Abstract
Acinetobacter baumannii possesses a tremendous potential to thrive under hostile conditions. To learn more about its survival strategy and capacity to persist in the environment, we studied the effect of temperature, nutrient deprivation and dryness on the long-term survival of two A. baumannii strains (ATCC 19606T and a clinical isolate). Our results revealed that both strains show a great persistence under stress that appears to involve a bust-and-boom strategy. Bacterial survival was differentially affected by temperature and physical environment: Desiccation favored cell resistance to stress at 20 and 37 °C, while survival in aqueous environments was temperature dependent and led to changes in several cellular characteristics. In addition, we tested the ability of the A. baumannii ATCC 19606T strain to form biofilms by monitoring the expression of adhesion-/biofilm-related genes (ompA, bfmR and csuAB). The observed downregulation of these genes suggests that the potential difficulties to adhere to solid surfaces and form biofilms likely limit the capacity of starved cells to spread and colonize abiotic surfaces.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microorganisms in the environment are often exposed to adverse physicochemical conditions. To increase their capacity of persistence and survival in hostile environments, bacteria can trigger a number of adaptation mechanisms. Rittershaus et al. (2013) highlighted three major strategies enabling to cope with the growth-limiting stress: bust-and-boom, cellular quiescence and true dormancy (sporulation). This last strategy can be neglected for non-spore-forming bacteria.
In the bust-and-boom model, most cells die upon starvation, and the few survivors subsist at the expense of the dead cells. These remaining cells have been defined as persisters (Zhang 2014). In the second model, cellular quiescence/dormancy or acquisition of viable but non-culturable (VBNC) state represents a common strategy enabling many non-differentiating bacteria to survive under various adverse conditions (Roszak and Colwell 1987; Barcina and Arana 2009; Oliver 2010). The entry into the VBNC state suggests that cells are unable to grow on the media that are normally used to culture them, although they retain metabolic activity and can possibly act as pathogens (Karunasagar and Karunasagar 2005), thus representing a hidden risk to public health. Moreover, since the VBNC response can occur simultaneously with the bust-and-boom strategy, Arana et al. (2007) suggested that VBNC cells could serve as a source of nutrients to sustain the survival of the persisting culturable cells until environmental conditions improve.
Over the last decade, Acinetobacter baumannii has emerged as an important nosocomial pathogen that has become endemic in some hospitals, causing serious opportunistic infections (Towner 2009; Lambiase et al. 2012; Roca et al. 2012). Hospital outbreaks of A. baumannii have been frequently reported worldwide from many medical, surgical and neonatal intensive care and burn units (Diancourt et al. 2010; Roca et al. 2012). Moreover, the emergence and rapid spread of multidrug-resistant isolates becomes a great concern because very few therapeutic options remain effective against them (Kempf et al. 2012; Eveillard et al. 2013).
Due to its clinical significance, during the last years several studies have contributed to understanding the pathogenesis of this organism. Although it is evident that this opportunistic pathogen has developed several mechanisms that control its persistence and spread, very little is known about the survival strategies that could explain the high persistence of this pathogen in adverse environments. Gayoso et al. (2014) have suggested that the bust-and-boom model could explain the recurrent outbreaks of A. baumannii found in intensive care units; however, the viability of non-culturable cells was not examined to assess the possible involvement of the VBNC state.
On the other hand, the environmental survival of A. baumannii has been suggested to involve its ability to colonize biotic and abiotic surfaces (Espinal et al. 2012). Among the biofilm-related virulence determinants, the pilus usher–chaperone assembly system (encoded by the csuAB–A–B–C–D–E gene cluster) and the outer membrane protein OmpA appear to play the major role. The expression of the csuAB–A–B–C–D–E gene cluster is controlled by a two-component regulatory system comprising the sensor kinase BfmS and the response regulator BfmR encoded by the bfmRS operon (Tomaras et al. 2008; Gaddy and Actis 2009). Inactivation of bfmR results in a loss of expression of the csuAB operon and the subsequent inhibition of both pili production and biofilm formation on abiotic surfaces (Tomaras et al. 2008). OmpA is essential for attachment to biotic and abiotic surfaces and biofilm formation (Choi et al. 2008; Lee et al. 2008).
Study and characterization of the changes occurring during the long-term starvation process can contribute to a better understanding of A. baumannii survival in the environment and its persistence strategies, which could provide key information for its control. The aim of this study was to examine the effect of temperature and nutrient-deprived environments on A. baumannii ATCC 19606T (model strain) and on a clinical isolate to assess how environmental conditions affect A. baumannii survival and persistence. Moreover, we also analyzed how these stressful conditions affect the expression of genes (ompA, bfmR and csuAB) implicated in adhesion and biofilm formation and known to play essential roles in colonization of abiotic environments.
Materials and methods
Acinetobacter baumannii strain and inocula preparation
Two Acinetobacter baumannii strains were used in this study: American Type Culture Collection (ATCC) strain 19606T and a clinical isolate of A. baumannii, strain 06-2790, obtained from a skin ulcer of a patient at the Hospital Universitario Marqués de Valdecilla (Prof. J Ramos-Vivas, Santander, Spain). Both strains were separately grown overnight at 37 °C in Mueller–Hinton (MH) broth. Cells were transferred to fresh MH broth and grown at 37 °C with shaking (120 rpm) to stationary phase. Cells were collected by centrifugation (4500 g for 15 min) and washed three times with sterile saline solution (0.9 % w/v NaCl). Finally, the pellets were suspended in sterile saline solution.
Survival assays
Acinetobacter baumannii cells from stationary phase were incubated under nutrient deprivation at 20 or 37 °C in liquid or solid environments. Starvation was implemented by incubating cells in sterile saline solution or on cellulose acetate filters (Whatman, GE Healthcare Life Sciences) sterilized by a 20-min exposure to UV-C (approximately 253.7 nm; 70 mW/cm2).
For survival assays in the aqueous environment, experiments were carried out in Erlenmeyer flasks containing 300 ml of sterile saline solution inoculated to reach a density of 108 cells/ml. To avoid organic residues, the glass flasks were first cleaned with acid, rinsed with deionized water and kept at 250 °C for 24 h.
For survival assays on solid surfaces, cellulose acetate filters were inoculated with A. baumannii by filtering cellular suspensions (described above) to a density of approximately 108 cells/cm2. The filters were incubated in sterile Petri dishes at 20 or 37 °C. Ambient humidity inside of Petri dishes was measured with a Fisher Scientific™ Traceable™ Digital Hygrometer/Thermometer (Fisher Scientific) and maintained at a relatively low level (21–27 %) at 20 and 37 °C. For each strain, the samples for subsequent analyses consisted of three randomly chosen filters that were individually placed in 10 ml of sterile saline solution and vigorously shaken for 2 min.
For the survival assays from both liquid and solid environments, subsamples were periodically collected to enumerate total, viable and culturable bacteria and analyzed directly their ability to adhere. Moreover, for A. baumannii ATCC 19606T, the expression of ompA, bfmR and csuAB genes by reverse transcription quantitative PCR (RT-qPCR) was analyzed.
Bacterial counts
The total bacterial count (TBC) was determined by means of the standard acridine orange staining as described by (Hobbie et al. 1977). Viable bacteria were estimated as bacteria with intact cytoplasmic membranes (MEMB+). These MEMB+ bacteria were counted with the Live/Dead BacLight™ kit (Invitrogen Life Technologies) (Joux et al. 1997). Culturability, expressed as colony-forming units (CFUs), was determined by spreading aliquots of A. baumannii cells on Mueller–Hinton agar followed by incubation at 37 °C for 24 h.
Scanning electron microscopy (SEM)
A. baumannii ATCC 19606T cells, periodically collected from survival assays in aqueous environments, were fixed with 2 % glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4). The suspensions of the fixed cells were filtered through 0.22-µm-pore-size membrane filters (GTTP02500 filters, Merck Millipore). The filters and the attached A. baumannii cells were further dehydrated by applying a series of increasing ethanol concentrations (30, 50, 70, 90 and 100 %). The filters were then overlaid with 1 ml of hexamethyldisilazane, incubated for 5 min and air-dried. Finally, the samples were coated with gold, and imaging was carried out by analyzing samples in a Hitachi S4800 scanning electron microscope.
Quantification of cell adherence to solid surfaces
To assess cell adherence, we employed the microtiter dish assay (O’Toole and Kolter 1998) with some modifications. Aliquots of cell suspensions (1 ml each), periodically collected from survival experiments, were added into the wells of sterile 24-well polystyrene plates (Corning Inc.) and incubated for 48 h at 37 °C without shaking. Sterile saline solution was used as a negative control. After incubation, plates were washed three times with sterile water and stained with 200 µl of 0.5 % (w/v) crystal violet solution. Then, plates were newly washed with sterile water (three times), and the dye bound to the attached cells was solubilized in 0.5 ml of 95 % ethanol. The optical density (OD) of each well was measured at 595 nm using an automated Synergy™ HT Multi-Detection Microplate Reader (BioTek Instruments, Inc., Vermont). During the survival experiments, cells were classified according to Stepanović et al. (2000). The cutoff OD (ODc) was defined as a value equal to three standard deviations above the mean OD of the negative control, and the following categories were established: OD ≤ ODC, non-adherent; ODc < OD ≤ (2 × ODc), weakly adherent; (2 × ODc) < OD ≤ (4 × ODc), moderately adherent; and (4 × ODc) < OD, strongly adherent cells.
Along the survival experiments, the variation in the ability to adhere was expressed with an arbitrary value, OD decline values which were calculated by subtracting the respective initial OD from those obtained in each survival period.
Statistical data analysis
Statistical tests were carried out with the Stat View program (Abacus Concept, Inc.). All of the results presented are means of at least three experiments, and the coefficients of variation between replicate experiments were less than 12 %. The differences between the means were detected by a one-way analysis of variances. Probabilities less than or equal to 0.05 were considered significant. Logarithmic transformation for bacterial counts was used.
RNA preparation and reverse transcription quantitative PCR (RT-qPCR) analysis
Samples of cells collected from the survival experiments of A. baumannii ATCC 19606T (aqueous and dry conditions) were mixed with stop solution (5 % phenol in ethanol) at a ratio of 8:1 and incubated on ice for 15–20 min, and the cells were collected by centrifugation (15 min, 4 °C, 4400 g). The pelleted cells were used to isolate total RNA using the Trizol® Max™ Bacterial RNA Isolation Kit (Ambion). Each RNA sample was further purified using the PureLink® RNA Mini Kit (Ambion) and treated with DNase I (Invitrogen). RNA samples were subjected to RT-qPCR at the General Genomic Service (SGIker) of the University of Basque Country (Spain). Namely, after verifying RNA quality and integrity by Lab-chip technology on an Agilent 2100 Bioanalyzer with Agilent RNA 6000 Nano Chips, RNA was used for cDNA synthesis using AffinityScript Multiple Temperature cDNA Synthesis Kit (Agilent Technologies).
The expression of three different genes related to adhesion or biofilm formation (ompA, bfmR and csuAB) was analyzed by RT-qPCR. Specific primer pairs were designed using PrimerQuest tool available at IDT Integrated DNA Technologies, using universal primer design parameters for qPCR. Primer secondary structure analysis (hairpin and primer dimer formation analysis) was verified with OligoAnalyzer tool, from IDT Integrated DNA Technologies, and primer specificity was checked using BLAST (NCBI). Specific primer pairs were designed for four candidate reference genes potentially suitable for normalization purposes: gltA, gyrB, cpn60 and gdhB (Bartual et al. 2005; Hamouda et al. 2010). The target and candidate reference gene specific primers used for RT-qPCR are listed in Table 1.
The RT-qPCR was carried out using Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies), and 10 ng of cDNA and 500 nM of primers were added per reaction. RT-qPCR experiments were performed in 384-well plates in a 7900HT Fast Real-Time PCR System (Applied Biosystems) to determine the relative changes in the level of three transcripts. The PCR amplification conditions were 3 min at 95 °C followed by 40 cycles of 95 °C for 5 s and 60 °C for 20 s. The specificity of the primer pairs was verified by melting curve analysis following the last amplification cycle. No-template controls and minus reverse transcriptase controls (samples in which no reverse transcriptase was added) were also included. Having found no reference candidate genes with low variability, the amount of cDNA, quantified fluorometrically by Qubit 2.0 (Invitrogen), was used for data normalization. All reactions were carried out in triplicate for three biological replicates.
RT-qPCR data analysis
The analysis of RT-qPCR results was carried out using the SDS 2.4 software (Applied Biosystems). The qPCR efficiency calculation and correction, and data normalization with respect to the amount of cDNA were done using GenEx version 5.4 software (MultiD). Then, the relative changes in mRNA expression levels were determined with respect to the initial amounts of cDNA obtained by RT, and the significance of differences was assessed using Student’s t test (p < 0.05).
Results
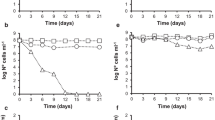
The effects of temperature (20 or 37 °C) and desiccation on culturability, viability and integrity of A. baumannii under nutrient deprivation are summarized in Fig. 1. For both strains, the total bacterial count (TBC) did not change during permanence in aqueous environments (Fig. 1a, b, e, f) or on solid surfaces (Fig. 1c, d, g, h), suggesting that A. baumannii cells preserved their integrity throughout the course of the experiment (at least 30 days). Moreover, culturability and viability also remained unchanged except for the populations incubated at 37 °C in saline solution (Fig. 1b, f). In this last case, the numbers of viable and culturable A. baumannii ATCC 19606T cells decreased by 100-fold between 5 and 12 days of incubation (Fig. 1b) and A. baumannii clinical isolate 06-2790 by 15-fold during the same period (Fig. 1f). The concurrent loss of viability and culturability indicates that a fraction of population presented cellular membrane injured and the VBNC state was not induced under these conditions. For both strains, at the end of the experimental period, there were approximately 105 viable and culturable cells/ml.
Evolution of Acinetobacter baumannii ATCC 19606T (a–d) and clinical isolate 06-2790 (e–h) populations maintained in aqueous environment at 20 °C (a, e) and 37 °C (b, f) and on dry surfaces at 20 °C (c, g) and 37 °C (d, h). Total (filled circle), viable (gray square) and culturable (inverted triangle) cells
Scanning electron microscopy (SEM) analysis of A. baumannii ATCC 19606T populations during survival experiments (Fig. 2) revealed some cells with altered cell morphology (distorted cells). These cells were only observed in populations maintained in saline solution at 37 °C for ten or more days (Fig. 2c).
Scanning electron microscopy (SEM) analysis of the morphology and integrity of Acinetobacter baumannii ATCC 19606T cells maintained in saline solution (aqueous environment). Panel a shows cells sampled at the beginning of the experiments and panels b and c after more than 10 days of permanence at 20 °C and 37 °C, respectively
To learn more about the putative survival strategies employed by A. baumannii during its persistence in saline solutions, we also estimated the ability of populations maintained in aqueous and dry environments to adhere to solid surfaces (Fig. 3a). We found that the A. baumannii ATCC 19606T strain behaves as a moderately adherent strain (OD = 0.224 ± 0.048) and its ability to adhere to solid surfaces progressively decreased along the survival period in both environments, being also temperature dependent (Fig. 3a). In the experiments in saline solution, this ability diminished quickly resulting in the complete loss of adhesiveness after 5 days (data not shown). Under dry conditions, the adhesiveness was also reduced but the populations retained a weak capacity to adhere at least for 15 days (data not shown). A. baumannii clinical isolate 06-2790, a strongly adherent strain (OD = 0.529 ± 0.065), showed a similar behavior (Fig. 3b).
Time-dependent changes in the ability of Acinetobacter baumannii ATCC 19606T (a) and clinical isolate 06-2790 (b) populations maintained in aqueous environment (filled rectangle) and under dry conditions (open rectangle) to adhere to solid surfaces at 20 °C and 37 °C. Relative values were calculated with respect to the initial time (0 days)
During the survival experiments, concentration of total RNA per cell varied with conditions (Table 2). In general, despite slight fluctuations, the amount of total RNA recovered per cell in A. baumannii ATCC 19606T populations maintained on the dry surfaces was constant over time. In contrast, variations in the amount of RNA were more evident in planktonic cells, especially at 37 °C, where the quantity of RNA recovered at the end of the experiment (after 8 days) was only 6 % when compared to the amount isolated from the cells present at the beginning of the experiments (Table 2).
The effects of temperature and desiccation under nutrient deprivation on the expression of the ompA, bfmR and csuAB genes are shown in Fig. 4. Regardless of the experimental conditions, for A. baumannii model strain, the expression of ompA and bfmR genes decreased progressively (p ≤ 0.05) from the beginning of the experiments. After 8 days of experimentation, lower expression was detected in cells exposed to dry environments at 37 °C. Both ompA and bfmR genes were downregulated by over threefold with respect to the initial values. csuAB gene showed greater variability in expression depending on environment. In contrast to slight variations detected for populations maintained in saline solution at 20 and 37 °C, the csuAB gene was readily downregulated when the populations were maintained on dry surfaces.
Time-dependent changes in the expression level of the ompA, bfmR and csuAB genes determined for Acinetobacter baumannii ATCC 19606T populations maintained in aqueous environment (filled rectangle) and under dry conditions (open rectangle) at 20 and 37 °C. Relative values were calculated with respect to the initial time (0 days)
Discussion
In its habitats ranging from natural ecosystems to medical devices and clinical surfaces, A. baumannii is often exposed to adverse conditions (nutrient deprivation, desiccation or non-optimal temperatures). To better characterize its ability to persist under stress (potentially facilitating infection outbreaks), we studied A. baumannii adaptation to starvation.
We found that its survival in liquid environments was clearly temperature dependent. Populations of A. baumannii ATCC 19606T and clinical isolate 06-2790 both incubated at 37 °C displayed a loss of viability and culturability (Fig. 1b, f), resulting in a large fraction of non-culturable non-viable cells [i.e., cells showing altered cytoplasmic membrane permeability (Joux et al. 1997)] and the appearance of distorted cells (Fig. 2c). The loss of viability and physiological functions at 37 °C seems to correlate with a profound decrease in the amount of total RNA in the stressed cells (Table 2), thus indicating that this temperature can be detrimental under certain starvation conditions. As a result, only a small fraction (ca. 0.2 %) of the whole population remained culturable after 30 days. In contrast, no loss of culturability was observed when A. baumannii ATCC 19606T was exposed to lower temperature (20 °C) at least for 30 days. The moderate decrease in the RNA content observed after 2 and 8 days could be associated with the overall decrease in the metabolism, thereby helping to save energy in favor of cell survival (Lever et al. 2015). Although there is no published work addressing the survival of A. baumannii in aqueous environments, several studies that were previously carried out on other mesophilic bacteria, including Escherichia coli and Pseudomonas fluorescens (Arana et al. 2010), support our observation that lower temperatures could be beneficial for prolong persistence of mesophilic bacteria under starvation.
Unlike survival in saline solutions, experiments with A. baumannii cells retained on dry surfaces revealed that the resistance to stress and survival under such conditions was even better. In other words, the viability and culturability of A. baumannii population did not experience significant changes for at least one month, and total RNA content remained constant for 8 days regardless of the temperature. Although several works (Wendt et al. 1997; Jawad et al. 1998; Espinal et al. 2012; Gayoso et al. 2014) have highlighted that A. baumannii survives long time on dry surfaces, these studies generally associate the resistance to desiccation to the longtime presence of only some culturable surviving bacteria. In this sense, our results differ from those obtained by Gayoso et al. (2014) and Wendt et al. (1997), who described the negative effect of dry conditions. With reference to these reports, it is even more remarkable that, in this study, the bacteria on dry surfaces at 37 °C could survive longer (and with higher percentage of survivors) than their planktonic counterparts. Webster et al. (2000) observed that a local strain of Acinetobacter species could persist on common clinical surfaces for relatively long periods, and this could partly explain that during the outbreaks the same A baumannii strain(s) continues to be recovered from both patients and environment. This result is consistent with our data and those obtained in other studies like the one conducted by Houang et al. (1998). The latter described the persistence (without loss of culturability) of a clinical isolate of A. baumannii maintained over 30 days at 22–24 °C under desiccation.
Different strategies have been envisaged to explain the mechanisms that allow bacteria to face stress conditions. The acquisition of the VBNC phenotype by Acinetobacter has been demonstrated only for A. calcoaceticus in aquatic systems (Lemke and Leff 2006). Gayoso et al. (2014) indicated that A. baumannii strain AbH12O-A2 maintained under prolonged desiccation conditions becomes dormant, a definition that corresponds to the VBNC state (Rittershaus et al. 2013), and that a few surviving cells embedded in biofilm followed a bust-and-boom strategy. In our work, populations of A. baumannii starved at 37 °C in a liquid environment (the only condition we found to decrease culturability) did not enter the VBNC state. This indicates that transition to the dormant state was not the major strategy for cells to maintain viability under these conditions. In contrast, the bust-and-boom strategy is compatible with our results: The persistence and subsequent colonization of other environments would be facilitated by mechanisms that allow the survival of the most persistent cells at the expense of dying cells.
Several authors have demonstrated that biofilm formation is closely linked to stress responses (Hall-Stoodley et al. 2004; Landini 2009) as it can increase the capacity to withstand environmental challenges, including desiccation (Costerton et al. 1999; Espinal et al. 2012; Marks et al. 2014). We observed that the survivability of A. baumannii strains used in this study was clearly dependent on the cell distribution in the environment (free cells in saline solution vs retained on filters). While cells on solid surfaces showed a high capacity for survival under all conditions, the response to aqueous environments was variable and dependent on temperature. We also found that planktonic bacteria under starvation possessed a lower adhesiveness in contrast to those retained on filters, thus indicating that non-planktonic bacteria could be more stable per se. Moreover, the expression of biofilm-related genes declined under all conditions, even though A. baumannii maintained viability for a long time. This suggests that starved populations could find difficulties to adhere to solid surfaces and develop biofilms and, therefore, were unable to spread and colonize abiotic surfaces. As suggested by Chang et al. (2014), the downregulation of genes playing less important roles in cell survival (e.g., ompA, bfmR and csuAB) should allow to save the energy required for the expression of those genes involved in adaption to environmental stress. Moreover, the low expression level of ompA, bfmR and csuAB does not seem to completely prevent cell adhesion and subsequent biofilm formation and therefore could explain why bacteria maintained under water-free conditions still preserved their adhesiveness, especially at 37 °C.
In summary, A. baumannii shows a great persistence under stress, although starvation and physical environment differentially affect its survival. The dissemination of this pathogen seems to be based on the ability of the bulk of the bacterial population to tolerate and overcome the negative effects of stress factors. We found that the adaptation process did not initiate the entry of A. baumannii cells into the VBNC state. Moreover, the populations maintained on dry surfaces were especially resistant to stress and were able to preserve not only cell culturability but also the cellular appearance and nearly unaltered RNA content. In contrast, the survival of bacteria in aqueous environments is temperature dependent and involves changes in viability, morphology and RNA content, as well as a loss of the ability to adhere to solid surfaces. Thus, while being a common stress factor, dryness seems to have a protective role for A. baumannii cells.
References
Arana I, Orruño M, Pérez-Pascual D, Seco C, Muela A, Barcina I (2007) Inability of Escherichia coli to resuscitate from the viable but nonculturable state. FEMS Microbiol Ecol 62:1–11. doi:10.1111/j.1574-6941.2007.00362.x
Arana I, Muela A, Orruño M, Seco C, Garaizabal I, Barcina I (2010) Effect of temperature and starvation upon survival strategies of Pseudomonas fluorescens CHA0: comparison with Escherichia coli. FEMS Microbiol Ecol 74:500–509. doi:10.1111/j.1574-6941.2010.00979.x
Barcina I, Arana I (2009) The viable but nonculturable phenotype: a crossroads in the life-cycle of non-differentiating bacteria? Rev Environ Sci Bio/Technol 3:245–255. doi:10.1007/s11157-009-9159-x
Bartual SG, Seifert H, Hippler C, Domínguez-Luzon MA, Wisplinghoff H, Rodríguez-Valera F (2005) Development of a Multilocus Sequence Typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390. doi:10.1128/JCM.43.9.4382-4390.2005
Chang KC, Kuo HY, Tang CY, Chang CW, Lu CW, Liu CC, Lin HR, Chen KH, Liou ML (2014) Transcriptome profiling in imipenem-selected Acinetobacter baumannii. BMC Genom 15:815. doi:10.1186/1471-2164-15-815
Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, Chae JP, Yoo SM, Lee JC (2008) Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol 10:309–319. doi:10.1111/j.1462-5822.2007.01041.x
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi:10.1126/science.284.5418.1318
Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S (2010) The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi:10.1371/journal.pone.0010034
Espinal P, Martí S, Vila J (2012) Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect 80:56–60. doi:10.1016/j.jhin.2011.08.013
Eveillard M, Kempf M, Belmonte O, Pailhoriés H, Joly-Guillou ML (2013) Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int J Infect Dis 17:e802–e805. doi:10.1016/j.ijid.2013.03.021
Gaddy JA, Actis LA (2009) Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol 4:273–278. doi:10.2217/fmb.09.5
Gayoso CM, Mateos J, Méndez JA, Fernández-Puente P, Rumbo C, Tomás M, Martínez de Ilarduya O, Bou G (2014) Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J Proteome Res 13:460–476. doi:10.1021/pr400603f
Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nature Rev Microbiol 2:95–108. doi:10.1038/nrmicro821
Hamouda A, Evans BA, Towner KJ, Amyes SGB (2010) Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla OXA-51-like genes. J Clin Microbiol 48:2476–2483. doi:10.1128/JCM.02431-09
Hobbie JE, Daley RJ, Jasper S (1977) Use of nucleopore filters for counting bacteria by epifluorescence microscopy. Appl Environ Microbiol 33:1225–1228
Houang ETS, Sormunen RT, Lai L, Chan CY, Leong ASY (1998) Effect of desiccation on the ultrastructural appearances of Acinetobacter baumannii and Acinetobacter lwoffii. J Clin Pathol 51:786–788. doi:10.1136/jcp.51.10.786
Jawad A, Snelling AM, Heritage J, Hawkey PM (1998) Exceptional desiccation tolerance of Acinetobacter radioresistens. J Hosp Infect 39:235–240. doi:10.1016/S0195-6701(98)90263-8
Joux F, Lebaron P, Troussellier M (1997) Succession of cellular states in Salmonella typhimurium population during starvation in artificial seawater microcosms. FEMS Microbiol Ecol 22:65–76. doi:10.1111/j.1574-6941.1997.tb00357.x
Karunasagar I, Karunasagar I (2005) Retention of pathogenicity in viable nonculturable pathogens. In: Belkin S, Colwell RR (eds) Oceans and health: pathogens in the marine environment. Springer, New York, pp 361–371
Kempf M, Rolain JM, Azza S, Diene S, Joly-Guillou ML, Dubourg JG, Colson P, Papazian L, Richet H, Fournier PE, Ribeiro A, Raoult D (2012) Investigation of Acinetobacter baumannii resistance to carbapenems in Marseille hospitals, south of France: a transition from an epidemic to an endemic situation. APMIS 121:64–71. doi:10.1111/j.1600-0463.2012.02935.x
Lambiase A, Piazza O, Rossano F, Del Pezzo M, Tufano R, Catania MR (2012) Persistence of carbapenem-resistant Acinetobacter baumannii strains in an Italian intensive care unit during a forty-six month study period. New Microbiol 35:199–206
Landini P (2009) Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Res Microbiol 60:259–266. doi:10.1016/j.resmic.2009.03.001
Lee HW, Koh YM, Kim J, Lee JC, Lee YC, Seol SY, Cho DT, Kim J (2008) Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin Microbiol Infect 14:49–54. doi:10.1111/j.1469-0691.2007.01842.x
Lemke M, Leff L (2006) Culturability of stream bacteria: assemblage and population level responses. Microb Ecol 51:365–374. doi:10.1007/s00248-006-9026-z
Lever MA, Rogers KL, Lloyd KG, Overmann J, Schink B, Thauer RK, Hoehler TM, Jørgensen BB (2015) Life under extreme energy limitation: a synthesis of laboratory-and field-based investigations. FEMS Microbiol Rev. doi:10.1093/femsre/fuv020
Marks LR, Reddinger RM, Hakansson AP (2014) Biofilm formation enhances fomite survival of Streptococcus pneumoniae and Streptococcus pyogenes. Infect Immun 82:1141–1146. doi:10.1128/IAI.01310-13
Oliver JD (2010) Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34:415–425. doi:10.1111/j.1574-6976.2009.00200.x
O’Toole GA, Kolter R (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi:10.1046/j.1365-2958.1998.00797.x
Rittershaus ESC, Baek SH, Sassetti CM (2013) The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe 13:643–651. doi:10.1016/j.chom.2013.05.012
Roca D, Espinal P, Vila-Farres X, Vila J (2012) The Acinetobacter baumannii oxymoron: comensal hospital dweller turned pan-drug resistant menace. Front Microbiol 3:1–30. doi:10.3389/fmicb.2012.00148
Roszak DB, Colwell RR (1987) Survival strategies of bacteria in the natural environment. Microbiol Rev 51:365–379
Stepanović S, Vuković D, Dakić I, Savić B, Svabić-Vlahović M (2000) A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40:175–179. doi:10.1016/S0167-7012(00)00122-6
Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA (2008) Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409. doi:10.1099/mic.0.2008/019471-0
Towner KJ (2009) Acinetobacter: an old friend, but a new enemy. J Hosp Infect 73:355–363. doi:10.1016/j.jhin.2009.03.032
Webster C, Towner KJ, Humphreys H (2000) Survival of Acinetobacter on three clinically related inanimate surfaces. Infect Control Hosp Epidemiol 21:246. doi:10.1086/503214
Wendt C, Dietze B, Dietz E, Rüden H (1997) Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol 35:1394–1397
Zhang Y (2014) Persisters, persistent infections and the Yin-Yang model. Emerg Microb Infect 3:e3. doi:10.1038/emi.2014.3
Acknowledgments
Real-time PCR analysis and scanning electron microscopy of A. baumannii preparations were performed at the Advanced Research Core Facilities (SGIker) of the University of Basque Country UPV/EHU (Gene Expression Unit of the Genomics Core Facility and Analytical and High-Resolution Microscopy in Biomedicine Service). This work was supported by the research projects S-PE12UN59 and S-PE13UN059 from the Basque Government and EHU13/57 from the Basque Country University (UPV/EHU), as well as Basque Government Predoctoral Grants BFI-2011-84 to Z. Bravo and BFI-2011-85 to C. Parada. V.R. Kaberdin was supported by IKERBASQUE (Basque Foundation for Science). Authors thank Prof. J. Ramos-Vivas (Department of Microbiology, Hospital Universitario Marqués de Valdecilla-IDIVAL, Santander, Spain) for providing them with the clinical Acinetobacter baumannii isolate.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Rights and permissions
About this article
Cite this article
Bravo, Z., Orruño, M., Parada, C. et al. The long-term survival of Acinetobacter baumannii ATCC 19606T under nutrient-deprived conditions does not require the entry into the viable but non-culturable state. Arch Microbiol 198, 399–407 (2016). https://doi.org/10.1007/s00203-016-1200-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-016-1200-1