Abstract
A Gram-stain-negative, facultative anaerobic strain, designated WSJ-3T, was isolated from soil. Phylogenetic analyses based on 16S rRNA gene sequences indicated that strain WSJ-3T belongs to genus Sediminibacterium and exhibits the highest sequence similarities to Sediminibacterium roseum SYL130T (97.0%), Sediminibacterium goheungense DSM 28323T (96.9%), Sediminibacterium aquarii AA5T (96.7%), and Sediminibacterium salmoneum NBRC 103935T (95.2%). The average nucleotide identity values of strain WSJ-3T/S. roseum SYL130T and strain WSJ-3T/S. goheungense DSM 28323T are 72.2% and 70.4%, respectively, and digital DNA–DNA hybridization values for these are 19.2% and 19.1%, respectively. Strain WSJ-3T has a genome size of 3.88 Mb, with a DNA G + C content of 50.1 mol% and comprises of 3263 predicted genes. A phylogenetic tree constructed using the genomic core protein coding sequences revealed that strain WSJ-3T clusters with S. roseum SYL130T. Strain WSJ-3T has menaquinone-7 as the only respiratory quinone and phosphatidylethanolamine, three unidentified phospholipids, four unidentified aminophospholipids, two unidentified aminolipids, and three unidentified lipids as the polar lipids. The major fatty acids of strain WSJ-3T are iso-C15:0, iso-C17:0 3-OH, and iso-C15:1 G. On the basis of the polyphasic results, the isolate represents a novel species of the genus Sediminibacterium, for which the name Sediminibacterium soli sp. nov. is proposed. The type strain is WSJ-3T (= KCTC 72839T = CCTCC AB 2019408T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genus Sediminibacterium belongs to phylum Bacteroidetes, class Chitinophagia, order Chitinophagales, and family Chitinophagaceae, and was established in 2008 with Sediminibacterium salmoneum as the type species (Qu and Yuan 2008). Currently, there are seven Sediminibacterium species (https://lpsn.dsmz.de/genus/sediminibacterium). Microbial community studies revealed that the certain members of genus Sediminibacterium may be associated with type 2 diabetes mellitus (Qiu et al. 2019) and carbon uptake during aerobic vinyl chloride biodegradation (Wilson et al. 2016), but the functions of Sediminibacterium strains are still poorly understood. The discovery of a new species of Sediminibacterium enriches the diversity of this genus and provides theoretical basis for further genomic, genetics and applicable researches.
Here, a novel Sediminibacterium strain WSJ-3T was isolated and used for polyphasic analyses. Although the strain belongs to the genus Sediminibacterium, it differs from the other types in several genetic, phenotypic, and chemotaxonomic traits. Therefore, it is proposed that a new species name of Sediminibacterium soli sp. nov. should be established.
Materials and methods
Sample source and strain isolation
Strain WSJ-3T was isolated from soil surrounding Zhongwei New Material Co. Ltd. in Tongren city, Guizhou province, P. R. China, with geographical coordinates of 26° 36′ 34.35″ N, 106° 42′ 28.07″ E. The soil sample has a pH of approximately 7.0. The sampled soil was suspended in 0.85% saline solution at 1% and shaking for 30 min at 28 °C. After setting for 2 h, the sample was spread on Reasoner’s 2A (R2A) agar. Then it was incubated at 28 °C for 7 days. A yellow-orange colony, designated WSJ-3T, was isolated. After several sub-cultivation cycles, the purified strain was obtained and stored at − 80 °C in a 25% glycerol suspension.
Phylogenetic analysis
The nearly complete 16S rRNA gene sequence of strain WSJ-3T was amplified from the genomic DNA using conserved primers 5′-AGAGTTTGATCCTGGCTCAG-3′ (27F) and 5′-GGTTACCTTGTTACGACTT-3′ (1492R) (Fan et al. 2010). The 16S rRNA gene sequence (1490 bp) was obtained using pGEM-T (Promega) vector (Wu et al. 2019) and compared with the sequences available in EzTaxon-e server using EzBioClould (Yoon et al. 2017a). It was revealed that this sequence was identical to the full-length 16S rRNA gene sequence extracted from the draft genome. MEGA 6.0 (Tamura et al. 2013) was used to construct neighbor-joining (NJ) (Saitou and Nei 1987), maximum-likelihood (ML) (Felsenstein 1981), and minimum-evolution (ME) (Rzhetsky and Nei 1992) trees with the bootstrap of 1000 replications (Felsenstein 1985) while an algorithm of Kimura’s two-parameter model (Kimura 1979) was used to calculate evolutionary distances. To further explore the relationship among strain WSJ-3T and its related strains, the NJ phylogenetic tree based on the genomic core multiproteins was constructed using up-to-date bacterial core gene set and pipeline (UBCG) (Na et al. 2018).
Genome sequencing and analysis
Since strain WSJ-3T exhibited the highest 16S rRNA gene sequence similarity to S. roseum SYL130T (97.0%), the draft genomes of the two strains were sequenced by Wuhan Frasergen Bioinformatics Co., Ltd. The genomic DNA was extracted and randomly fragmented using Covaris ultrasonic crusher, and a shotgun library was constructed using TruSeq DNA Sample Prepare Kit (Vazyme Biotech). Pair-end sequencing was performed using the Illumina HiseqX system and the sequenced reads were assembled using SPAdes v3.11.1 (http://cab.spbu.ru/software/spades/). The draft genomes of strain WSJ-3T and S. roseum SYL130T were submitted to NCBI and annotated using Prokaryotic Genome Annotation Pipeline.
To detect the relationship among strain WSJ-3T and these strains, average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) were performed. The ANI values among strain WSJ-3T and strains S. roseum SYL130T and S. goheungense DSM 28323T were analyzed using the web version of the ANI calculator (http://www.ezbiocloud.net/tools/ani) (Yoon et al. 2017b) whereas digital DNA–DNA hybridization (dDDH) was analyzed by a webserver (http://ggdc.dsmz.de/ggdc.php) (Meier-Kolthoff et al. 2013). Cluster of Orthologous Groups of proteins (COG) (http://weizhong-lab.ucsd.edu/webMGA/server/cog/) was used to analyze the protein functional categories with an E value is 10−10 (Tatusov et al. 2000). The genome sequences of strain WSJ-3T, S. roseum SYL130T, S. goheungense DSM 28323T, and S. salmoneum NBRC 103935T were analyzed using CGView Server (http://stothard.afns.ualberta.ca/cgview_server/) to obtain the graphical circular map (Grant and Stothard 2008). To construct the Venn diagram, OrthoFinder was used to extract the homologous proteins (Emms and Kelly 2015), after that, Excel was used for statistics and charting. The metabolic pathways associated with physiology and biochemical characters were analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis (https://www.kegg.jp/) (Du et al. 2014).
Morphological and physiological analysis
To conduct morphology, physiology and biochemistry analysis, strain WSJ-3T and other related strains S. roseum SYL130T, S. goheungense KCTC 23945T, S. aquarii JCM 31013T and S. salmoneum NBRC 103935T were cultured in R2A broth or on R2A agar at 28 °C, unless otherwise indicated. For cell morphology observation, scanning electron microscopy (SEM) (JSM-6390; JEOL) and transmission electron microscopy (TEM) (H-7650; Hitachi) were used. For SEM, cells cultured in R2A broth for 3 d were collected by centrifugation at 5000 rpm for 5 min. The cell pellets were washed thrice with 0.1 M PBS (pH 7.4) and fixed in 1 mL 2.5% glutaraldehyde overnight. Once fixed, the pellets were washed thrice using 0.1 M PBS and dehydrated with increasing ethanol concentrations (30%, 50% and 70%) and the cells were freeze-dried using a vacuum. For TEM observation, the cells grown on R2A agar for 5 days were suspended in 0.85% saline solution. Gram stain was performed using a Gram-staining kit (Jian-cheng Biotech, China), motility was observed using 0.3% agar, and gliding motility was tested with the hanging-drop method (Bernardet et al. 2002). Growth was tested at different temperatures (4, 15, 20, 28, 37, 42 and 45 °C), pH levels (4.0, 5.0, 6.0, 7.0, 8.0 and 9.0) (Wu et al. 2019) and NaCl concentrations (0, 0.5, 1.0, 1.5, 2.0, 3.0, 4.0 and 5.0%). The growth of strain WSJ-3T was also observed in different media (LB, Luria–Bertani; NB, nutrient broth; 1/10TSB, tryptic soy broth). The anaerobic growth was assessed on R2A agar in an anaerobic chamber, where the O2 had been removed using O2-adsorbing agent (Anaero-Pack, Mitsubishi Gas Chemical) for 2 weeks. Its production of flexirubin-type pigments was measured with 20% (w/v) KOH (Bernardet et al. 2002), oxidase activity was tested using 1% (w/v) tetramethyl-β-phenylenediamine (Cappuccino and Sherman 1987), and catalase activity was determined by observing bubble production using 3% H2O2. The hydrolysis of casein, gelatin, starch, cellulose and Tween 20, 40 and 60, and the production of H2S and indole were tested as depicted as Smibert and Krieg (Smibert and Krieg 1994). The utilization of sole carbon and acid production were performed using traditional methods (Dong and Cai 2001). Antibiotic susceptibility was determined based on the Kirby-Bauer method, which included the use of (μg/per disc, unless otherwise stated) ampicillin (10 μg), carbenicillin (100 μg), cefoperazone (75 μg), cefradine (30 μg), cefuroxime (30 μg), chloramphenicol (5 μg), doxycycline (30 μg), erythromycin (15 μg), gentamincin (10 μg), kanamycin (30 μg), minocycline (30 μg), neomycin (30 μg), novobiocin (30 μg), penicillin G (10 U), rifampicin (5 μg), streptomycin (30 μg), tetracycline (30 μg), and vancomycin (30 μg). For additional biochemical characterization, API 20 NE and API ZYM (bio-Mérieux, France) were detected at 28 °C for 4 days and 6 h, respectively, according to the manufacturer’s instructions.
Chemotaxonomic analysis
The respiratory quinones of strain WSJ-3T were detected using HPLC (Xie and Yokota 2003) and its polar lipids were analyzed by two-dimensional TLC (O’Donnell et al. 1982). The whole-cell fatty acids of strain WSJ-3T and the other four type strains were extracted when these strains were cultured to logarithmic phase and determined using the Sherlock Microbial Identification System (version 6.1 and TSBA library version 6.1) (Sasser 1990).
Results and discussion
Phylogeny analysis
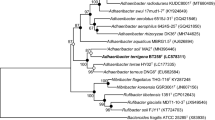
The 16S rRNA gene sequence of strain WSJ-3T shows the highest sequence similarities to S. roseum SYL130T (97.0%), S. goheungense DSM 28323T (96.9%), S. aquarii AA5T (96.7%), and S. salmoneum NBRC 103935T (95.2%). As shown in Fig. 1, a phylogenetic tree based on the NJ method indicated that strain WSJ-3T is grouped in the same branch with S. roseum SYL130T. Similar topologies are also observed in the ML (Fig. S1) and ME (Fig. S2) trees. In addition, the phylogenetic analysis based on the genomic core protein coding sequences (Fig. 2) also shows that strain WSJ-3T clusters with S. roseum SYL130T.
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences, exhibiting the relationship between strain WSJ-3T and other members of genus Sediminibacterium. Bootstraps values of > 50% are shown at branch points. Filled circles represent the corresponding nodes that were consistent to that in maximum-likelihood and minimum-evolution trees. Flavobacterium limi THG-AG6.4T was used as an outgroup. Bar 0.02 substitutions per nucleotide position
Neighbor-joining phylogenetic tree based on genomic core protein sequences showing the relationship between strain WSJ-3T and the most closely strains of genus Sediminibacterium. Bootstrap values (> 50%) based on 1000 replications are shown at branch nodes. Flavobacterium limi THG-AG6.4T was used as an outgroup. Bar, 0.05 substitutions per nucleotide position
Genome characterization
The genomic information of strains WSJ-3T (JAACJR000000000) and S. roseum SYL130T (JAACJS000000000) are listed in Table S1, and the quality of genome sequences meet the standards for the taxonomy of prokaryotes (Chun et al. 2018). The ANI values for WSJ-3T/S. roseum SYL130T and WSJ-3T/S. goheungense DSM 28323T are 72.21% and 70.4%, respectively; the dDDH values are 19.2% and 19.1%, respectively, which are both significantly lower than the thresholds (95% for ANI and 70% for dDDH) for prokaryotic species delineation (Wayne et al. 1988; Chun et al. 2018). The distribution of proteins into COGs functional categories are shown in Table S2. The KEGG analysis indicates that strain WSJ-3T has putative genes for gliding, and assimilation of maltose and xylose, which are consistent with the physiological and biochemical characters (Table S3). Fig. S3 shows the Venn diagram of the total number of core proteins and species-specific proteins among the genomes of strain WSJ-3T and its related strains. Although strain WSJ-3T shares the highest number of core protein numbers with its most closely related strain S. roseum SYL130T, it has 411 specific proteins (Table S4). The graphical circular map of the comparison among strain WSJ-3T and the related strains is provided in Fig. S4. In blast rings 3, 4 and 5, the height and density of the bars reflect the degree of protein similarity, which demonstrates the differences among strain WSJ-3T and the closely related type strains at amino acid level.
Morphological and physiological characteristics
Strain WSJ-3T is Gram-stain-negative, facultative anaerobic, and non-flagellated cells (0.3–0.5 × 1.0–2.0 μm) (Fig. S5). It has the ability of gliding motility and does not produce flexirubin pigment and H2S. Strain WSJ-3T is negative for the hydrolysis of cellulose, starch, and Tween series, and resistant to ampicillin, carbenicillin, erythromycin and penicillin G.
Other physiological characteristics are summarized in the Species description, and the differences among strain WSJ-3T and the related strains are listed in Table 1. Strain WSJ-3T exhibits different characteristics from the other strains in the hydrolysis of Tween 60, the utilization of lactose, and the activities of trypsin and β-glucuronidase (Table 1). The negative reactions from the API ZYM and API 20 NE tests are listed in Table S5.
Chemotaxonomic characteristics
For strain WSJ-3T, menaquinone-7 is the only respiratory quinone. Phosphatidylethanolamine, three unidentified phospholipids, four unidentified aminophospholipids, two unidentified aminolipids, and three unidentified lipids are the polar lipids (Fig. S6). The major fatty acids contain iso-C15:0, iso-C17:0 3-OH and iso-C15:1 G (> 10%) (Table 2). These results are similar to the characters of Sediminibacterium strains (Qu and Yuan 2008; Kim et al. 2013; Lee et al. 2013; Albert et al. 2014; Kang et al. 2014; Kim et al. 2016; Song et al. 2017).
In terms of phenotypic and chemotaxonomic characters, strain WSJ-3T has many common features of the Sediminibacterium strains, which indicates that it is a member of the genus Sediminibacterium. However, the strain shows unique traits in the hydrolysis of Tween 60, the utilization of lactose, the enzyme activities of trypsin and β-glucuronidase. The Venn diagram and graphical circular map based on comparison among strain WSJ-3T and the related strains indicate that strain WSJ-3T is distinct at protein level. Thus, it further indicates that strain WSJ-3T represents a novel species of the genus, for which the name Sediminibacterium soli sp. nov. is proposed.
Description of Sediminibacterium soli sp. nov.
Sediminibacterium soli sp. nov. (so’li. L. gen. n. soli of soil).
Gram-stain-negative, facultatively anaerobic, and non-flagellated rods. Colonies are smooth, circular and yellow-orange growing on R2A agar. Flexirubin pigment is not produced, and motile by gliding. Grow at 15–37 °C, at pH 5.0–7.0 and without NaCl addition. Grow in R2A and NB media but not in LB and 1/10 TSB. Positive for catalase and oxidase, but negative for the production of H2S and the hydrolysis of casein, carboxymethyl cellulose, starch, tween 20, tween 40 and tween 60. Maltose and xylose can be utilized, but d-arabinose, l-alanine, d-fructose, glucose, glycogen, histidine, lactose, melibiose, raffinose, l-rhamnose, d-ribose, salicin, l-serine, d-sorbitol, sucrose, trehalose and trisodium acid cannot be used. Acid is produced from glucose and lactose, but not maltose, d-mannitol, l-rhamnose, d-ribose, d-sorbitol, sucrose, trehalose and xylose. In API ZYM, positive for acid phosphatase, N-acetyl-β-glucosaminidase, alkaline phosphatase, cystine arylamidase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, β-glucuronidase, leucine arylamidase, α-mannosidase, naphthol-AS-BI-phosphohydrolase and valine arylamidase. The menaquinone is menaquinone 7, and the major polar lipids are phosphatidylethanolamine, unidentified phospholipids, unidentified aminophospholipids, unidentified aminolipids and unidentified lipids, and the major fatty acids are iso-C15:0, iso-C17:0 3-OH and iso-C15:1 G. The genomic DNA G + C content of the type strain is 50.1 mol%.
The type strain Sediminibacterium soli WSJ-3T (= KCTC 72839T = CCTCC AB 2019408T) was isolated from soil nearby Zhongwei New Material Co. Ltd, Tongren city, Guizhou province, P. R. China. The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence and the draft genome sequence of strain WSJ-3T are MT299774 and JAACJR000000000, respectively.
References
Albert RA, Zitomer D, Dollhopf M, Schauer-Gimenez AE, Struble C, King M, Son S, Langer S, Busse HJ (2014) Proposal of Vibrionimonas magnilacihabitans gen. nov., sp. nov., a curved Gram-stain-negative bacterium isolated from lake water. Int J Syst Evol Microbiol 64:613–620
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Cappuccino JG, Sherman N (1987) Microbiology, a laboratory manual, 6th edn. Pearson Education, Inc., Benjamin Cummings, San Francisco
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Dong XZ, Cai MY (2001) Determinative manual for routine bacteriology. Scientific Press, Beijing
Du J, Yuan Z, Ma Z, Song J, Xie X, Chen Y (2014) KEGG-PATH: kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol BioSyst 10:2441–2447
Emms DM, Kelly S (2015) OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:157
Fan H, Su C, Wang Y, Yao J, Zhao K, Wang Y, Wang G (2010) Sedimentary arsenite-oxidizing and arsenate-reducing bacteria associated with high arsenic groundwater from Shanyin, Northwestern China. J Appl Microbiol 105:529–539
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Grant J, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184
Kang H, Kim H, Lee BI, Joung Y, Joh K (2014) Sediminibacterium goheungense sp. nov., isolated from a freshwater reservoir. Int J Syst Evol Microbiol 64:1328–1333
Kim YJ, Nguyen NL, Weon HY, Yang DC (2013) Sediminibacterium ginsengisoli sp. nov., isolated from soil of a ginseng field, and emended descriptions of the genus Sediminibacterium and of Sediminibacterium salmoneum. Int J Syst Evol Microbiol 63:905–912
Kim Y, Kim B, Kang K, Ahn T-Y (2016) Sediminibacterium aquarii sp. nov., isolated from sediment in a fishbowl. Int J Syst Evol Microbiol 66:4501–4505
Kimura M (1979) A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lee DG, Park JM, Kang H, Hong SY, Lee KR, Chang HB, Trujillo ME (2013) Asinibacterium lactis gen. nov., sp. nov., a member of the family Chitinophagaceae, isolated from donkey (Equus asinus) milk. Int J Syst Evol Microbiol 63:3180–3185
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformat 14:60
Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J (2018) UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:280–285
O’Donnell AG, Goodfellow M, Minnikin DE (1982) Lipids in the classification of Nocardioides: reclassification of Arthrobacter simplex (Jensen) lochhead in the genus Nocardioides (Prauser) emend. Arch Microbiol 133:323–329
Qiu J, Zhou H, Jing Y, Dong C (2019) Association between blood microbiome and type 2 diabetes mellitus: a nested case-control study. J Clin Lab Anal 33:e22842
Qu JH, Yuan HL (2008) Sediminibacterium salmoneum gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from sediment of a eutrophic reservoir. Int J Syst Evol Microbiol 58:2191–2194
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 9:945–967
Saitou NNM, Nei MC (1987) The neighbor-joining method-a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. Newark, pp 1–7
Smibert RM, Krieg NR (1994) Phenotypic characterization. In methods for general and molecular bacteriology, pp 611–651
Song Y, Jia J, Liu D, Choi L, Wang G, Li M (2017) Sediminibacterium roseum sp. nov., isolated from sewage sediment. Int J Syst Evol Microbiol 67:4674–4679
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1988) International committee on systematic bacteriology announcement of the report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Syst Appl Microbiol 268:433–434
Wilson FP, Liu X, Mattes TE, Cupples AM (2016) Nocardioides, Sediminibacterium, Aquabacterium, Variovorax, and Pseudomonas linked to carbon uptake during aerobic vinyl chloride biodegradation. Environ Sci Pollut Res Int 23:19062–19070
Wu S, Xia X, Zhou Z, Wang D, Wang G (2019) Nocardioides gansuensis sp. nov., isolated from geopark soil. Int J Syst Evol Microbiol 69:390–396
Xie CH, Yokota A (2003) Phylogenetic analyses of Lampropedia hyalina based on the 16S rRNA gene sequence. J Gen Appl Microbiol 49:345–349
Yoon SH, Ha SM, Kwon S, Lim J, Chun J (2017a) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017b) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Acknowledgements
The authors are grateful to Professor Dr. Bernhard Schink for the Latin construction of the species name. We acknowledge the Guangdong Institute of Microbiology for the analyses of respiratory quinone and polar lipids.
Funding
This work was supported by the National Natural Science Foundation of China (31670108).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence and the draft genome sequence of strain WSJ-3T are MT299774 and JAACJR000000000, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, S., Zhong, L., Liao, S. et al. Sediminibacterium soli sp. nov., isolated from soil. Arch Microbiol 203, 967–973 (2021). https://doi.org/10.1007/s00203-020-02089-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-02089-2