et al. 2004) and currently includes 49 validly described species, including synonyms: (http://www.bacterio.net/pseudoalteromonas.html). Pseudoalteromonads are distributed worldwide and occur among dominant culturable bacteria in the marine environment (Glöckner et al. 1999). They are isolated from seawater (Wu et al. 2017) and from fouling biofilms on stones and other nonliving objects (Park et al. 2005). Members of the genus Pseudoalteromonas are often associated with various aquatic organisms: mollusks (Ivanova et al. 2002; Donovan et al. 2009), micro- (Skerratt et al. 2002) and macroalgae (Radjasa et al. 2009), and fish (Nelson and Ghiorse 1999). Interest in these bacteria first arose due to their extreme prevalence in the marine environment and second due to their ability to synthesize a great number of metabolites that were active toward viruses, bacteria, fungi, macro-, and microalgae (Holmström and Kjelleberg1999), and tumor cells (Zheng et al. 2005, 2006). Large clades have been identified within the genus Pseudoalteromonas, which include nonpigmented species (e.g., P. haloplanktis) and pigmented species (e.g., P. piscicida, P. tunicate) (Gauthier et al. 1995). It should be noted that synthesis of biologically active substances is typical mainly of pigmented strains, but there is information about the production of the antifungal compound isatin by a nonpigmented strain of P. issachenkonii (Ivanova et al. 2014).
Like other Pseudoalteromonas, P. piscicida synthesizes active substances processing cytotoxic, antibacterial, algicidal activity (Bowman 2007; Harvey et al. 2016; Whalen et al. 2018). Some of these antimicrobial constituents are associated with pigments (Offret et al. 2016). Whole-genome sequencing of P. piscicida strains isolated from seawater also confirmed the presence DNA sites encoding a number of proteolytic enzymes with antibacterial properties (Richards et al. 2017).
Showing a broad range of biological activities, P. piscicida is considered as an antifouling organism. It should be noted that P. piscicida has been used as a probiotic, as the other species of the genus Pseudoalteromonas (Pham et al. 2014; Hamsah et al. 2017). However, in the literature, there is information reflecting the pathogenic potential of P. piscicida, which can serve as a barrier to the widespread use of this species of bacteria in aquaculture. P. piscicida is known as fish pathogen (Bowman 2007). Flavobacterium piscicida, which was later reclassified as Pseudoalteromonas piscicida (Gauthier et al. 1995), was a causative agent that induced the mass mortality of marine fish in Florida in the 1950s (Austin and Austin 2007). P. piscicida caused death of eggs of damselfish species, yellow clown fish, and staghorn damselfish, which are popular ornamental fish in marine (Nelson and Ghiorse 1999). In this regard, study of the pathways by which P. piscicida strains interact with the host organism and other components of marine ecosystems is of particular relevance.
In our study, a culturable strain of bacterium with high antimicrobial activity was isolated from the hemal fluid of the bivalve mollusk Modiolus kurilensis (F. R. Bernard, 1983). The goal of the present work was to identify the species of the bacterium, and also to determine the biological activity and possible pathogenic potential of this culturable strain. A pathogenicity analysis was carried out on adult mature individuals of the bivalves M. kurilensis and Mytilus edulis, embryos and larvae of M. edulis, as well as based on the highly sensitive sea urchin embryonic test (SET), which is widely used for bioassaying toxicity of different classes of xenobiotics, including inorganics, organics, different complex mixtures, and natural products (Pagano et al. 2017).
Materials and methods
Isolation and characteristic of strain 2202
The P. piscicida strain 2202 was isolated from M. kurilensis collected manually by divers at a depth of 3 m in Vostok Bay, Peter the Great Bay, Sea of Japan, in July 2017. The collected animals were placed in sterile containers and immediately delivered to the laboratory. Bivalves’ shells were flamed and opened aseptically. The hemal fluid was taken for inoculation; a 0.1 mL aliquot was transferred onto an agar medium and spread over the surface with a spatula. All tools, glassware, and seawater used for isolating cultures were sterilized by autoclaving. For cultivation, we used Marine agar 2216 (Difco). Dishes with inoculated media were kept at room temperature for 5 days. Separate colonies of different morphotypes after triple passage were taken for further studies. Cell morphology was examined in Gram-stained smears under a Prima Star light microscope (Carl Zeiss).
For scanning electron microscopy, pieces of agar with biofilm on their surface were fixed in 2.5% glutaraldehyde on cacodylate buffer (CB), pH 7.3, for one day, then washed with CB, postfixed with 1% OsO4 on CB, dehydrated in a series of alcohol solutions with increasing concentrations, then acetone, critical-point dried using BAL TEC CPD 030 (Irchel), coated with chromium on a Quorum Q150T ES (Quorum Technologies), and analyzed under a Sigma VP scanning electron microscope (Carl Zeiss). For transmission electron microscopy, the suspension of bacteria, prefixed with 2.5% glutaraldehyde on CB (for 2 h), and washed with CB and distilled water (to remove salts), was applied onto a single slot grids with Butvar film and examined using a Libra 120 transmission electron microscope (Carl Zeiss).
The properties of the isolated strains, including the presence of arginine dihydrolase, were identified according to Moeller (1955). Hydrolysis of gelatine, starch, and urease, glucose oxidation and fermentation by Hugh and Leifson (1953), reduction of nitrates, presence of cytochrome oxidase, catalase, the production of indole and the utilization of citrate, acid formation from lactose, maltose, xylose, and mannitol were determined by the methods of Smibert and Krieg (1994). The ability to grow in various NaCl concentrations was determined in a medium containing 5.0 g peptone, 2.5 g yeast extract, 1.0 g glucose, 0.2 g K2HPO4, 0.05 g MgSO4, 1000 mL distilled water, and 15.0 g agar with 0–10% NaCl. Growth at different temperatures (4–42 °C) was tested using Marine agar 2216 (Difco). Additional phenotypic characteristics of the strain were determined using an API 20NE test system (bioMérieus).
The strain 2202 of P. piscicida is stored at –80 °C in the LiCONiC STC Compact ULT automated biobanking system of Marine Biobank, NSCMB FEB RAS, under accession no. LV1003019546 (see http://marbank.dvo.ru/index.php/en/). In addition, the bacterial strain is stored at – 85 °C in cryotubes with seawater containing 1% peptone, 30% glycerol, and MgSO4 at a concentration of 5 g/L in the Museum of Marine Heterotrophic Bacteria, NSCMB FEB RAS.
The sensitivity of the strain 2202 to antibiotics was tested using the method of diffusion in Mueller–Hinton agar (Difco), enriched in 2% NaCl; the results are interpreted according to CLSI (2006). Sensitivity to ampicillin (10 µg/disk), carbenicillin (100 µg/disk), oxacillin (5 µg/disk), tetracycline (30 µg/disk), oleandomycin (15 µg/disk), cefazolin (30 µg/disk), cefotaxime (30 µg/disk), ceftazidime (30 µg/disk), streptomycin (25 µg/disk), gentamicin (10 µg/disk), lincomycin (15 µg/disk), ciprofloxacin (5 µg/disk), chloramphenicol (30 µg/disk), and polymyxin B (300 U) was assessed by the diffusion method with Oxoid disks.
The analysis of the antimicrobial activity of the isolates towards the type strains of the test cultures of Escherichia coli ATCC 15,034, Bacillus subtilis BKMB501, Candida albicans KMM 455, Pseudomonas aeruginosa KMM 433, and Staphylococcus aureus ATCC 21,027 was carried out as described earlier (Beleneva et al. 2013). It is a set of typical microbial strains that covers a wide range of microorganisms, and is successfully used to characterize a new freshly isolated strain (Harjodh et al. 2019).
Study of biological activity of strain 2202
To study interaction of strain 2202 with other bacterial species, we obtained a mature biofilm of test cultures of the type strains, E. coli ATCC 15,034 and B. subtilis BKMB501. For this, an overnight culture on Tryptic soy broth (Difco) in an amount of 0.1 mL was spread over the surface of Tryptic soy agar TSA (Difco) and incubated during 3 days. Then 0.2 mL of suspension of strain 2202 grown during a day on Marine Broth 2216 (Difco) was spread over its surface, and cultivated for another day. After the formation of pigmented colonies of strain 2202, a fragment of biofilm with a colony on its surface was fixed with 2.5% glutaraldehyde on CB. Then, the material for scanning electron microscopy was treated as described above.
For bioassay of the pathogenic properties of strain 2202, bacteria were grown for 3 days in the liquid Youschimizu–Kimura medium (Y–K) with the following composition: peptone (5.0 g), yeast extract (2.0 g), glucose (1.0 g), К2HPO4 (0.2 g), MgSO4·7H2O (0.1 g), seawater (1000 mL), pH 7.8–8.0 (Youschimizu and Kimura 1976). The strain 2202 was grown on an Exella E5 shaker (New Brunswick, USA) at 100 rpm, centrifuged at 4000 rpm for 20 min and diluted with sterile sea water (SSW) to a concentration of 106 cells/mL. The stain 2202 was added in 5-L flasks with SSW and 20 adult M. kurilensis and 5-L flasks with SSW and 20 adult M. edulis to a final concentration of 103 cells/mL. The control groups of M. kurilensis and M. edulis individuals paced in similar conditions were exposed to a suspension of E. coli with a final concentration of 103 cells/mL. The exposure time was 3 days. Then, the animals were kept in tanks with regularly replaced and aerated water during two months.
In a parallel series of experiments, a suspension of strain 2202 was introduced into the tissue of the adductor muscle of adult individuals of M. kurilensis and M. edulis. In the experimental groups, 10 mL of a suspension of strain 2202 (1010 cells/mL) in sterile seawater was injected in 20 individuals of each species. In the control groups, a suspension of the bacterium E. coli (1010 cells/mL) in sterile seawater was injected in 20 individuals of M. kurilensis and M. edulis. Then, the animals were kept in tanks with regularly replaced and aerated water during 14 days. The material for genetic analysis was fixed in 96% ethanol.
We conducted an experiment with 150–200 embryos in 24-well plates with 2 mL liquid added to each well. Each variant of the experiment was performed in triplicate. The embryos of Strongylocentrotus nudus and M. edulis were placed in sterile seawater containing strain 2202 at a concentration of 103 cells/mL (variant 1) and 5 × 103 cells/mL (variant 2) as well as E. coli at a concentration of 103 cells/mL; as a control of development, embryos were placed in SSW. The state of the material was evaluated under a binocular microscope at 4 h after the beginning of the experiment. The culture of embryos was fixed at 24 h (for M. edulis) and 48 h (S. nudus) after the beginning of the experiment, and also when they reached the stages of trochophore (M. edulis) and pluteus (S. nudus) in the control wells. The samples were fixed with 2.5% glutaraldehyde in seawater (pH 7.2) for 60 min and then rinsed several times in sterile seawater for 24 h. The material was stored in 70% ethanol.
We measured the linear dimensions of embryos (length and width) in the AxioVizion software installed on an Axiovert (Carl Zeiss) microscope with a AxioCam HRc (Carl Zeiss) digital camera. The difference between the embryo length and width values was used in statistical analysis. For calculations, we took 100 embryos from each variant of the experiment: embryos developing in sterile sea water (SSW), in SSW with the addition of E. coli, and in SSW with the addition of strain 2202. All experiments were set up in at least in triplicates; the results are presented as mean ± SD. Statistical analysis was performed using the Statistica 7 software package. Student’s t test was applied to determine significance of differences between groups. With a p value of 0.005, the differences were considered statistically significant.
The experimental larvae of S. nudus (48 h of cleavage) and M. edulis (24 h) after being exposed to strain 2202 and the control larvae kept in SSW were fixed with 4% paraformaldehyde (Sigma) in 0.1 M PBS, pH 7.5, for 60 min at 4 °C and rinsed three times with cold PBS. The material was stored in PBS with 0.03% NaN3 at 4 °C. For α-tubulin detection, rabbit polyclonal Anti-alpha Tubulin antibody (Abcam) was used. Secondary antirabbit antibody labeled Alexa 546 (Molecular Probes). For actin visualization, the FITC-labeled phalloidin (Molecular Probes) was used. To reduce nonspecific binding, the samples were incubated overnight in a blocking solution containing 10% normal goat serum (Sigma), 0.25% BSA, 0.1% Triton X-100, and 0.03% NaN3 in PBS. For simultaneous detection of α-tubulin and actin, fixed larvae were first incubated for 12 h at 10 °C in the blocking solution with the antialpha Tubulin antibody (diluted 1/500) and then, after being washed in PBS (3 × 10 min), were incubated with the secondary antirabbit antibody diluted 1/1000 and FITC-labeled phalloidin (diluted 1/300) for 2 h at room temperature. The specimens were then washed and embedded in the Vectashield mounting medium (Vector Laboratories) containing 0.1 μg/mL DAPI to reveal nuclei. For negative controls, primary antibodies were omitted from the staining protocol. The specimens were analyzed under a Zeiss LSM 780 confocal laser scanning microscope equipped with a high sensitivity GaAsP detector (with a 25 × oil immersion objective lens). From 25 to 64 thin optical sections of the specimens were scanned through along the Z axis with a stepping size of 0.35–0.45 mm. The depth (Z range) of the scanned areas varied between 10 and 40 mm. The resulting laser confocal stacks of optical sections were processed using the ZEN-2010 imaging software. For 3D reconstructions, the ImageJ (NIH) software was used.
Analysis of 16S rRNA
DNA was extracted from bacterial culture using a GeneJET Genomic DNA Purification Kit (ThermoFisher Scientific) under conditions recommended by the manufacturer. The quality of extracted DNA was tested in 1% agarose gel in 0.5 × TRIS–borate buffer. PCR of 16S rRNA fragment of the bacterial genome was conducted using the 11F (5’-GTTTGATCMTGGCTCAG-3’) and 1492R primers (5’-TACGGYTACCTTGTTACGACTT-3’) (Lane 1991) in 12.5 µL reaction mixture under the following conditions: 10 ng of DNA, 0.25 µM of each primer; 0.2 mM of each dNTPs; 1 units of HotStart Taq polymerase (Evrogen); 1 × buffer (75 mM Tris–HCl (pH 8.8 at 25 °C), 20 mM (NH4)2SO4, 0.01% Tween 20, and 1.5 mM MgCl2). Reaction parameters were as follows: initial denaturation, 95 °C for 3 min; 35 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min 30 s; elongation щa PCR products, 72 °C for 5 min; cooling to 12 °C. PCR products were tested by electrophoresis in agarose gel and then cleaned with ExoSap to remove residues of primers and triphosphates. Sequencing reaction was performed in both directions using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc.) in conditions recommended by the manufacturer. To clarify the sequence ends (5’- and 3’-), sequencing reaction was conducted with the 342R (5’-CTGCTGCSYCCCGTAG-3’) and 907F primers (5’-AAACTYAAAKGAATTGACGG-3’) (Lane 1991) with the PCR products obtained in the reaction with the 11F and 1492R primers. Sequencing was performed on a GA3500 genetic analyzer (Applied Biosystems Inc.). The obtained sequence was deposited in the genetic database (NCBI) under the accession number MH150834.
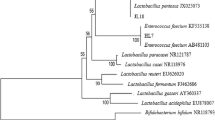
A phylogenetic tree was constructed to confirm the species affiliation of strain 2202. Several closely related species to strain 2202 were taken from the gene bank for the construction of a phylogenetic tree. Pseudoalteromonas luteoviolacea strain NCIMB 1893 T (X82144) was used as an out group.
The 16S rRNA sequences were aligned using the MAFFT algorithm (Katoh et al. 2002; Katoh and Standley 2013) in the Geneious R11 software (https://www.geneious.com/). The MEGA X program (Kumar et al. 2018) showed the model K2 + G (gamma shape = 0.0500) as the best model of nucleotide substitution. This model was used in the MEGA X program for building the ML and NJ trees. To construct the Bayesian tree, we used the Geneious program with nucleotide substitution model calculated in the jModelTest v. 2.1.7 program. We used the following parameters: chain length was 10,000,000 iterations (Total Chain Length); selection of each 500th sample (Subsample Freq.); a total of 19,500 samples were analyzed (Samples Analyzed). In this case, the model HKY + I (p-inv = 0.9320) was selected.
Results
The bacterial strain 2202 capable of growing on an artificial medium was isolated from the hemal fluid of the bivalve mollusk M. kurilensis. On the second day of growth on Marine agar 2216, strain 2202 formed yellow-orange translucent convex colonies 3 mm in diameter; subsequently, their color grew darker, and the pigment diffused into the culture medium. In Gram-stained smears, the bacteria were represented by nonspore-bearing, thin, curved rods 1.2–1.5 µm in length and 0.6–0.8 µm in width with the Gram-negative type of cell wall.
The TEM method showed that, when grown in a liquid medium, bacteria exhibited rod-like shapes that were each ca. 2–3 µm long and 0.6 µm wide. Cells bore 1–2 polar-located flagella (Fig. 1A). On a solid substrate, at 24 h post inoculation, the bacteria had rod-like shapes of ca. 2–3 µm in length and 0.6 µm in diameter without flagella (Fig. 1B), but bearing microvesicles with a size of 60–100 nm on the surface (Fig. 1C). The bacteria formed a biofilm, and were interconnected by a network of long contact processes, pili (Fig. 1D).
The structure of strain 2202 at 24 h of cultivation: (A) a motile rod with two flagella in a liquid medium; (B) biofilm of rods without flagella, interconnected by a network of contact processes (black arrows) on a solid substrate; (C) vesicles on the surface of bacteria; (D) long contact processes (black arrows) and clusters of microvesicles (white arrow) in an activated colony
Biochemically, the isolate was characterized as oxidase positive; it hydrolyzed esculin and gelatin, but did not hydrolyze starch and urease and formed acid from glucose. Arginine dihydrolase, nitrate reductase, β-galactosidase, and indole formation were absent. The strain utilized glucose, mannose, maltose, citrate, and malate. Seawater was required for growth, and the strain grew in the medium with a NaCl content of 0.5 to 10%. The strain showed resistance to oxacillin, ceftazidime, tetracycline, and lincomycin, and was sensitive to ampicillin, streptomycin, gentamicin, ciprofloxacin, oleandomycin, chloramphenicol, and rifampicin. The strain exhibited a medium sensitivity to the rest of the tested antibiotics.
Tests with type strains showed that the strain 2202 exhibited antimicrobial activity against S. aureus, C. albicans, and B. subtilis, but not against E. coli and P. aeruginosa.
The bactericidal properties of strain 2202 were studied on the example of its interaction with Gram-positive bacterium B. subtilis and Gram-negative bacterium E. coli precultivated on a solid substrate for 3 days (Fig. 2A, D). The bacteria B. subtilis and E. coli formed a biofilm consisting of rod-shaped individuals interconnected by a network of contact processes and immersed in a structureless matrix.
Interaction of strain 2202 with B. subtilis (A–C) and E. coli (D–F). (A) A biofilm formed by B. subtilis within 3 days of cultivation. (B) Colonies of strain 2202 at 24 h of cultivation on the biofilm of B. subtilis; the foamy structure (white arrows) is located in the zone of contact between bacteria of the two species; in colonies of strain 2202, there are atypical, elongated rods (inset, scale bar is 2 µm). (C) Microvesicles on the surface of bacteria of strain 2202 and forming the foamy structure (white arrows). (D) Biofilm formed by E. coli on day 3 of cultivation. (E,F) Colonies of strain 2202 represented by typical, short rods (white arrows) at 24 h of cultivation on biofilm of E. coli; the foamy structure at the interface between bacteria of two species is absent
On day 1 post-inoculation, strain 2202 formed colonies of cells on the surface of B. subtilis biofilm that were interconnected by a contact pili (Fig. 2B). Along with typical rods with a length of ca. 2–3 µm and width of 0.6 µm, some cells reached a length of 6 µm (Fig. 2B, inset). On their surface, the bacteria had microvesicles with a diameter of approximately 60–100 nm (Fig. 2C). An extensive foam structure was observed at the interface between the two adjoining bacteria: the species B. subtilis and strain 2202 (Fig. 2B, C, white arrows). The foam structure was composed of microvesicles having a size of ca. 60–100 nm, formed as a result of activity of strain 2202 cells.
On day 1 post-inoculation, strain 2202 formed colonies of rods up to 2 µm in length and ca. 0.6 µm in diameter on the surface of the E. coli biofilm, and the rods were interconnected by contact pili (Fig. 2E, F, white arrows). The foamy zone at the site of contact between the bacteria of the two species was absent. Small clusters of microvesicles were present both over the colonies of strain 2202 and in the area of E. coli biofilm; similar formations were present in the control culture (Fig. 2A, D).
To test the pathogenic properties of strain 2202, we carried out a series of experiments using adult, sexually mature individuals of M. kurilensis and M. edulis as well as embryos and larvae of M. edulis and S. nudus.
The sterile seawater that contained adult, mature individuals of M. kurilensis and M. edulis was supplemented by a suspension of strain 2202 bacteria. After 3 days, the animals were placed in aerated tanks with regularly replaced seawater. Subsequent observations during 2 months revealed no signs of pathology in the animals.
When a suspension of strain 2202 bacteria was injected into the adductor muscle of adult, mature individuals of M. kurilensis and M. edulis, mortality of a part of the individuals was observed on day 5 post-injection. A total of 13 out of the 20 M. kurilensis individuals (65%) and 15 out of the 20 M. edulis individuals (75%) in the experimental groups died within 14 days. The presence of strain 2202 in tissues of the dead individuals was confirmed by sequencing 16S rRNA. The animals in the control groups showed no signs of pathology throughout the experiment.
In a series of experiments with M. edulis embryos as a test system, the embryos immediately after fertilization were placed in SSW containing strain 2202 at a two different concentrations (1 × 103 and 5 × 103 cells/mL) as well as E. coli at a concentration of 103 cells/mL; development in SSW was the control. After 24 h, larvae in the SSW and in the medium containing E. coli reached the trochophore stage (Fig. 3A, B). In variant 1 of the experiment with strain 2202, a part of larvae was destroyed, and the rest developed with anomalies, which were manifested as abnormal shape and structure (Fig. 3C) and reliable increase in the variations in their linear dimensions versus control. The ratio of the larger and smaller diameters of embryos was: 1, 23 ± 0, 07 (development in the presence of 2202), 1, 05 ± 0, 04 (development in the presence of E. coli), 1, 04 ± 0, 03 (development in sterile seawater).
Larvae of M. edulis, 24 h post-fertilization: (A) development in sterile seawater; (B) in sterile seawater containing E. coli (103 cells/mL); (C) in sterile seawater containing bacteria of strain 2202 at a concentration of 103 cells/mL; (D) in sterile seawater containing bacteria of strain 2202 at a concentration of 5 × 103 cells/mL. Larvae of S. nudus, 48 h post fertilization: (E) development in sterile seawater; (F) in sterile seawater containing E. coli at a concentration of 103 cells/mL; (G) in sterile seawater containing bacteria of strain 2202 at a concentration of 103 cells/mL
In variant 2 of the experiment, with the increase in concentration of strain 2202 bacteria in the medium, the development was inhibited at the blastula stage, and a part of embryos disintegrated into blastomeres (Fig. 3D).
In a series of experiments using embryos of the sea urchin S. nudus as a test system, they were immediately placed in SSW containing bacteria of strain 2202 and E. coli bacteria after fertilization; development in SSW was the control. At 48 h after the beginning of the experiment, the larvae reached the pluteus stage in all three variants of experiments (Fig. 3E–G).
An immunocytochemical analysis using antibodies against α-tubulin and phalloidin staining for actin showed that at 24 h post-fertilization M. edulis larvae developing in SSW had clusters of tubulin-containing structures in the area bands of cilia and that their basal bodies were stained. Moreover, larvae cells were weakly and diffusely phalloidin-stained for cytoskeletal actin. At the vegetative pole of the larvae, there were intensely colored, actin-positive, presumably muscle cells (Fig. 4A). In the medium containing bacteria of strain 2202 (variant 1 of the experiment), the structure of the larval cytoskeleton varied: tubulin was diffusely distributed in the cytoplasm of larval cells without forming dense aggregations, while actin filaments in larval cells formed globules. Intensely colored actin-positive cells were absent (Fig. 4B).
Immunocytochemical staining for α-tubulin (red fluorescence) and actin (green fluorescence) of larval M. edulis at 24 h post-fertilization (A, B) and S. nudus at 48 h post-fertilization (C, D) developing in sterile seawater containing bacteria of strain 2202 (A, C) and in sterile seawater (B, D). DNA of nuclei is post-stained with DAPI (blue fluorescence). The scale bar corresponds to 20 µm
No abnormalities were found in sea urchin embryos developing in a medium containing strain 2202 when stained with antibodies against α-tubulin and phalloidin staining for actin (Fig. 4C, D).
To determine the species of strain 2202, we carried out a BLAST analysis of the obtained sequence of 16S rRNA gene with the available sequences in the genetic data bank. We observed a complete identity of our sequence with the 16S rRNA gene sequences of some lineages of P. piscicida. The results of phylogenetic reconstructions are presented (Fig. 5). As a base, we selected the NJ tree constructed in the MEGA X program. Supports for NJ/ML/BA trees are shown in nodes. It was established that strain 2202 occurs in the clade P. piscicida. The sequence of its 16S rRNA was completely identical to the sequences KF880965, NR_114190, CP011924; differs by one nucleotide from CP031761, CP021646; and two nucleotides from KY366347, belonging to different lineages of P. piscicida. Using the EzTaxon service (https://www.ezbiocloud.net/identify) for taxa analysis showed the greatest similarity to P. piscicida JCM 20779T (CP011925). With 100% overlap of our sequence with CP011925, the similarity was 99.93%.
Phylogenetic tree (based on NJ tree) inferred from comparison of 16S rRNA gene sequences of bacteria of the genus Pseudoalteromonas. The values of bootstrap support (NJ/ML > 70%) and Bayesian a posteriori probability (BA > 0.9) are shown at tree nodes. The strain 2202 of Pseudoalteromonas sp. is highlighted in bold
Discussion
The hemal fluid of healthy bivalves is known to contain a community of symbiotic bacteria, referred to as hemomicrobiota, as a part of the mollusks microbiome, that includes bacteria that exhibit an antimicrobial, predatory-like activity (Desriac et al. 2014; Offret et al. 2019). Thus, animal microbiomes can harbor active bacterial predators, which may regulate microbiome structure and protect the host by consuming potential pathogens (Papaleo et al. 2012; Welsh et al. 2016). These bacteria form the so-called “microbial shield” which is considered as a component of the defense system in mollusks (Desriac et al. 2010; Oelschlaeger 2010). Most of the culturable strains of hemomicrobiota are common inhabitants of marine ecosystems that enter the host organism from the environment. As a result, the composition of hemomicrobiota is largely determined by the species diversity of microorganisms in the habitat and varies in concordance with it depending on changes in environmental parameters (Wendling et al. 2014). Thus, this component of the mollusk’s defense system is most labile, which allows the host organism to respond successfully to external challenges.
Among the culturable strains of hemolymph bacteria, members of the genus Pseudoalteromonas show the highest antimicrobial activity (Desriac et al. 2014). Members of the genus exhibit predatory-like behavior towards a wide range of gram-negative and gram-positive bacteria, including pathogens (Richards et al. 2017; Tang et al. 2020).
It allows to use the members of the genus Pseudoalteromonas probiotics that increase mollusks’ resistance to pathogens in aquaculture (Offret et al. 2019), which makes the search for new culturable strains of symbiotic microorganisms from this group, as well as description of their properties and pathways of interaction with other organisms, especially relevant.
We isolated a culturable strain of bacteria with high antimicrobial activity from the hemal fluid of mature healthy individuals of M. kurilensis.
To identify the species of the bacterial strain we used the method of partial genetic sequencing. An analysis of the 16S rRNA gene sequence of strain 2202 showed its 100% identity to the analogous sequences from strains KF880965, NR_114190 and CP011924 of P. piscicida, which allows us to classify strain 2202 as one of the lines of P. piscicida. The biochemical characteristics of the isolate also match those of the species P. piscicida (Brenner et al. 2005). Thus, the bacterium P. piscicida has been isolated, for the first time, from the hemal fluid of representatives of bivalves.
P. piscicida is known as a species that produces a pigment with an activity against some of pathogenic microorganism and marine bacteria. In particular, the β-carboline alkaloid noharman inhibits S. aureus (Zheng et al. 2005). P. piscicida caused growth inhibition and death of Vibrio vulnificus, V. parahaemolyticus, V. cholerae, Photobacterium damselae, and Shewanella algae. Inhibition also occurred on lawns of S. aureus but not on E. coli O157:H7 or Salmonella enterica serovar typhimurium (Richards et al. 2017).
According to our data the strain 2202 is a pigment producing. When cultivated on a solid substrate, the bacteria form pigmented colonies that are yellow orange in color. It was found that the isolate obtained by us exhibited activity against S. aureus, C. albicans, and B. subtilis and was not active against E. coli.
The mode of interaction of P. piscicida with antagonistic bacterial species is characterized as predator-like (Richards et al. 2017). It was found that due to the action of proteolytic enzymes produced by P. piscicida, holes are formed in the membrane of a competitive bacterium species V. parahaemolyticus. The presence of encoding regions for putative proteases associated with antimicrobial activity was shown using the example of genome-wide DNA sequencing of P. piscicida strain DE2-B, including the genes for 11 serine proteases and 7 metalloproteases. DNA from the two other strains (i.e., DE1-A and DE2-A) contains 6 and 7 serine proteases (Richards et al. 2019).
To visualize the features of interaction of strain 2202 with representatives of Gram-positive bacteria, we conducted joint cultivation of these bacteria with a competitive species B. subtilis and a neutral species E. coli as a control. When cultivated on the surface of a biofilm formed by B. subtilis and E. coli, the strain 2202 formed microcolonies. The individuals in microcolonies were merged contact pili. Also, there were long pili of an alternative type which contributed to the rapid mutual relocation of bacterial cells within the forming biofilm (Pohlschroder and Esquivel 2015). Single microvesicles were present on the bacterial surface in all variants of the experiment. However, the presence of an extensive foamy structure over the colonies 2202 during its interaction with the competitive species B. subtilis is associated with the bactericidal activity of the strain. Previously, the role of vesicles in the antimicrobial system of three P. piscicida strains, (i.e., DE1-A, DE2-A, and DE2-B) was investigated on the example of interaction with a competitive bacterium species, V. parahaemolyticus (Richards et al. 2017). The direct transfer of lytic vesicles from the surface of the P. piscicidae strains to the surface of Vibrio cells, with subsequent digestion of holes in the Vibrio cell walls takes place.
In the case of strain 2202, holes on the surface of B. subtilis were extremely rare. Instead of this, there was a fragmentation of components of the B. subtilis biofilm as well as the formation of contact pili between individuals of two species. This difference may be related to the individual characteristics of the competing bacterial species used in the experiment, and may be due to variations in the molecular mechanisms that provide these competitive interactions.
In particular, it was shown that Pseudoalteromonas can prey on Gram-positive bacteria by secreting metalloprotease pseudoalterin, and thus degrading the PG in their cell wall. At the same time, pseudoalterin has no killing activity against Gram-negative bacteria (Tang et al. 2020). The most likely, strain 2202 used a strategy called group or ‘wolfpack’ predation during the interaction with competing bacterial species. A prerequisite for this collaborative type of hunting is a quorum of predatory cells, which pool hydrolytic enzymes, proteases, or nucleases to lyse and feed on nearby prey (Martin 2002). Group predation occurs primarily in bacteria, which also exhibit social role-playing behavior, gliding mobility, and complex communication systems, which is characteristic of Pseudoalteromonas.
With rare exceptions, Pseudoalteromonas strains are associated with healthy animals or algae. To date, few strains are known as pathogenic or opportunistic. P. piscicida is known as fish pathogen (Austin 1993; Gauthier et al. 1995; Bowman 2007). P. agarivoran NW4327 has been recently reported as pathogenic for the sponge Rhopaloeides odorabile (Choudhury et al. 2015).
As regards strain 2202, the lack of signs of pathology in the individuals of M. kurilensis whose hemal fluid contained the isolated strain is an evidence of tolerance of the host organism to the associated bacteria. The introduction of a suspension of strain 2202 bacteria into the seawater where M. kurilensis and M. edulis were kept also did not cause pathology in the mollusks either. The bactericidal activity of strain 2202 allows us to consider it as a presumable probiotic that can contribute to improving the survivability of the host species, as has been shown for other members of the genus Pseudoalteromonas (Desriac et al. 2014). Being a component of the in marine ecosystems, the bacterium P. piscicida is a facultative symbiont that enters the host organism from the environment. The most probable way of settlement is activation of bacterial phagocytosis by cells of the integumentary epithelium (Zhukova and Eliseikina 2012). Although being within specialized vacuoles, microorganisms resistant to the action of lysosomal enzymes overcome the epithelial barrier and get into the hemolymph of mollusk. The ability to persist in the hemolymph while avoiding the effect of protective factors exhibited by Pseudoalteromonas species is explained by the chemical structure of their lipopolysaccharide (LPS), which is composed of a pentaacylated lipid A instead of a hexaacylated one (Carillo et al. 2011). Such a structural difference may provide an advantage to evade PRRs-mediated recognition (Offret et al. 2016).
Owing to the introduction of a suspension of strain 2202 bacteria into the bivalve’s muscle, the integrity of host cells become broken, which triggers manifestation of the pathogenic potential of the strain, showing characteristics of opportunistic microflora in this case.
Thus, being a component of normal hemomicrobiota, strain 2202 implements various strategies of symbiosis depending on external conditions, from mutualism to parasitism leading to death of the host organism. This allows us to characterize strain 2202 as a member of opportunistic microflora in M. kurilensis.
In experiments using embryos and larvae of M. edulis with pathogenicity assays, the larvae reached the trochophore stage within 24 h post-fertilization in the control. Cytoskeletal actin was detected in all larval cells; in addition, at the larval vegetative pole, there were pioneer muscle cells with high actin content, which indicated the onset of larval muscle ring differentiation (Dyachuk and Odintsova 2009). In case of exposure to high concentrations of strain 2202 bacteria, development was inhibited at the blastula stage. At lower concentrations, bacterial metabolites disturbed the normal development of M. edulis. In particular, the linear dimensions and the architecture of larvae changed, ciliary cords did not form, and the muscle pioneer cells at the vegetative pole of larvae were absent. The structure of cytoskeletal actin in larval cells was disrupted, which indicates a disturbance of the process of actin filament assembly. A portion of the larvae were destroyed, apparently, as a result of exposure to a set of bacteria’s proteolytic enzymes. The observed anomalies indicate a toxic effect of secondary metabolites of strain 2202 on the processes of cell differentiation during the early development of M. edulis. Moreover, under natural conditions, the proteolytic enzymes produced by P. piscicida may affect the ability of M. edulis larvae to settle. It has been shown that proteolytic enzymes produced by members of the genus Pseudoalteromonas dissolve the protein used by invertebrate larvae to attach to the substrate; thus, preventing their settlement (Dobretsov et al. 2007). Metabolites of the Pseudoalteromonas species include the substances that stimulate apoptosis and cause cessation of cell proliferation, differentiation, and migration (Zheng et al. 2005; Seipp et al. 2006). The production of substances that affect the cell cycle, as well as proteolytic enzymes, determines the ability of members of the genus Pseudoalteromonas to regulate settlement and metamorphosis of invertebrate larvae. Moreover, the pattern of such interaction is species-specific, both inhibiting and facilitating settlement (Dobretsov et al. 2007; Beleneva et al. 2015).
A test system based on the sea urchin embryos and larvae (SET) for assessing toxicity is universal and highly sensitive. Toxicity of the natural isolates of fluorescent pseudomonads was analyzed using embryos and larvae of the sea urchin S. nudus (Beleneva et al. 2015). SET is widely applied to bioassay the toxicity of different classes of xenobiotics, including inorganics, organics, different complex mixtures, and natural products (Pagano et al. 2017). An assessment of the pathogenic potential of strain 2202 using SET showed those metabolites of these bacteria neither manifest toxicity nor disturb the early development of the sea urchin S. nudus, which confirms the species-specific effects of metabolites of strain 2202 bacteria towards larvae of marine invertebrates.
Conclusions
A bacterium P. piscicida (stain 2202) has been isolated, for the first time, from the hemal fluid of the bivalve mollusk M. kurilensis. Being part of the normal microflora in the hemolymph of this bivalve and having bactericidal properties, the strain can contribute to increase in the survival rate of the host species; however, when external conditions change, it shows features characteristic of opportunistic microflora. Strain 2202 disturbs the early development of M. edulis but does not exhibit toxicity in SET. Thus, the bactericidal properties of the strain and the pattern of interaction with marine invertebrates is species-specific, which must be taken into consideration when using this microorganism to address various biotechnological issues such as, for example, developing antifouling agents, searching for sources of novel bactericidal substances or as a probiotic in aquaculture.
References
Austin B (1993) Environmental issues in the control of bacterial diseases of farmed fish. In: Pullin RSV, Rosenthal H, Maclean JL (ed) Environment and Aquaculture in Developing Countries ICLARM (International Center for Living Aquatic Resources Management) Conference Proceedings 31. ICLARM, Manila, Philippines (now, WorldFish Center, Penang, Malaysia) and Deutsche Gesellschaft für Technische Zusammenarbeit. Germany, Eschborn. pp 237–251.
Austin B, Austin DA (2007) Bacterial fish pathogens: disease of farmed and wild fish, 4th edn. Springer Science and Business Media, UK
Beleneva IA, Kukhlevsky AD, Kharchenko UV (2013) Antimicrobial activity of heterotrophic bacterial strains of marine origin. Jundishapur J Microbiol 6:166–175
Beleneva IA, Shamshurina EV, Eliseikina MG (2015) Assessment of the toxic effect exerted by fluorescent pseudomonads on embryos and larvae of the sea urchin Strongylocentrotus nudus. Ecotoxicol Environ Saf 115:263–271
Bowman JP (2007) Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar Drugs 5:220–241
Brenner DJ, Krieg NR, Staley JT, Garrity GM (2005) Bergey's Manual of Systematic Bacteriology, 2nd Edition, Vol. 2 (The Proteobacteria), part C (The Alpha-, Beta-, Delta-, and Epsilonproteobacteria). Springer, New York.
Carillo S, Pieretti G, Parrilli E, Tutino ML, Gemma S, Molteni M, Lanzetta R, Parrilli M, Corsaro MM (2011) Structural Investigation and Biological Activity of the Lipooligosaccharide from the Psychrophilic Bacterium Pseudoalteromonas haloplanktis TAB 23. Chem Eur J 17:7053–7060
Choudhury JD, Pramanik A, Webster NS, Llewellyn LE, Gachhui R, Mukherjee J (2015) The pathogen of the great barrier reef sponge Rhopaloeides odorabile is a new strain of Pseudoalteromonas agarivorans containing abundant and diverse virulence-related genes. Mar Biotechnol 17:463–478
Desriac F, Defer D, Bourgougnon N, Brillet B, Le Chevalier P, Fleury Y (2010) Bacteriocin as weapons in the marine animal-associated bacteria warfare: inventory and potential applications as an aquaculture probiotic. Mar Drugs 8:1153–1177
Desriac F, Le Chevalier P, Brillet B, Leguerinel I, Thuillier B, Paillard C, Fleury Y (2014) Exploring the hologenome concept in marine bivalvia: haemolymph microbiota as a pertinent source of probiotics for aquaculture. FEMS Microbiol Lett 350(1):107–116
Dobretsov S, Xiong H, Xu Y, Levin A, Qian P-Y (2007) Novel antifoulants: inhibition of larval attachment by proteases. Mar Biotechnol 9:388–397
Donovan CJ, Garduno RA, Kalmokoff M, Ku JC, Quilliam MA, Gill TA (2009) Pseudoalteromonas bacteria are capable of degrading paralytic shellfish toxins. Appl Environ Microbiol 75(21):6919–6923
Dyachuk V, Odintsova N (2009) Development of the larval muscle system in the mussel Mytilus trossulus (Mollusca, Bivalvia). Dev Growth Differ 51:69–79
Gauthier G, Gauthier M, Christen R (1995) Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (Emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int J Syst Bacteriol 45:755–761
Glöckner FO, Fuchs BM, Amann R (1999) Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol 65:3721–3726
Hamsah W, Alimuddin YM, Zairin MZ (2017) The nutritional value of Artemia sp. enriched with the probiotic Pseudoalteromonas piscicida and the prebiotic mannan-oligosaccharide. AACL Bioflux 10:8–17
Harjodh S, Manpreet K, Manoj J, Sunita M, Hemraj N, Anil K (2019) Antimicrobial properties of the novel bacterial isolate Paenibacilllus sp. SMB1 from a halo-alkaline lake in India. Sci Rep 9:11561. https://doi.org/10.1038/s41598-019-47879-x
Harvey E, Deering R, Rowley C, El Gamal A, Schorn M, Moore B, Whalen K (2016) A bacterial quorum-sensing precursor induces mortality in the marine Coccolithophore, Emiliania Huxleyi. Front Microbiol. https://doi.org/10.3389/fmicb.2016.00059
Holmström C, Kjelleberg S (1999) Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extra-cellular agents. FEMS Microbiol Ecol 30:285–293
Hugh R, Leifson E (1953) The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various gram-negative bacteria. J Bacteriol 66:24–26
Ivanova EP, Gorshkova NM, Sawabe T, Hayashi K, Kalinovskaya NI, Lysenko AM, Zhukova NV, Nicolau DV, Kuznetsova TA, Mikhailov VV, Christen R (2002) Pseudomonas extremorientalis sp. nov., isolated from a drinking water reservoir. Int J Syst Evol Microbiol 52:2113–2120
Ivanova EP, Flavier S, Christen R (2004) Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int J Syst Evol Microbiol 54:1773–1788
Ivanova EP, Ng HJ, Webb HK (2014) The Family Pseudoalteromonadaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes. Springer, Berlin, pp 575–582
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2:3059–3066
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lane DJ (1991) 16S/23S sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematic. John Wiley & Sons, Chichester, pp 115–175
Martin MO (2002) Predatory prokaryotes: an emerging research opportunity. J Mol Microbiol Biotechnol 4:467–478
Moeller V (1955) Simplified tests for some amino acid decarboxylases and for the arginine dihydrolase system. Acta Pathol Microbiol Scand 36:158–172
Nelson EJ, Ghiorse WC (1999) Isolation and identification of Pseudoalteromonas piscicida strain Cura-d associated with diseased damselfish (Pomacentridae) eggs. J Fish Dis 22:253–260
Oelschlaeger TA (2010) Mechanisms of probiotic actions—a review. Int J Med Microbiol 300(1):57–62
Offret C, Desriac F, Le Chevalier P, Mounier J, Jégou C, Fleury Y (2016) Spotlight on antimicrobial metabolites from the marine bacteria Pseudoalteromonas: Chemodiversity and ecological significance. Mar Drugs 14:129
Offret C, Jegou C, Mounier J, Fleury Y, Le Chevalier P (2019) New insights into the haemo- and coelo-microbiota with antimicrobial activities from Echinodermata and Mollusca. J Appl Microbiol 126(4):1023–1031
Pagano G, Guida M, Trifuoggi M, Thomas P, Palumbo A, Romano G, Oral R (2017) Sea urchin bioassays in toxicity testing: I. inorganics, organics, complex mixtures and natural products. Expert Opin Environ Biol 6:1
Papaleo MC, Fondi M, Maida I, Perrin E, Lo Giudice A, Michaud L, Mangano S, Bartolucci G, Romoli R, Fani R (2012) Sponge-associated microbial Antarctic communities exhibiting antimicrobial activity against Burkholderia cepacia complex bacteria. Biotechnol Adv 30:272–293
Park YD, Baik KS, Yi H, Bae KS, Chun J (2005) Pseudoalteromonas byunsanensis sp. nov., isolated from tidal flat sediment in Korea. Int J Syst Evol Microbiol 55:2519–2523
Pham D, Ansquer D, Chevalier A, Dauga C, Peyramale A, Wabete N, Labreuche Y (2014) Selection and characterization of potential probiotic bacteria for Litopenaeus stylirostris shrimp hatcheries in New Caledonia. Aquaculture 432:475–482
Pohlschroder M, Esquivel RN (2015) Archaeal type IV pili and their involvement in biofilm formation. Front Microbiol 6:190
Radjasa OK, Limantara L, Sabdono A (2009) Antibacterial activity of a pigment producing-bacterium associated with Halimeda sp. from a land-locked marine lake Kakaban. Indonesia. J Coast Dev 12(2):100–104
Richards GP, Watson MA, Needleman DS, Uknalis J, Boyd EF, Fay JP (2017) Mechanisms for Pseudoalteromonas piscicida - induced killing of vibrios and other bacterial pathogens. Appl Environ Microbiol 83:e00175-e217
Richards GP, Needleman DS, Watson MA, Polson SW (2019) Whole-genome sequences of two Pseudoalteromonas piscicida strains, DE1-A and DE2-A, with strong antibacterial activity against Vibrio vulnificus. Microbiol Resour Announc 8:e01451-e1518
Seipp S, Wittig K, Stiening B, Böttger A, Leitz T (2006) Metamorphosis of Hydractinia echinata (Cnidaria) is caspase-dependent. Int J Dev Biol 50:63–70
Skerratt JH, Bowman JP, Hallegraeff G, James S, Nichols PD (2002) Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar Ecol Prog Ser 244:1–15
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC, pp 607–655
Tang BL, Yang J, Chen X-L, Wang P, Zhao H-L, Su H-N, Zhang Y-Z (2020) A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria. Nat Commun. https://doi.org/10.1038/s41467-019-14133-x
Whalen KE, Kirby C, Nicholson RM, O’Reilly M, Moore BS, Harvey EL (2018) The chemical cue tetrabromopyrrole induces rapid cellular stress and mortality in phytoplankton. Sci Rep. https://doi.org/10.1038/s41598-018-33945-3
Welsh RM, Zaneveld JR, Rosales SM, Payet JP, Burkepile DE, Thurber RV (2016) Bacterial predation in a marine host-associated microbiome. ISME J 10(6):1540–1544. https://doi.org/10.1038/ismej.2015.219
Wendling C, Batista F, Wegner K (2014) Persistence, seasonal dynamics and pathogenic potential of vibrio communities from Pacific oyster hemolymph. PLoS ONE 9(4):e94256
Wu YH, Cheng H, Xu L, Jin XB, Wang CS, Xu XW (2017) Physiological and genomic features of a novel violacein-producing bacterium isolated from surface seawater. PLoS ONE 12:e0179997
Youschimizu M, Kimura T (1976) Study on the intestinal microflora of salmonids. Fish Pathol 10:243–259
Zheng L, Chen H, Han X, Lin W, Yan X (2005) Antimicrobial screening and active compound isolation from marine bacterium NJ6-3-1 associated with the sponge Hymeniacidon perleve. World J Microbiol Biotechnol 21:201–206
Zheng L, Yan X, Han X, Chen H, Lin W, Lee FS, Wang X (2006) Identification of norharman as the cytotoxic compound produced by the sponge (Hymeniacidon perleve)-associated marine bacterium Pseudoalteromonas piscicida and its apoptotic effect on cancer cells. Biotechnol Appl Biochem 44:135–142
Zhukova N, Eliseikina M (2012) Symbiotic bacteria in the nudibranch mollusk Dendrodoris nigra: Fatty acid composition and ultrastructure analysis. Mar Biol 159:1783–1794
Acknowledgements
The work was performed in the “Far Eastern Center of Electron Microscopy” (National Scientific Center of Marine Biology, Far Eastern Branch, Russian Academy of Sciences, Vladivostok, Russia).
Funding
This research received no external funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Eliseikina, M.G., Beleneva, I.A., Kukhlevsky, A.D. et al. Identification and analysis of the biological activity of the new strain of Pseudoalteromonas piscicida isolated from the hemal fluid of the bivalve Modiolus kurilensis (F. R. Bernard, 1983). Arch Microbiol 203, 4461–4473 (2021). https://doi.org/10.1007/s00203-021-02432-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02432-1