Abstract
Anaerobic digestion, a recently hot technology to produce biogases especially methane generation for biofuel from wastewater, is considered an effective explanation for energy crisis and global pollution threat. A complex microbiome population is present in sludge, which plays an important role in the digestion of complex polymer into simple monomers. 16S rRNA approaches simply are not enough for amplification due to the involvement of extreme complex population. However, Illumina sequencing is a recent powerful technology to reveal the entire microbiome structure and methane generation pathways in anaerobic digestion. Metagenomic sequencing was tested to reveal the microbial structure of a digested sludge from a local wastewater treatment plant in Beijing. The Illumina HiSeq program was used to extract about 5 GB of data for metagenomic analysis. The classification investigation revealed about 97.64% dominancy of bacteria while 1.78% were detected to be archaea using MG-RAST server. The most abundant bacterial communities were reported to be Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria. Furthermore, the important microbiome involved in methane generation was revealed. The dominant methanogens were detected (Methanosaeta and Methanosarcina), with affiliation of dominant genes involved in acetoclastic methanogenesis in a digesting sludge. The metagenomic analysis showed that microbial structure and methane generation pathways were successfully dissected in an anaerobic digester.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many innovations and alterations have been made in methodologies in the past decades for wastewater treatment improvement but activated sludge has recently caught the attention for the biological treatment of wastewater (van Loosdrecht and Brdjanovic 2014; Guo et al. 2013). However, considerable volume of sludge is produced throughout wastewater treatment that needs further processing which needs more than 50% of the overall processing in wastewater treatment plants (Canales et al. 1994). Anaerobic digestion is broadly utilized to lower the extent of sludge to diminish pathogens and more methane generation is one of the key and effective explanations for environmental pollution and energy shortage problems (Appels et al. 2008). Previous reports have shown that anaerobic system can transform about 50% of organic waste to methane, an impressive source of biofuel, producing 36.5 MJ/m3 in agitation (Amani et al. 2010).
As we know, anaerobic digestion of sludge involves extremely complicated microbiome that has an important role in sludge treatment and methane generation capacity. Several molecular diagnostic tools and methodologies including gel electrophoresis, fluorescent in situ hybridization, 16S rRNA gene and other marker gene sequencing have been reported to reveal microbiome structure in anaerobic scheme (Vanwonterghem et al. 2014a, b). However, these tools are not enough to provide the complete information of the whole genome belonging to a genetically diversified and complicated microbiome structure in an anaerobic system.
Furthermore, the access to the base of clone library sequencing of the 16S rRNA gene for green analysis of useful microbiome may give unpredictable results because of their inherent bias of elaboration (Ye et al. 2012; Aird et al. 2011). Illumina sequencing and 454 pyrosequencing technologies are high-throughput sequencing methods that have been proved recently as innovative and bright methods to reveal highly complicated microbial genome (Bragg and Tyson 2014; Albertsen et al. 2003). Many reports have been published so far on microbiome of anaerobic system using 454 pyrosequencing (Wong et al. 2013; Li et al. 2013; Sundberg et al. 2013). Illumina sequencing has reported greater significance and is cost friendly in comparison with 454 pyrosequencing to evaluate microbial community structure (Mardis 2008; Glenn 2011). It has been used to reveal microbial population from soil (Mackelprang et al. 2011), ocean (Mason et al. 2014), human gut (Qin et al. 2010) and digested sludge (Albertsen et al. 2006; Ju et al. 2014). However, a very small attempt has been made to evaluate in detail the microbial community including practical biome structure using Illumina (Yang et al. 2014). Furthermore, any little accomplishment made to analyze the dominant methane-producing pathway in anaerobic digestion sludge is still ambiguous.
The purpose of this research is to dissect the microbial structure and identify metagenomic community composition, exposing useful characteristics of anaerobic digester. For this target, DNA was extricated from sludge, and high-throughput metagenomic sequencing was performed on the Illumina HiSeq 2000 platform. The complex microbial population, useful traits, and metabolic pathways were evaluated in detail. Fundamental microorganisms engaged in all degradation steps including hydrolysis, acidogenesis, acetogenesis and methane generation were extensively evaluated. In addition, the desirable genes affiliated with methanogenic pathways were displayed. The current study focusses on dominant microbial traits in anaerobic digester, enhancing the establishment of more powerful anaerobic systems to generate maximum methane.
Materials and methods
Bioreactor’s outline
The sludge sample was obtained from an anaerobic digester from a local wastewater treatment plant in Beijing, China. This plant delights an average flow of about one million m3/day and serves approximately 2,400,000 people in Beijing city. The pH of the sludge was found to be around 7.8, while the total suspended solids (TSS) and volatile suspended solids (VSS) of the sludge were 8.66 g/L, and 4.86 g/L, respectively. A laboratory-scale continuous stirred-tank reactor with a working volume of 1 L (total 2 L) was operated at 35 °C for more than 2 months. The anaerobic reactor was fully stirred by magnetic stirring. The SRT of the reactors was set for 2 days and OLR was 10 gVS/L day by feeding synthetic substrate (15 g glucose diluted in 500 mL synthetic medium). The pH of the reactor was controlled by a pH monitor with the addition of HCl (2 M) and NaOH (2 M) according to the need. The reactor was fed and drawn off once a day and the draw-off effluent was used for analysis. Gas samples were taken from the reactor via a small sampling port located at the top of the reactor while effluent sample was taken from pull-off materials like syringes every day. Standard method was used to measure TS, VS, TSS and VSS (APHA 1998).
Sampling and DNA extraction
A total of 100% ethanol was infused immediately at a ratio of 1:1 (volume/volume) after collection of samples, and then shifted to lab and reserved at − 20 °C before the extraction of DNA for experiments. Genomic DNA were extracted after collection of samples from the plant. About 2–3 mL sample was centrifuged at 4600g for 2 min to obtain the pellet by discarding the supernatant. Fast-DNA SPIN Kit for Soil (QBIOgene Inc., Carlsbad, CA, USA) was used for the extraction of DNA, according to the instruction of the manufacturer. Gel electrophoresis was used to analyze the standard of DNA. Qubit Fluorometer (Thermo, USA) was used to find out the concentration of DNA which was detected to be 740 ng/μL.
DNA library construction
Illumina HiSeq 2000 platform was used for the metagenomic sequencing at School of medicine Tsinghua University Beijing, China. About 200–300 ng/μL of extracted genomic DNA was used for further processing. Covaris S2 Ultrasonicator was used for the DNA fragmentation followed by processing of end reparation, A-tailing, adapter ligation, DNA size selection, PCR and purification on the instruction base of Illumina HiSeq 2000. The base-calling pipeline was used to produce the raw fluorescence images and call sequences. The max 5.0 Gb reads of the sample were practiced by metagenomic data bank. The reads were shortened using a minimum 30 quality score, a minimum read length of 32 bp and allowing no ambiguous nucleotides. The given parameters were adopted for overlapping is: about 20 nucleotide length of the overlap region was needed and allowed at least two mismatches.
Bioinformatic analysis
Metagenomic rapid annotation (MG-RAST) server (v3.1) was used for the annotation of non-assembled DNA. MG-RAST facilitates metabolic and phylogenetic reorganization and performs analysis of protein similarities, such as function annotation division (Meyer et al. 2008). About 5.0 Gb of DNA databank was used for metagenomic investigation in the current research (MG-RAST, Temporary ID: 20170705). Best hit classification at the 10−5E value cutoff was used for calculating classification profiling based on MG-RAST databases. The taxonomy profiling including phyla, orders, families and genus was evaluated in detail for annotations. Hierarchical classification at E value cutoff of 10−5 was used for the gene annotation of functional profile (Yang et al. 2014). Majority of the genes were classified successfully into the hierarchical metabolic groups.
The whole short read sequences were elucidated against the databases of Clusters of Orthologous Groups of proteins (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using BLASTP with a cutoff E value of 10−5 to find out the gene traits involved in the microbial community of the anaerobic system (Kanehisa et al. 2006; Tatusov et al. 2000). The hit numbers of sequences correlating methanogenic pathway-involving enzymes were calculated in a brief evaluation from anaerobic digestion sludge. The ‘KEGG’ computing program was used for the pathway’s evaluation (Mitra et al. 2011; Huson et al. 2011). The proteins, formyl-methanofuran dehydrogenase (FmdA), formyl-methanofuran-tetrahydromethanopterin N-formyltransferase (FTR), methyl-coenzyme M reductase alpha subunit (McrA), acetate kinase (AckA) and phosphate acetyltransferase (PTA), perform key acts in known methanogenesis pathways, but provide inadequate sequences in the KEGG and NOG datasets in this debate. BLASTX results were evaluated manually by inquiring code words based on NCBI for high accuracy, where genes entitling dominant similarity were found from GenBank. The recovered genes involved in methanogenesis were confirmed by coordinating the matched sequences manually, contrary to NCBI database with E value cutoff of 10−10 using BLAST.
Results and discussion
Operational performance
The fermenter was fed with digested sludge in anaerobic conditions with glucose and water including nutrient solutions. The optimized temperature for growth condition was kept about 35 °C, as sludge contains usually mesophilic bacteria. The fermenter in anaerobic condition showed an amazing performance; sample evaluation by analytical methods reported organic acid destruction (average 47%) and destruction of pathogens (above 85%). The total organic acids found in the fermenter’s effluent was positively reduced (below 200 mmol/L) (Fig. 1), illustrating that the sludge is useful and works actively in transformation of volatile fatty acids into methane (biogas). The total average generation of methane was reported to be about 500 mL/day (Fig. 2).
Microbial configuration in fermenter
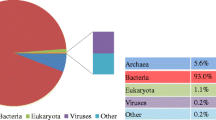
Comprehensively, the Illumina sequencing produced more than 5.0 Gb of short reads from the DNA of filtered sample from the anaerobic fermenter. To explore microbiome configuration, explanation of classification was investigated by best hit classification at the cutoff E value 10−5 with least of 50 bp sequence order on the base of whole dataset MG-RAST source (Yang et al. 2014). After analysis, bacteria were reported as the master domain, contributing 97.64% to the fermenter sludge DNA sequence as shown in Fig. 3. Furthermore, the concentration ratio of archaea reported (1.78%) was found in lower quantity as compared with the previously published studies (Yang et al. 2014), i.e., in their fermenters, the archaea abundance was reported to be less than 4.7%. A fewer short read sequences were found from eukaryotes and viruses contributing 0.52% and 0.02%, respectively, from the sludge sample. The whole domain contribution details are shown in Fig. 3.
To expose in more detail the microbiome structure taken from digesting sludge, various degrees of evaluation were investigated including phylum, class, order, family and genus level as described in Fig. 4. The extreme highest number of bacteria was found to be Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria reporting 37.36%, 28.11%, 20.76%, and 7.09% of the total bacterial reads, respectively, at phylum level. The group of bacteria belonging to Proteobacteria is considered an important bacterium in degradation of all kinds of glucoses due to the presence of alpha-, beta-, gamma-, and delta-proteobacteria group (Ariesyady et al. 2007). A huge number of Firmicutes were detected in phylum level classification, which are syntrophic bacteria, that can convert different organic acids usually reported in various kinds of fermenters (Garcia-Peña et al. 2011). Clostridia and Bacilli in Firmicutes phylum are found in large quantities and make a big class of Firmicutes, considered to play a big role in the hydrolysis of volatile fatty acids in most of the digesting sludge. The dominant groups of classes detected in the phylum level classification were Bacteroidia, Cytophagia and Flavobacteriia. The percent rate was detected to be higher for Bacteroidia as compared to other classes. The family belonging to Bacteroidaceae from the class of Bacteroidetes is an outstanding fermentative member found in sludge which plays an important role in the hydrolysis of organic substances and produces volatile fatty acids, CO2 and H2 at the time of digestion procedures (Traversi et al. 2012).
The Methanobacteriales were found to be one of the highly abundant bacteria in class level classification accounting for 282,623 (1.66%) of the total bacteria present inside the digesting sludge as shown in Fig. 4. These Methanobacteriales are correlated with the Methanosaeta and Methanosarcina abundance (discussed below in detail). There are more than 3000 various taxa reported at genus level classification, exhibiting diversified microbiota in digesting sludge. Among these 3000 taxa, the dominant 30 taxa were highlighted in the sludge sample as shown in Fig. 5. About 10 taxa have been found to be higher than 1–2% in the sample. Bacteroides and Clostridium were found to be the most dominant bacteria at genus level in the sludge detected as the dominant taxon in the previously published reports (Pelletier et al. 2008). The said bacteria are considered a hydrogen-generating syntrophic bacterium that broadly exists in several fermenters.
Many published reports have revealed microbiome structure from various biogas plants recently via metagenomic sequencing technology (Nelson et al. 2011). The present research revealed that Actinobacteria was detected as the dominant bacterium in phylum taxonomy, followed by Bacteroidetes, Firmicutes and Proteobacteria, which are rational with the past conducted study (Yang et al. 2014), where microbiome characterization of a digested sludge from wastewater treatment plant was performed using llumina sequencing. Earlier, published study reported that a broad range of Proteobacteria, Chloroflexi and Spirochaetes was found in biogas plant via 454 pyrosequencing of 16S rRNA gene sequences (Sundberg et al. 2013). In addition, several effective cultures relating to phyla Chloroflexi and Proteobacteria were found in anaerobic fermenter fed with various feedstocks using 16S rRNA gene sequencing (Nelson et al. 2011). This different microbial community structure might be affiliated with different operational conditions of the sludge that has powerful effect on the structure of microbiome (Ziganshin et al. 2013; Regueiro et al. 2012; Vanwonterghem et al. 2014a, b).
Global gene functional profiles
The total short reads were illustrated based on grading of COG and KEGG databases, to expose the characteristic features of the microbiome from the digesting sludge (Fig. 6). Illustration for the distribution of functional category of COG exhibited that 47.75% were associated with metabolism, 18.81% reads were related to housekeeping genes attached in information storage and processing, 17.10% of reads were assigned to cellular processes and signaling while 16.34% reads were poorly characterized of the total reads (Supporting file). The achieved conclusion was compared with the previously published reports, where almost 28% reads were associated with COG categories while greater quantities of reads were involved in metabolism (Li et al. 2013). The most generous metabolic category was carbohydrate metabolism (16.81%), followed by amino acid metabolism (10.37%) and protein metabolism (7.45%) in metabolism category (Table 1). These mentioned metabolic categories are well allied with transformation of sludge into methane generation from digested anaerobic fermentation process (Yang et al. 2014).
The genes engaged in the degradation of amino acids were found in short reads corresponding to the synthesis of valine, leucine and isoleucine, metabolism of threonine, serine and glycine, and metabolism of methionine and cysteine (results not shown), considered three important dominant groups. These mentioned amino acids are frequently associated with Stickland reactions. There are mainly two types of mechanism where amino acids can be degraded: (1) amino acid couple degraded via the Stickland reaction, and (2) individual amino acid can be fermented in a procedure that needs the assistance of hydrogen-consuming biome (Ramsay and Pullammanappallil 2001). The order level classification shows that Clostridiales are one of the most predominant bacteria, acknowledging that the Stickland reaction is only detected with the species of Clostridiales in past published reports (Ramsay and Pullammanappallil 2001). There are several types of proteins which are engaged in the degeneration of amino acids including cystine reductase and cysteine synthase, found with highest short read digits. The discussed conclusion above is affiliated with the best acidogenic performance.
Many short read-related genes involved in metabolism of carbohydrates were reported such as gluconeogenesis, AMP salvage pathway, EMP enolase pathway, glucokinase pathway, glycogen catalysis pathway, glycolysis pathway, ribose isomerase pathway, pyruvate dehydrogenase pathway, pyruvate formate lyase, TCA cycle, malate dehydrogenase, succinate dehydrogenase, fumarate dehydrogenase, glycoside hydrolase and lactate dehydrogenase as shown in Fig. 7. The detection of this annotation assures the existence of a wide variety of species related to carbohydrate metabolisms in the digesting fermenter (Yang et al. 2014).
Carbohydrate metabolism. Blue (on left)—AMP salvage, dark green—carbon fixation pathways, dark blue—Embden–Meyerhof–Parnas pathway, brown—malate metabolism, yellow-green + blue—glucogenesis, purple—glycogen metabolism, green-yellow—glycolysis and oxidative pentose phosphate pathway, grayish purple—pentose phosphate metabolism, light brown—pyrophosphate, light green—pyruvate metabolism, red—TCA cycle, light red—acetate (2 columns), alcohol (1 column), aldehyde (1 column) formate (1 column), lactate (2 columns), light brown—amylolytic degradation (2 columns), cellulose degradation (1 column), and glycosyl hydrolase (colour figure online)
In subsystem level classification in MG-RAST, majority of reads were found for carbohydrates followed by protein metabolism, amino acids, proteins, vitamins, RNA, DNA, cell wall, nucleotide and fatty acid metabolisms as discussed above Table 1. The category of level 2 classification of carbohydrates was further evaluated via metagenomic study. The level 2 subsystem exhibits that carbohydrate and one-carbon metabolisms are found to be the highest as shown in Fig. 8. The main carbohydrate metabolism is used to determine the assimilation of routes for pushing carbon molecules into the cell (Papagianni 2012). This process consists of a series of reactions to transform substrates in the active precursors including acetyl-CoA and pyruvate 6-phosphate, converting to produce the cell biomass (Noor et al. 2010). The second most abundant function was detected to be the one-carbon metabolism of 2.73% of the described carbohydrate. This type of metabolism transforms complicated organic material to elementary carbon alloy, which plays fundamental roles in methanogenesis (Ferry 1999).
Leading microorganisms in digestion process
Overall there are four phases that exist in the digestion operation, i.e., hydrolysis, fermentation, acetogenesis and methanogenesis (Sandoval et al. 2009). Different microbiome engage in every related stage and work together for the maximum generation of methane in the anaerobic fermentation. After applying metagenomic sequencing analysis, the data revealed that the genus Halanaerobiales was reported to be dominant in defusing long-chain molecules to monomers, at the early step. Afterwards, the principal bacteria belonging to Bacteroidaceae family and class Clostridia detected in the current research execute well the process of acidogenesis at the second step, generating various types of organic acids, carbon dioxide and hydrogen. Later, acetic acid was generated from these products using acetogenic bacteria present in the digesting sludge by CO2 reduction with acetyl-CoA synthase pathway. Published literature has reported over 100 acetogenic species (Ragsdale and Pierce 2008). Our predicted results found that Bacteroides, Thermoanaerobacter, Clostridium and Eubacterium are the dominant acetogenic bacteria (Supplementary file), comparable with the previously published literature (Vanwonterghem et al. 2014a, b).
The final step of anaerobic fermentation is methane generation, where methanogenic bacteria play a very important role in methane generation. Methanogens are classified into five orders including Methanomicrobiales, Methanobacteriales, Methanococcales, Methanopyrales and Methanosarcinales. The principal genus engaged in methane generation pathway was further explored on the base of metagenomic data as shown in Fig. 9. The conclusion here exhibited that the top five highest methane-generating bacteria are Methanosaeta, Methanosarcina, Methanospirillum, Methanosarcina and Methanoculleus in the digesting sludge sample. There are only two acetogens for acetic acid production among these including Methanosaeta and Methanosarcina. Methanosaeta is used more specifically against acetic acid, while Methanosarcina has a generalized machinery to generate methane (Liu et al. 2008). Methane-generating hydrogenotrophs reduce carbon dioxide and methane with hydrogen as the elementary electron donor. A desperate member of hydrogenotrophs were detected including Methanospirillum, and Methanoculleus in the sample; however, their wealth was reported lesser in comparison with acetoclastic members. The methylotrophs including Methanococcoides and Methanohalophilus were detected in lowest reads in the sludge sample.
Gene profiles in methanogenesis
The relevant genes for the methanogenic pathway were diagnosed from KEGG annotation as indicated schematically in Fig. 10. As it is already reported, there are principally three major identified pathways for methane generation such as acetoclastic, hydrogenotrophic and methylotrophic pathways (Liu et al. 2008). Carbon dioxide is reduced to methane via several steps including formyl, methylene, and methyl levels in hydrogenotrophs. From methyl level, the methyl group is converted to coenzyme M, making methyl-CoM and then methane, by methyl-coenzyme M reductase (Mcr) at last stage (blue in Fig. 10). In the acetoclastic pathway, Methanosarcina’s exploit acetate kinase (AK)–phosphotransacetylase (PTA) system to convert acetate to acetyl-CoA, while Methanosaeta utilizes adenosine monophosphate (AMP)-forming acetyl-CoA synthetase. The acetyl-CoA is transformed into methyl group followed by methane via enzymes of Cdh, Mtr and Mcr (red in Fig. 10). The methyl groups from methylated compounds are moved to methanogenic pathway and generate methane by Mcr reductase in methylotrophic pathway (green in Fig. 10). Hence, the genes encoding enzyme-related short reads for acetoclastic pathway are on top as compared with the rest of the pathways. The maximum reads found for AckA and PTA are 439 and 166 hits, respectively, while for FmdA and FTR, 69 and 52 hits, respectively.
The genes involved in methylotrophs were found at the bottom as compared with the hydrogenotrophic pathway. The results achieved in the current research study revealed that acetoclastic pathway is probably the prominent pathway of methane generation in the digesting sludge in anaerobic digestion processes (Yu et al. 2005). It should be distinguished that the genes read were evaluated on the level of DNA, instead of meta-transcriptomics or meta-proteomics (RNA or protein level), which is needed in the future study to investigate further functional traits. Further investigations will be required for the understanding of differential changes and the mechanisms of these microbial communities at variable stable and unstable operational rates. This might also provide an insight into the efficiency of the energetic classification for these dominant microorganisms, especially the Methanosarcina and Methanosaeta in the AD reactors.
Conclusions
The complete microbiome structure was anatomized, and the principal pathways involved in methane generation were evaluated by metagenomic sequencing technology in this study. Taxonomic evaluation showed that Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria are the four major bacterial groups in the sludge sample. For methane generation in the sludge, there are different microbiome detected at various stages including hydrolysis, fermentation, acetogenesis and methanogenesis. The families Bacteroidaceae and Clostridia are major consortia in the fermentation stage. Thermoanaerobacter, Clostridium and Eubacterium are the three major groups of acetogens reported, which play a key role in acetate production. The genes encoding enzyme-related short reads for the acetoclastic pathway are on top as compared with the rest of the pathways. The maximum reads found for AckA and PTA are 439 and 166 hits, respectively, while for FmdA and FTR, 69 and 52 hits, respectively. It should be noted that the gene reads were evaluated at the level of DNA, instead of meta-transcriptomics or meta-proteomics (RNA or protein level), which is needed in the future study to investigate further functional traits.
References
Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C (2011) Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol 12:18
Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH (2003) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538
Albertsen M, Hansen LBS, Saunders AM, Nielsen PH, Nielsen KL (2006) A metagenome of a full-scale microbial community carrying out enhanced biological phosphorus removal. ISME J 6:1094–1106
Amani T, Nosrati M, Sreekrishnan TR (2010) Anaerobic digestion from the viewpoint of microbiological, chemical, and operational aspects—a review. Environ Rev 18:255–278
APHA (1998) Standard methods for the examination of water and wastewater, 20th edn. APHA, Washington
Appels L, Baeyens J, Degreve J, Dewil R (2008) Principles and potential of the anaerobic digestion of waste-activated sludge. Prog Energ Combust 34:755–781
Ariesyady HD, Ito T, Okabe S (2007) Functional bacterial and archaeal community structures of major trophic groups in a full-scale anaerobic sludge digester. Water Res 41:1554–1568
Bragg L, Tyson GW (2014) Metagenomics using next-generation sequencing. In: Paulsen IT, Holmes AJ (eds) Environmental microbiology: methods and protocols. Methods in molecular biology, vol 1096, 2nd edn. Humana Press, New York City, pp 183–201
Canales A, Pareilleux A, Rols JL, Goma G, Huyard A (1994) Decreased sludge production strategy for domestic wastewater treatment. Water Sci Technol 30:97–106
Ferry JG (1999) Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol Rev 23:13–38
Garcia-Peña EI, Parameswaran P, Kang DW, Canul-Chan M, Krajmalnik-Brown R (2011) Anaerobic digestion and co-digestion processes of vegetable and fruit residues: process and microbial ecology. Bioresour Technol 102:9447–9455
Glenn TC (2011) Field guide to next-generation DNA sequencers. Mol Ecol Resour 11:759–769
Guo J, Peng Y, Wang S, Ma B, Ge S, Wang Z (2013) Pathways and organisms involved in ammonia oxidation and nitrous oxide emission. Crit Rev Environ Sci Technol 43:2213–2296
Huson D, Mitra S, Ruscheweyh H, Weber N, Schuster S (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21:1552–1560
Ju F, Guo F, Ye L, Xia Y, Zhang T (2014) Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ Microbiol Rep 6:80–89
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:354–357
Li A, Chu Y, Wang X, Ren L, Yu J, Liu X (2013) A pyrosequencing-based metagenomic study of methane-producing microbial community in solid-state biogas reactor. Biotechnol Biofuels 6:3
Liu Y, Whitman WB, Wiegel J, Maier RJ, Adams MWW (eds) (2008) Incredible anaerobes: from physiology to genomics to fuels, vol 1125. HighWire Press, New York, pp 171–189
Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ (2011) Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368-U120
Mardis ER (2008) The impact of next-generation sequencing technology on genetics. Trends Genet 24:133–141
Mason OU, Scott NM, Gonzalez A, Robbins-Pianka A, Baelum J, Kimbrel J (2014) Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME 8:1464–1475
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386
Mitra S, Rupek P, Richter DC, Urich T, Gilbert JA, Meyer F (2011) Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinformatics 12:21
Nelson MC, Morrison M, Yu Z (2011) A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour Technol 102:3730–3739
Noor E, Eden E, Milo R, Alon U (2010) Central carbon metabolism as a minimal biochemical walk between precursors for biomass and energy. Mol Cell 39:809–820
Papagianni M (2012) Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb Cell Fact 11:50
Pelletier E, Kreimeyer A, Bocs S, Rouy Z, Gyapay G, Chouari R (2008) “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol 190:2572–2579
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59-U70
Ragsdale SW, Pierce E (2008) Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta Proteins Proteom 1784:1873–1898
Ramsay IR, Pullammanappallil PC (2001) Protein degradation during anaerobic wastewater treatment: derivation of stoichiometry. Biodegradation 12:247–257
Regueiro L, Veiga P, Figueroa M, Alonso-Gutierrez J, Stams AJM, Lema JM (2012) Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol Res 167:581–589
Sandoval LCJ, Vergara MM, De Carreno AM, Castillo MEF (2009) Microbiological characterization and specific methanogenic activity of anaerobe sludges used in urban solid waste treatment. Waste Manag 29:704–711
Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH (2013) 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol 85:612–626
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36
Traversi D, Villa S, Lorenzi E, Degan R, Gilli G (2012) Application of a real-time qPCR method to measure the methanogen concentration during anaerobic digestion as an indicator of biogas production capacity. J Environ Manag 111:173–177
van Loosdrecht MCM, Brdjanovic D (2014) Anticipating the next century of wastewater treatment. Science 344:1452–1453
Vanwonterghem I, Jensen PD, Ho DP, Batstone DJ, Tyson GW (2014a) Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr Opin Biotechnol 27:55–64
Vanwonterghem I, Jensen PD, Dennis PG, Hugenholtz P, Rabaey K, Tyson GW (2014b) Deterministic processes guide long-term synchronised population dynamics in replicate anaerobic digesters. ISME 8:2015–2028
Wong MT, Zhang D, Li J, Hui RKH, Tun HM, Brar MS (2013) Towards a metagenomic understanding on enhanced biomethane production from waste activated sludge after pH 10 pretreatment. Biotechnol Biofuels 6:38
Yang Y, Yu K, Xia Y, Lau FTK, Tang DTW, Fung WC (2014) Metagenomic analysis of sludge from full-scale anaerobic digesters operated in municipal wastewater treatment plants. Appl Microbiol Biot 98:5709–5718
Ye L, Zhang T, Wang TT, Fang ZW (2012) Microbial structures, functions, and metabolic pathways in wastewater treatment bioreactors revealed using high-throughput sequencing. Environ Sci Technol 46:13244–13252
Yu Y, Lee C, Hwang S (2005) Analysis of community structures in anaerobic processes using a quantitative real-time PCR method. Water Sci Technol 52:85–91
Ziganshin AM, Liebetrau J, Proeter J, Kleinsteuber S (2013) Microbial community structure and dynamics during anaerobic digestion of various agricultural waste materials. Appl Microbiol Biotechnol 97:5161–5174
Acknowledgements
This work was supported by Major Science and Technology Program for Water Pollution Control and Treatment of China (Grant no. 2017ZX07102-004), and National Natural Science Foundation of China (Grant no. 21206084). We are grateful to School of Medicine, Tsinghua University for providing the tools of metagenomic Matlab.
Author information
Authors and Affiliations
Contributions
All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that there is no conflict of interest.
Additional information
Communicated by Kristina Beblo-Vranesevic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ali, N., Gong, H., Liu, X. et al. Evaluation of bacterial association in methane generation pathways of an anaerobic digesting sludge via metagenomic sequencing. Arch Microbiol 202, 31–41 (2020). https://doi.org/10.1007/s00203-019-01716-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01716-x