Abstract
Multidrug-resistant (MDR) and hypervirulent Klebsiella pneumoniae (hvKp) clones have become a major threat to global public health. The clonal group 258 (CG258) is considered a high-risk CG and the K. pneumoniae strains belonging to it are often multi-resistant and to spread mainly in the hospital environment. This study aimed to characterize the antimicrobial resistance profile, virulence factors, and the clonal relationships among 13 K. pneumoniae strains belonging to CG258 from patients admitted to a tertiary hospital in Teresina, in the state of Piauí, northeastern Brazil. Ten strains were classified as MDR and three as extensively drug-resistant (XDR). Three different β-lactamase-encoding genes (blaKPC, blaOXA-1-like, and blaCTX-M-Gp1) and six virulence genes (fimH, ycfM, mrkD, entB, ybtS, and kfu) were detected. Moreover, two hypermucoviscous K. pneumoniae strains and one capsular K-type 2 were found. Multilocus sequence typing analysis revealed ten different sequence types (STs) (ST14, ST17, ST20, ST29, ST45, ST101, ST268, ST1800, ST3995, and ST3996) belonging to CG258, being two (ST3995 and ST3996) described for the first time in this study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Klebsiella pneumoniae is a Gram-negative bacillus, usually encapsulated, nonmotile bacterium and belongs to the Enterobacteriaceae family. It is an opportunistic pathogen that can cause several infections and is among the most common nosocomial pathogens worldwide (Wyres et al. 2020). K. pneumoniae strains can contain a wide range of virulence and antimicrobial resistance factors (Ashurst and Dawson 2020). Several virulence factors further increase the severity of infections by K. pneumoniae, such as siderophores that are systems of chelating molecules that can competitively eliminate iron from host proteins and other sources; capsules, including their overproduction characterized by the hypermucoviscous (HM) phenotype; among others (Paczosa and Mecsas 2016).
The presence of clinically relevant antimicrobial resistance genes (ARGs), including extended-spectrum β-lactamases (ESBLs) and carbapenemases-encoding genes, can limit therapeutic options, making it difficult to treat infections in affected patients (Piperaki et al. 2017). Multidrug-resistant (MDR) and hypervirulent K. pneumoniae (hvKp) clones have become major global health problems (Wyres et al. 2020).
This study aimed to determine the pathogenic potential, antimicrobial resistance profile, and the clonal relationships among K. pneumoniae strains belonging to CG258 from patients admitted to hospitals in northeastern Brazil.
Materials and methods
Bacterial strains
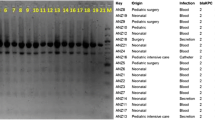
This study included 13 K. pneumoniae strains isolated from different patients admitted to a tertiary hospital in Teresina, a Brazilian municipality located in the northeast region, being the capital of the state of Piauí, with an estimated population of 868,075 inhabitants. The strains were randomly isolated between August and September 2013 from several sources (catheter tip, hemoculture, purulent secretion, tracheal secretion, and urine) (Table 1). The strains were identified by the VITEK 2 system (bioMérieux, France) and confirmed by the 16S rRNA sequencing, after amplification and sequencing with the primers fD1 (5'-AGAGTTTGATCCTGGCTCAG-3') and rP2 (5'-ACGGCTACCTTGTTACGACTT-3'), according to Weisburg et al. (1991). The strains were maintained at − 80 °C in 15% glycerol for subsequent experiments.
Hypermucoviscosity test
The HM phenotype was investigated using the string test according to Wiskur et al. (2008). The strains were inoculated on Mueller–Hinton agar (Oxoid Ltd., United Kingdom) and incubated for 18 h at 37 °C. After bacterial growth, using a bacteriological loop, an isolated colony was touched and raised vertically. The HM phenotype was considered positive when there was the formation of a viscous filament ≥ 5 mm.
Antimicrobial susceptibility testing
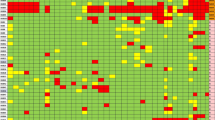
Antimicrobial susceptibility testing was realized by the disk diffusion method on Mueller–Hinton agar (Oxoid Ltd., United Kingdom) following the recommendations of the Clinical Laboratory Standards Institute (CLSI 2020) and the Brazilian Committee on Antimicrobial Susceptibility Testing (BrCAST 2020). For this approach, 40 different antimicrobial disks (Oxoid Ltd., United Kingdom) recommended for Enterobacteriales were used. The susceptibility for colistin (COL) was determined by the broth microdilution method (CLSI 2020). Each strain was considered susceptible or non-susceptible (either intermediate or resistant) to each antimicrobial tested. The antimicrobials tested are shown in Fig. 1.
Frequency of non-susceptibility found among the 13 strains of K. pneumoniae studied. Each color represents a class of antimicrobials. Antimicrobials tested: amoxicillin-clavulanate (AMC), amikacin (AMI), ampicillin-sulbactam (APS), aztreonam (ATM), ceftazidime (CAZ), cefaclor (CFC), cefixime (CFM), cefoxitin (CFO), cefazolin (CFZ), ciprofloxacin (CIP), chloramphenicol (CLO), colistin (COL), cefepime (CPM), ceftriaxone (CRO), cefuroxime (CRX), ceftaroline (CTL), cefotetan (CTT), cefotaxime (CTX), doripenem (DOR), doxycycline (DOX), ertapenem (ERT), streptomycin (EST), fosfomicin (FOS), gentamicin (GEN), imipenem (IPM), levofloxacin (LEV), lomefloxacin (LMX), minocycline (MIN), meropenem (MPM), nalidixic acid (NAL), netilmicin (NET), nitrofurantoin (NIT), norfloxacin (NOR), ofloxacin (OFX), piperacillin-tazobactam (PIT), sulphonamide (SUL), trimethoprim-sulfamethoxazole (SUT), ticarcillin-clavulanate (TAC), tetracycline (TET), tobramycin (TOB), and trimethoprim (TRI)
The strains Escherichia coli ATCC® 25922 and Pseudomonas aeruginosa ATCC® 27853 were used as controls in these experiments. Based on the susceptibility profile, the K. pneumoniae strains were classified as MDR, extensively drug-resistant (XDR), or pandrug-resistant (PDR) according to Magiorakos et al. (2012).
Detection of virulence genes and ARGs by polymerase chain reaction (PCR)
The strains were subjected to conventional PCR assays to detect K1 (magA, K1 capsule-specific wzy gene) and K2 capsular serotypes and nine gene encoding virulence factors: fimH and mrkD (adhesins), ycfM (lipopolysaccharide), entB, iutA, ybtS, and kfu (iron acquisition systems), allS (allantoin metabolism), and rmpA (capsular serotype and HM regulator phenotype) (Compain et al. 2014; Fang et al. 2004; Yu et al. 2007). Moreover, 17 ARGs were investigated, including 16 β-lactamase-encoding genes (blaGES, blaIMP, blaKPC, blaNDM, blaOXA-1-like, blaOXA-48-like, blaVIM, blaSPM, blaGIM, blaCTX-M-Gp1, blaCTX-M-Gp2, blaCTX-M-Gp8, blaCTX-M-Gp9, blaCMY-2, blaVEB, and blaBEL) and the COL resistance gene mcr-1 (Clímaco et al. 2013; Dallenne et al. 2010; Ellington et al. 2007; Liu et al. 2016; Peirano et al. 2011; Pitout et al. 2005; Poirel et al. 2010).
One amplicon from each gene found was randomly selected to confirm its identity by sequencing using an automated sequencer (ABI 3500xL Genetic Analyzer; Applied Biosystems, USA). The obtained sequences were compared with those available in GenBank using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The ycfM gene, although confirmed by sequencing, was not deposited due to its small size amplicon (< 200 bp). In addition, all wzi genes were directly analyzed and deposited, when necessary, in the K. pneumoniae MLST database (https://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html).
Determination of capsular type
The capsular type (K-type) of the strains was determined by amplification and sequencing of the wzi gene as described by Brisse et al. (2013). The wzi gene was amplified by PCR and the amplicons were sequenced using the automated sequencer (ABI 3500xL Genetic Analyzer; Applied Biosystems, USA). Subsequently, the obtained sequences were submitted to the K. pneumoniae MLST database (https://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html) to determine the alleles corresponding to each K-type and K-locus of the strains.
Multilocus sequence typing (MLST)
The strains were evaluated by the MLST technique using protocol 2 of the K. pneumoniae MLST database (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). To show the clonal relationships among sequence types (STs) and clonal groups (CGs), goeBURST 1.2.1 version (http://www.phyloviz.net/goeburst) was used.
Results and discussion
Antimicrobial resistance profiles
The majority of strains were non-susceptible to nitrofurantoin (NIT), ciprofloxacin (CIP), and trimethoprim-sulfamethoxazole (SUT) (n = 13, 92.8%), the other members of the class of folate pathway antagonists also demonstrated high percentages, such as sulphonamide (SUL) and trimethoprim (TRI) (both 85.7%). Similar results were obtained for the tetracyclines class, such as doxycycline (DOX), tetracycline (TET) (both 85.7%), and minocycline (MIN) (78.6%). All strains were susceptible to cefotetan (CTT). Almost all were susceptible to COL since the minimal inhibitory concentration (MIC) found was < 1 mg/L, except for the KpPi151 strain that showed resistance to COL (MIC 32 mg/L). However, the mcr-1 gene was not found in this strain, indicating that resistance to COL is due to other genes or mechanisms not investigated in this study, as described by El-Sayed et al. (2020). Figure 1 shows the percentage of non-susceptibility of each antimicrobial.
A significant and worrying level of multidrug resistance strains was found, being ten strains classified as MDR and three strains (KpPi150, KpPi156, and KpPi159) as XDR (Table 1). None of the strains were classified as PDR. Among these strains, seven (KpPi145, KpPi146, KpPi147, KpPi149, KpPi151, KpPi155, and KpPi156) did not present any of the investigated ARGs, suggesting that other genes or mechanisms not researched in this study may be involved, such as efflux pumps, gene target modifiers, and changes in antimicrobial targets by mutations (Blair et al. 2015). The ideal to fully characterize the ARGs among the strains would be to characterize their resistomes by whole genome sequencing, which was not the aim of the study. Many K. pneumoniae strains with multiple antimicrobial resistance have been reported around Brazil. MDR and XDR K. pneumoniae strains have been found in the north (Ferreira et al. 2019), northeast (Aires et al. 2017), southeast (Braun et al. 2018), and south (Nava et al. 2019). In a study by Gonçalves et al. (2017), a total of 26 strains of K. pneumoniae, including 14 MDR, 7 XDR, and 3 PDR strains were found in a university hospital in southern Brazil, corroborating with our results.
Presence of ARGs and the relationship with antimicrobial resistance profiles
Three different ARGs were detected among the strains, including blaOXA-1-like, blaCTX-M-Gp1, and blaKPC (GenBank accession numbers MT330307, MT330309, and MT330311). The blaOXA-1-like was detected in four strains—KpPi148, KpPi152, KpPi153, and KpPi159 (Table 1). According to Sugumar et al. (2014), the production of OXA-1 and other β-lactamases results in resistance to ampicillin, piperacillin, ticarcillin, and cephalosporins; in fact, this profile was observed in our results. Four strains (KpPi148, KpPi152, KpPi153, and KpPi159) demonstrated resistance to APS, PIT, TAC, CAZ, CFC, CFM, CFZ, CPM, CRO, CRX, CTL, CTX, and CFO, and the only exceptions were CFO for which two strains (KpPi148 and KpPi152) were susceptible and for CTT which all strains were susceptible. All tested antibiotics are shown with their abbreviations in Fig. 1.
The KpPi152 strain also has blaCTX-M-Gp1, being the unique strain to present this gene. Chagas et al. (2011) studied the diversity of genotypes in 38 CTX-M-producing K. pneumoniae from different Brazilian hospitals located in three southeastern cities and suggested that the intensive use of broad-spectrum cephalosporins could be responsible for the amount of blaCTX-M found. The KpPi152 strain presented resistance to cephalosporins CAZ, CFC, CFM, CFZ, CPM, CRO, CRX, CTL, and CTX.
Three strains (KpPi150, KpPi153, and KpPi157) harbored the blaKPC gene (Table 1). KPC-producing bacteria are closely related to infections associated with a high level of morbidity and mortality because carbapenem antimicrobials are generally not effective against these bacteria (Arnold et al. 2011). The KpPi153 strain presented resistance to all tested carbapenems (DOR, ERT, IPM, and MPM) and KpPi150 strain to IPM and MPM. Interestingly, although the KpPi157 strain presented the blaKPC gene, it was susceptible to carbapenems. On the other hand, as described by Marschall et al. (2009) and Villegas et al. (2006), the presence of the blaKPC gene may not always result in resistance in vitro to carbapenems, resulting in a possible failure to detect this phenotype during routine workup. Therefore, an accurate investigation to detect KPC and the real resistance to carbapenems is crucial to establish control of their silent dissemination in strains that remain susceptible in vitro (Marschall et al. 2009; Villegas et al. 2006).
Occurrence of virulence genes and HM phenotype
Several virulence genes were found among the strains. All strains presented the genes fimH and ycfM, followed by mrkD (92.3%), entB (69.2%), ybtS (53.8%), and kfu (38.5%) (GenBank accession numbers MT330315, MT330318, MT330321, MT330323, MT330325, and MT330326) (Table 1). The high prevalence of fimH, mrkD, and entB genes found in this study was already expected, since several studies have shown a high correlation of these genes in K. pneumoniae strains isolated from different sources and countries (Azevedo et al. 2019; Ferreira et al. 2019; Gonçalves et al. 2017). Similar results were obtained by Kuş et al. (2017), who demonstrated that the four virulence genes most detected in K. pneumoniae strains of nosocomial infections were entB (96.2%), ycfM (86.8%), mrkD (83%), and fimH (64.2%). The strains KpPi149 and KpPi150 presented HM phenotype in addition to being MDR and XDR, respectively. The rmpA and magA genes are associated with this phenotype and some studies have shown that magA is associated to the K1 capsule-specific wzy gene (Catalán-Nájera et al. 2017). However, none of these genes were detected in both strains. This characteristic has already been reported in some Klebsiella spp., including K. pneumoniae (Lee et al. 2010; Yu et al. 2006; Mohammed and Flayyih 2018). Recently, an HM Klebsiella variicola subsp. variicola without the genes rmpA and magA was described causing primary endodontic infection and none of these genes were detected (Nakamura-Silva et al. 2020). These studies corroborate our results, indicating that other genes may be involved in the HM phenotype.
Molecular typing of K. pneumoniae strains
MLST analysis showed ten different STs (ST14, ST17, ST20, ST29, ST45, ST101, ST268, ST1800, ST3995, and ST3996) (Table 1). Two STs (ST3995 and ST3996) were described for the first time in this study, since the strains presented new alleles for the tonB gene (allele 573 for ST3995 and allele 574 for ST3996). All STs were found to belong to CG258 (Fig. 2), which is considered a high-risk CG and is well known for spreading ARGs, mainly blaKPC and virulence genes worldwide (Pitout et al., 2015). CG258 and ST11 have been described as widespread in Brazil (Gonçalves et al. 2017), including outpatient infections (Azevedo et al. 2019).
The new ST3996 was found in an XDR strain (KpPi156) which is a single-locus variant (SLV) of ST11 (Fig. 2). Two MDR strains (KpPi146 and KpPi153) presented ST101, which is considered an emerging clone that has been identified worldwide and has the potential to become a persistent threat to global public health (Roe et al. 2019). Moreover, the Kpi153 strain presented the blaKPC gene (Table 1). These characteristics were also found in southern Brazil and Italy (Gonçalves et al. 2017; Roe et al. 2019). Another new ST found in this study was ST3995 that was described in the strains Kpi152 and Kpi159, which is also an SLV of ST101. The appearance of strains with the same ST in different patients and sources suggests the spread of these STs, a characteristic observed in ST3995, ST101, and ST17 (Table 1).
The Kpi145 strain presented ST268/KL20, which corroborates with several other studies that demonstrate an association between ST268 and K20 in K. pneumoniae strains (Table 1) (Liu et al. 2014; Lin et al. 2015; Yan et al. 2015, 2016; Zhang et al. 2016; Guo et al. 2017; Chen et al. 2017). The KpPi148 strain (ST14) harbors the blaOXA-1-like gene and other β-lactamase-encoding genes that have already been described in this ST, such as K. pneumoniae strains harboring blaCTX-M-15 causing neonatal sepsis in Tanzania (Mshana et al. 2013), and K. pneumoniae co-producing blaOXA-48 and blaNDM carbapenemases in Dubai (Moubareck et al. 2018). Furthermore, the KpPi148 strain presented the capsular serotype K2, which is frequently associated with invasive infections when found in hv clones (Wyres et al. 2019); however, ST14 is a non-hv clone and K. pneumoniae ST14/K2 strains have been found in several other studies (Brisse et al. 2009; Harada et al. 2018; Musicha et al. 2019).
The KpPi147 presented ST45, wzi allele 101, and associated K-type 24 (Table 1). These same characteristics were found in an outbreak caused by MCR-1-producing K. pneumoniae strains (24 strains of ST45 and one of ST1112) isolated from patients in a hospital in Porto, Portugal (Mendes et al. 2018). Moreover, in a study of ESBL-producing Enterobacteriaceae causing sepsis in neonates at a tertiary hospital in Tanzania, 18 out 38 K. pneumoniae strains showed ST45, being the most common ST among the strains; however, unlike KpPi147, all K. pneumoniae ST45 strains in the African study presented the blaCTX-M-15 gene (Marando et al. 2018). ST20 has already been reported causing outbreaks in neonatal wards. Between 2012 and 2013, Jin et al. (2015) described two outbreaks in China involving MDR K. pneumoniae ST20 and ST17 harboring blaNDM and blaKPC. In comparison, our strain KpPi155 (ST20) did not show any of these genes. In a recent review of the genomic population of K. pneumoniae, Wyres et al. (2020) described eight MDR global problem K. pneumoniae clones, including ST101 and ST20, which were found in our strains (Table 1).
The strains KpPi149 and KpPi150 presented ST17, both have wzi allele 141, similar virulence genes, and HM phenotype, but only Kpi150 presented the blaKPC gene and an XDR profile (Table 1). ST17 has also been shown to carry other ARGs in southern Brazil, such as blaOXA-370 and blaCTX-M-8 (Aires et al. 2016). In addition, ST17 was also found in a CTX-M-15-producing K. pneumoniae strain in Norway, which caused an outbreak in a neonatal intensive care unit and subsequent intestinal colonization of affected children for up to 2 years (Löhr et al. 2015). Some STs found in this study were described as causing neonatal outbreaks in other countries (Mshana et al. 2013; Marando et al. 2018; Jin et al. 2015; Löhr et al. 2015). However, in the present study, none of the strains were isolated from neonates, with the average age of the patients being 57 years old and the youngest a child of 3 years old (KpPi159) (Table 1).
In the present study, the strain KpPi151 presented ST29 with wzi allele 85 and K-type 30. Moura et al. (2017) analyzed the genome sequence of an ST29 HM/hv MDR CTX-M-15 K. pneumoniae strain isolated from human infection in southeastern Brazil; however, they found the wzi allele 19 and K-type 19 for this strain. To the best of our knowledge, there are no reports of the ST1800 of K. pneumoniae ST1800 (KpPi157) in the literature, where only two strains are deposited in the K. pneumoniae MLST database (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html), however, without any information about them. The wzi sequences submitted to the MLST database did not find alleles corresponding to K-type for the strains KpPi149, KpPi150, KpPi153, and KpPi155 (Table 1).
Conclusions
The results presented in this study raise worrying data related to multidrug resistance and virulence in K. pneumoniae strains isolated from inpatients in the northeast of Brazil. In addition, all STs found, including two new STs, belonged to CG258, an international high-risk CG. Therefore, this study contributes to epidemiological surveillance studies in monitoring MDR, XDR, and virulent clones pertaining to CG258 worldwide.
References
Aires CA, Rocha-de-Souza CM, Timm LN, Pereira PS, Carvalho-Assef AP, Asensi MD (2016) Early detection of OXA-370-producing Klebsiella pneumoniae ST17 co-harboring blaCTX-M-8 in Brazil. Diagn Microbiol Infect Dis 86:434–436. https://doi.org/10.1016/j.diagmicrobio.2016.09.007
Aires CA, Rybak MJ, Yim J, Pereira PS, Rocha-de-Souza CM, Albano RM, Cavalcanti VO, Carvalho-Assef APD, Gomes MZR, Asensi MD (2017) Genomic characterization of an extensively drug-resistant KPC-2-producing Klebsiella pneumoniae ST855 (CC258) only susceptible to ceftazidime-avibactam isolated in Brazil. Diagn Microbiol Infect Dis 89:324–327. https://doi.org/10.1016/j.diagmicrobio.2017.08.017
Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ (2011) Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J 104:40–45. https://doi.org/10.1097/SMJ.0b013e3181fd7d5a
Ashurst JV, Dawson A (2020) Klebsiella pneumonia. In: StatPearls. Treasure Island (FL): StatPearls Publishing. PMID: 30085546
Azevedo PAA, Furlan JPR, Gonçalves GB, Gomes CN, Goulart RS, Stehling EG, Pitondo-Silva A (2019) Molecular characterisation of multidrug-resistant Klebsiella pneumoniae belonging to CC258 isolated from outpatients with urinary tract infection in Brazil. J Glob Antimicrob Resist 18:74–79. https://doi.org/10.1016/j.jgar.2019.01.025
Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ (2015) Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. https://doi.org/10.1038/nrmicro3380
Braun G, Cayô R, Matos AP, de Mello FJ, Gales AC (2018) Temporal evolution of polymyxin B-resistant Klebsiella pneumoniae clones recovered from blood cultures in a teaching hospital during a 7-year period. Int J Antimicrob Agents 51:522–527. https://doi.org/10.1016/j.ijantimicag.2017.08.031
Brazilian Committee on Antimicrobial Susceptibility Testing—BrCAST version 10.0 EUCAST. http://brcast.org.br/documentos/. Accessed 08 Oct 2020
Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P (2009) Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS ONE 4:e4982. https://doi.org/10.1371/journal.pone.0004982
Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decré D (2013) wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 51(12):4073–4078. https://doi.org/10.1128/JCM.01924-13
Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H (2017) Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence 8:1111–1123. https://doi.org/10.1080/21505594.2017.1317412
Chagas TP, Alves RM, Vallim DC, Seki LM, Campos LC, Asensi MD (2011) Diversity of genotypes in CTX-M-producing Klebsiella pneumoniae isolated in different hospitals in Brazil. Braz J Infect Dis 15:420–425. https://doi.org/10.1016/s1413-8670(11)70222-7
Chen YT, Lai YC, Tan MC, Hsieh LY, Wang JT, Shiau YR, Wang HY, Lin AC, Lai JF, Huang IW, Lauderdale TS (2017) Prevalence and characteristics of pks genotoxin gene cluster-positive clinical Klebsiella pneumoniae strains in Taiwan. Sci Rep 7:43120. https://doi.org/10.1038/srep43120
Clímaco EC, Oliveira ML, Pitondo-Silva A, Oliveira MG, Medeiros M, Lincopan N, da Costa Darini AL (2013) Clonal groups 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in Southeast Brazil. Infect Genet Evol 19:127–133. https://doi.org/10.1016/j.meegid.2013.06.024
Clinical and Laboratory Standards Institute (CLSI) (2020) Performance standards for antimicrobial susceptibility testing, M100. 30th ed. M02-M07
Compain F, Babosan A, Brisse S, Genel N, Audo J, Ailloud F, Kassis-Chikhani N, Arlet G, Decré D (2014) Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol 52:4377–4380. https://doi.org/10.1128/JCM.02316-14
Dallenne C, Decré D, Favier C, Arlet G (2010) Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495 (Epub 2010 Jan 12 PMID: 20071363)
Ellington MJ, Kistler J, Livemore DM, Woodford N (2007) Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob 59:321–322. https://doi.org/10.1093/jac/dkl481
El-Sayed Ahmed MAE, Zhong LL, Shen C, Yang Y, Doi Y, Tian GB (2020) Colistin and its role in the Era of antibiotic resistance: an extended review (2000–2019). Emerg Microbes Infect. 9:868–885
Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT (2004) A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. https://doi.org/10.1084/jem.20030857
Ferreira RL, da Silva BCM, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, Brito MCA, Silva EML, Freire CCM, Cunha AF, Prancheyicius MCS (2019) High Prevalence of multidrug-resistant Klebsiella pneumoniae Harboring several virulence and β-Lactamase encoding genes in a Brazilian intensive care unit. Front Microbiol 9:3198. https://doi.org/10.3389/fmicb.2018.03198
Gonçalves GB, Furlan JPR, Vespero EC, Pelisson M, Stehling EG, Pitondo-Silva A (2017) Spread of multidrug-resistant high-risk Klebsiella pneumoniae clones in a tertiary hospital from southern Brazil. Infect Genet Evol 56:1–7. https://doi.org/10.1016/j.meegid.2017.10.011
Guo Y, Wang S, Zhan L, Jin Y, Duan J, Hao Z, Ly J, Qi X, Chen L, Kreiswirth BN, Wang L, Yu F (2017) Microbiological and clinical characteristics of Hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front Cell Infect Microbiol 7:24. https://doi.org/10.3389/fcimb.2017.00024
Harada S, Ishii Y, Saga T, Aoki K, Tateda K (2018) Molecular epidemiology of Klebsiella pneumoniae K1 and K2 isolates in Japan. Diagn Microbiol Infect Dis 91:354–359. https://doi.org/10.1016/j.diagmicrobio.2018.03.010
Jin Y, Shao C, Li J, Fan H, Bai Y, Wang Y (2015) Outbreak of multidrug resistant NDM-1-producing Klebsiella pneumoniae from a neonatal unit in Shandong Province, China. PLoS ONE 10:e0119571. https://doi.org/10.1371/journal.pone.0119571
Kuş H, Arslan U, Türk Dağı H, Fındık D (2017) Investigation of various virulence factors of Klebsiella pneumoniae strains isolated from nosocomial infections. Mikrobiyol Bul 51:329–339. https://doi.org/10.5578/mb.59716
Lee CH, Liu JW, Su LH, Chien CC, Li CC, Yang KD (2010) Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in Taiwan. Int J Infect Dis 14:e688–e692. https://doi.org/10.1016/j.ijid.2010.01.007
Lin YT, Wang YP, Wang FD, Fung CP (2015) Community on set Klebsiella pneumonia in Taiwan: clinical features of the disease and associated microbiological characteristics of isolates from pneumonia and nasopharynx. Front Microbiol 9:122. https://doi.org/10.3389/fmicb.2015.00122
Liu YM, Li BB, Zhang YY, Zhang W, Shen H, Li H, Cao B (2014) Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother 58:5379–5385. https://doi.org/10.1128/AAC.02523-14
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. https://doi.org/10.1016/S1473-3099(15)00424-7
Löhr IH, Hülter N, Bernhoff E, Johnsen PJ, Sundsfjord A, Naseer U (2015) Persistence of a pKPN3-like CTX-M-15-encoding IncFIIK plasmid in a Klebsiella pneumonia ST17 host during two years of intestinal colonization. PLoS ONE 10:e0116516. https://doi.org/10.1371/journal.pone.0116516
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Marando R, Seni J, Mirambo MM, Falgenhauer L, Moreni N, Mushi MF, Kayange N, Manyama F, Imirzalioglu C, Chakraborty T, Mshana SE (2018) Predictors of the extended-spectrum-beta lactamases producing Enterobacteriaceae neonatal sepsis at a tertiary hospital. Tanzania Int J Med Microbiol 308:803–811. https://doi.org/10.1016/j.ijmm.2018.06.012
Marschall J, Tibbetts RJ, Dunne WMJr, Frye JG, Fraser VJ, Warren DK, (2009) Presence of the KPC carbapenemase gene in Enterobacteriaceae causing bacteremia and its correlation with in vitro carbapenem susceptibility. J Clin Microbiol 47:239–241. https://doi.org/10.1128/JCM.02123-08
Mendes AC, Novais Â, Campos J, Rodrigues C, Santos C, Antunes P, Ramos H, Peixe L (2018) mcr-1 in Carbapenemase-Producing Klebsiella pneumoniae with Hospitalized Patients, Portugal, 2016–2017. Emerg Infect Dis 24:762–766. https://doi.org/10.3201/eid2404.171787
Mohammed ES, Flayyih MT (2018) Detection of rmpA and magA genes and hypermucoviscosity phenotype in Klebsiella pneumoniae isolated from water samples in compare with clinical isolates. Curr Res Microbiol Biotechnol 6:1424–1430
Moubareck CA, Mouftah SF, Pál T, Ghazawi A, Halat DH, Nabi A, AlSharhan MA, AlDeesi ZO, Peters CC, Celiloglu H, Sannegowda M, Sarkis DK, Sonnevend A (2018) Clonal emergence of Klebsiella pneumoniae ST14 co-producing OXA-48-type and NDM carbapenemases with high rate of colistin resistance in Dubai, United Arab Emirates. Int J Antimicrob Agents 52:90–95. https://doi.org/10.1016/j.ijantimicag.2018.03.003
Moura Q, Esposito F, Fernandes MR, Espinoza-Muñoz M, Souza T, Santos SR, Cerdeira LT, Cassettari V, Lincopan N (2017) Genome sequence analysis of a hypermucoviscous/hypervirulent and MDR CTX-M-15/K19/ST29 Klebsiella pneumoniae isolated from human infection. Pathog Dis. https://doi.org/10.1093/femspd/ftx121
Mshana SE, Hain T, Domann E, Lyamuya EF, Chakraborty T, Imirzalioglu C (2013) Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis 13:466. https://doi.org/10.1186/1471-2334-13-466
Musicha P, Msefula CL, Mather AE, Chaguza C, Cain AK, Peno C, Kallonen T, Khonga M, Denis B, Gray KJ, Heyderman RS, Thomson NR, Everett DB, Feasey NA (2019) Genomic analysis of Klebsiella pneumoniae isolates from Malawi reveals acquisition of multiple ESBL determinants across diverse lineages. J Antimicrob Chemother 74:1223–1232. https://doi.org/10.1093/jac/dkz032
Nakamura-Silva R, Macedo LMD, Cerdeira LT, Oliveira-Silva M, Silva-Sousa YTC, Pitondo-Silva A (2020) First report of hypermucoviscous Klebsiella variicola subsp variicola causing primary endodontic infection. Clin Microbiol Infect 27(2):303–304
Nava RG, Oliveira-Silva M, Nakamura-Silva R, Pitondo-Silva A, Vespero EC (2019) New sequence type in multidrug-resistant Klebsiella pneumoniae harboring the blaNDM-1-encoding gene in Brazil. Int J Infect Dis 79:101–103. https://doi.org/10.1016/j.ijid.2018.11.012
Paczosa MK, Mecsas J (2016) Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. https://doi.org/10.1128/MMBR.00078-15
Peirano G, Ahamed-Bentley J, Woodford N, Pitout JD (2011) New Delhi metallo-β-lactamase from traveler returning to Canada. Emer Infect Dis 17:242–244. https://doi.org/10.3201/eid1702.101313
Piperaki ET, Syrogiannopoulos GA, Tzouvelekis LS, Daikos GL (2017) Klebsiella pneumoniae: virulence, biofilm and antimicrobial resistance. Pediatr Infect Dis J 36:1002–1005. https://doi.org/10.1097/INF.0000000000001675
Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL (2005) Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol 43:3129–3135. https://doi.org/10.1128/JCM.43.7.3129-3135.200
Pitout JD, Nordmann P, Poirel L (2015) Carbapenemase-Producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother 59:5873–5884. https://doi.org/10.1128/AAC.01019-15
Poirel L, Docquier JD, Luca F, Verlinde A, Ide L, Rossolini GM, Nordmann P (2010) Bel-2, an extended-spectrum Beta-lactamase with increased activity toward expanded-spectrum cephalosporins in Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:533–535. https://doi.org/10.1128/AAC.00859-09
Roe CC, Vazquez AJ, Esposito EP, Zarrilli R, Sahl JW (2019) Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front Microbiol 10:542. https://doi.org/10.3389/fmicb.2019.00542
Sugumar M, Kumar KM, Manoharan A, Anbarasu A, Ramaiah S (2014) Detection of OXA-1 β-lactamase gene of Klebsiella pneumoniae from blood stream infections (BSI) by conventional PCR and in-silico analysis to understand the mechanism of OXA mediated resistance. PLoS ONE 9:e91800. https://doi.org/10.1371/journal.pone.0091800
Villegas MV, Lolans K, Correa A, Suarez CJ, Lopez JA, Vallejo M, Quinn JP (2006) First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother 50:2880–2882. https://doi.org/10.1128/AAC.00186-06
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Wiskur BJ, Hunt JJ, Callegan MC (2008) Hypermucoviscosity as a virulence factor in experimental Klebsiella pneumoniae endophthalmitis. Invest Ophthalmol vis Sci 49:4931–4938. https://doi.org/10.1167/iovs.08-2276
Wyres KL, Wick RR, Judd LM, Froumine R, Tokolyi A, Gorrie CL, Lam MMC, Duchêne S, Jenney A, Holt KE (2019) Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet 15:e1008114. https://doi.org/10.1371/journal.pgen.1008114
Wyres KL, Lam MMC, Holt KE (2020) Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. https://doi.org/10.1038/s41579-019-0315-1
Yan JJ, Zheng PX, Wang MC, Tsai SH, Wang LR, Wu JJ (2015) Allocation of Klebsiella pneumoniae bloodstream isolates into four distinct groups by ompK36 typing in a Taiwanese University Hospital. J Clin Microbiol 53:3256–3263. https://doi.org/10.1128/JCM.01152-15
Yan Q, Zhou M, Zou M, Liu WE (2016) Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur J Clin Microbiol Infect Dis 35:387–396. https://doi.org/10.1007/s10096-015-2551-2
Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC (2006) Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 42:1351–1358. https://doi.org/10.1086/503420
Yu W, Fung C, Ko W, Cheng K, Lee C, Chuang Y (2007) Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis 195:1235–1236. https://doi.org/10.1086/512686
Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Wang R, Wang H (2016) High prevalence of Hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother 60:6115–6120. https://doi.org/10.1128/AAC.01127-16
Acknowledgements
The authors also thank the team of curators from the Institut Pasteur MLST and genome MLST databases for curating the data and making them publicly available at http://bigsdb.pasteur.fr. We would like to thank John Carpenter, Ribeirão Preto, SP, Brazil for the English revision. R.N.S is a PhD fellow of CAPES, therefore, this study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [grant no 88882365161 and Finance Code 001]. J.P.R.F. is a PhD fellow of CAPES [grant no. 88882.180855/2018-01 and Finance Code 001] and FAPESP [grant no. 2018/01890-3].
Funding
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [grant no. 2013/22581-5].
Author information
Authors and Affiliations
Contributions
APS designed this study, contributed to the analysis and with intellectual experience. RNS carried out all experiments and analyzes of this study and wrote the manuscript. MOS contributed to DNA extraction, antimicrobial susceptibility testing, hypermucoviscosity test, and PCRs. CESM provided the bacterial collection. JPRF and EGS contributed to the analysis of results. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Ethical approval was received from the School of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo (Ribeirão Preto, SP, Brazil) [approval no. CEP/FCFRP 362; CAEE 36031914.9.0000.5403].
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nakamura-Silva, R., Oliveira-Silva, M., Furlan, J.P.R. et al. Characterization of multidrug-resistant and virulent Klebsiella pneumoniae strains belonging to the high-risk clonal group 258 (CG258) isolated from inpatients in northeastern Brazil. Arch Microbiol 203, 4351–4359 (2021). https://doi.org/10.1007/s00203-021-02425-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02425-0