Abstract
Cytoplasmic male sterility (CMS) is an increasingly important issue within the context of hybrid seed production. Its genetic framework is simple: S-cytoplasm for male sterility induction and dominant allele of the restorer-of-fertility gene (Rf) for suppression of S. However, breeders sometimes encounter a phenotype of CMS plants too complex to be explained via this simple model. The molecular basis of CMS provides clue to the mechanisms that underlie the expression of CMS. Mitochondria have been associated with S, and several unique ORFs to S-mitochondria are thought to be responsible for the induction of male sterility in various crops. Their functions are still the subject of debate, but they have been hypothesized to emit elements that trigger sterility. Rf suppresses the action of S by various mechanisms. Some Rfs, including those that encode the pentatricopeptide repeat (PPR) protein and other proteins, are now considered members of unique gene families that are specific to certain lineages. Additionally, they are thought to be complex loci in which several genes in a haplotype simultaneously counteract an S-cytoplasm and differences in the suite of genes in a haplotype can lead to multiple allelism including strong and weak Rf at phenotypic level. The stability of CMS is influenced by factors such as the environment, cytoplasm, and genetic background; the interaction of these factors is also important. In contrast, unstable CMS becomes inducible CMS if its expression can be controlled. CMS becomes environmentally sensitive in a genotype-dependent manner, suggesting the feasibility of controlling the expression of CMS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytoplasmic male sterility (CMS) is a maternally inherited trait that prevents a plant from forming pollen (Levings 1993; Schnable and Wise 1998). The genetic model of CMS includes cytoplasm having the ability to induce male sterility (designated as S) and non-inducible cytoplasm (N). Also included in this model is a nuclear gene that suppresses the action of S. This gene is termed restorer-of-fertility (Rf). In most cases, the dominant Rf allele restores pollen fertility; hence, the recessive rf allele is necessary for the induction of male sterility. The male sterile plant can receive pollen to set seeds because its female reproductive organ remains functional.
As hybrid maize became more significant in the early twentieth century (Duvick 2001), breeders anticipated applying this new breeding method to other crops. However, this attempt was hampered by the hermaphroditism expressed by crops (i.e., male and female organs are closely positioned in a flower), which results in the purity of the resultant crop being reduced. The idea that CMS can be used to emasculate seed parental line was proposed for several crops (Kaul 1988), and Jones (1943) was first to implement it in the onion. The hybrid expresses sterility if the pollen parental line is rfrf; hence, pollen parental line for hybrid seed production should be RfRf to secure pollination if the yield is a seed or fruit. Hybrid breeding through the use of CMS has now been widely introduced across many crops (Kim and Zhang 2018). Unfortunately, not all crop species are amenable to CMS-based breeding because of the absence of appropriate CMS. For CMS-based breeding to be implemented within the context of certain crops, S and both dominant and recessive alleles of Rf must first be characterized.
CMS has been reported in more than 150 plant species (Laser and Lersten 1972). Potential sources of CMS are as follows: In some crops such as onion, maize, and sugar beet, the CMS plant was discovered from breeding materials (Jones 1936; Owen 1945; Rogers and Edwardson 1952). A common strategy to identify CMS is an intraspecific cross between distantly related populations or an interspecific cross (Duvick 1965; Mikami et al. 1985; Marinković and Miller 1995). The rationale for the occurrence of a CMS plant from two normal fertile parents is that CMS is suppressed in one parent due to the dominant Rf allele but the Rf allele is segregated out from the offspring because of the recessive rf allele in the other parent. Overall, this strategy entails the elucidation of cryptic CMS (Touzet 2012) using crossing experiments. In a case in which the donor is sexually incompatible, CMS can be transferred via protoplast fusion followed by plant regeneration (e. g., Pelletier and Budar 2015). In plant reproductive biology, CMS is known as a principal cause of gynodioecy, a sexual system in which females and hermaphrodites co-occur in a population (Touzet 2012). Gynodioecy has been found in more than 18% of 449 angiosperm families (Dufay et al. 2014), from which CMS may be introduced into related crops.
Several efforts based on the above-mentioned strategy have been made to identify and introduce CMS into crops, and some have been a success (Bosemark 1998; Sattari et al 2007; Reddy et al 2008; Yamagishi and Bhat 2014; Huang et al 2014; Saxena et al 2018). However, not all CMS is practically useful because of the difficulty in handling the phenotypic instability that is accompanied by the implementation of this technique. For example, a caveat of Polima-type CMS in rapeseed is the reversion of pollen fertility under high temperatures in certain genotypes (Burns et al. 1991). Such issues can be overcome if we elucidate the mechanisms that underlie the expression of CMS. Here, we would like to review important advances and insightful studies rooted in exploring the molecular genetics of CMS and Rf, with an emphasis on recent findings. We stress that previously unappreciated aspects in this research field such as multiple allelism of Rf, effect of environmental factors, and their interactions should be considered for the successful application of CMS-based breeding across a wider range of crops. Previous reviews on the relevant subject may complement this review, allowing readers to delve deeper into CMS and Rf (Horn et al. 2014; Chen and Liu 2014; Gaborieau et al. 2016; Bohra et al. 2016; Chen et al. 2017; Kim and Zhang 2018; Kubo et al. 2020; Toriyama 2021; Xu et al. 2022).
S-cytoplasm is associated with mitochondria
Cytoplasmic inheritance has been associated with mitochondria, plastids, and infectious agents such as viruses (Mogensen 1996; Greiner et al. 2015; Vong et al. 2019). A CMS in fava bean was reported to be linked to cytoplasmic double-strand RNA (Pfeiffer 1998), but the vast majority of CMS have been shown to stem from mitochondria (Schnable and Wise 1998). Although plant mitochondrial genomes typically contain 20–40 protein-coding genes and 20–30 other genes (i.e., the number varies across species) (Møller et al. 2021), few mutations in these genes have been associated with CMS. A wild beet CMS termed G-type is an exception in which several mutations that truncate reading frames of mitochondrial genes such as cox2 and nad9 have been found (Ducos et al. 2001; Darracq et al. 2011). The association of non-coding RNA has been proposed in one case (Stone et al. 2017), and many cases of CMS have been associated with unique mitochondrial ORF that has the potential to encode a polypeptide absent from N-cytoplasm (hereafter termed S-orf). S-mitochondria contain the same suite of genuine mitochondrial genes as N-mitochondria (Satoh et al. 2004; Makarenko et al. 2019; Wang et al. 2020; Zhong et al. 2021). Generally, the S-orfs that have been identified thus far are not homologous to each other, instead, exhibit a few instances of interspecific similarities (e.g., Tang et al. 2017). One of the notable features is their patchy homology to mitochondrial genes such as genes for subunits of ATP synthase (Köhler et al. 1991; Handa et al. 1995; Tang et al. 1996). The rest of the ORF region (or sometimes the entire ORF region) is unique to each S-orf (Yamamoto et al. 2005; Kim et al. 2007).
Heterologous expression of S-orf with mitochondria-transit peptide in transgenic plants yielded a sterile male phenotype (Yamamoto et al. 2008; Wang et al. 2006; He et al. 1996; Jing et al. 2012; Yang et al. 2010; Luo et al. 2013; Jiang et al 2022), but male sterility phenotype is not always reproduced in similar experiments (Wintz et al. 1995; Chaumont et al. 1995; Kojima et al. 2010). On the other hand, pollen-fertile revertants have been obtained from CMS maize and common bean; their S-orfs are lost or suffer frame-shift mutation (Wise et al. 1987; Janska et al. 1998), supporting a notion that they are responsible for the male sterility phenotype.
Plant mitochondrial genome editing is now feasible through the deployment of mitoTALEN, a chimeric protein consisting of mitochondria-transit peptide and transcription activator-like effector nuclease (Kazama et al. 2019). Unlike CRISPR/Cas9 system, mitoTALEN does not require guide RNA (Kazama et al. 2019), whose import into mitochondria seems difficult. By using mitoTALEN, DNA double-strand breaks were introduced into S-orfs of rice, rapeseed, and tomato CMSs, which caused genome rearrangement resulting in the loss of these S-orfs from the mitochondrial genomes (Kazama et al. 2019; Omukai et al. 2021; Kuwabara et al. 2022; Takatsuka et al. 2022). Plants that lost S-orfs were male fertile, indicating that they are responsible for CMS. An interesting result was reported by Omukai et al. (2021); in this study, they produced rice plants depleted of orf352 which is associated with RT102 cytoplasm but the plants brought males that were partially fertile, instead of being fully fertile. Omukai et al. (2021) discussed that an additional unidentified mitochondrial gene is associated with the male sterility phenotype, implying that the CMS gene is not always a single ORF. The occurrence of multiple unique ORFs in addition to genuine genes is usual in plant mitochondria irrespective of their ability to induce male sterility (e.g., Marienfeld et al. 1997). It seems likely that most of such unique ORFs would not impinge upon the production of pollen, but each has the potential to evolve CMS-causing genes. This notion is consistent with the proposed evolutionary history of rice WA352c (for WA-type CMS), which appeared to have evolved from a unique but non-male sterility inducible ORF (Tang et al. 2017).
The location and expression of S-orf can be classified as illustrated in Fig. 1. In many cases, S-orfs are located upstream or downstream of mitochondrial genes and co-transcription of the S-orfs and the mitochondrial genes has been shown to occur frequently (Fig. 1a; Dewey et al. 1987; Rathburn and Hedgcoth 1991; Song and Hedgcoth 1994; Wang et al. 2006). S-orfs fused with mitochondrial genes have been reported in several plant species (Fig. 1b, c; Bailey-Serres et al. 1986; Yamamoto et al. 2005; Meyer et al. 2018), but their occurrence has been shown to be rarer than the co-transcription type. One may expect fusion protein from such gene organization; however, in sugar beet, the fusion protein seems to be processed into two independent proteins (Fig. 1b, Yamamoto et al. 2005). On the other hand, the fusion protein was detected in CMS sorghum (Fig. 1c, Bailey-Serres et al. 1986). Note that cox1 in wild beet with G-type CMS has a NH2-terminal extension that can be considered an S-orf, resulting in the detection of the fusion protein (Fig. 1c, Meyer et al. 2018). Fewer cases of solitary S-orf have been recorded (Fig. 1d; Yamamoto et al. 2008; Park et al. 2013). For these observations, we favor the notion that, because S-orf is an evolutionarily young gene, it does not have its own promoter and terminator sequences. Therefore, the easiest way to achieve S-orf expression is to capture the preexisting sequences. This possibility may be reflected in the fact that the co-transcription type S-orf has the highest frequency with regard to expression. Gene fusion may facilitate the expression of S-orf, but it would be necessary not to severely impair the function of the fused mitochondrial gene, which may make the emergence of fusion-type S-orf difficult; hence, this type is rarer. The expression of solitary S-orf is only possible if sequences necessary for its expression are available, but this may be evolutionarily rare scenario in the context of the plant mitochondrial genome.
Locations and expression mechanisms that underlie S-orfs in the mitochondrial genome. Striped and open boxes represent S-orfs and mitochondrial genes, respectively. S-orfs encode either chimeric sequence of partial mitochondrial gene and unknown origin, or their entire sequence is unknown origin. Curved lines represent transcripts. Striped and open circles represent translation products of S-orfs and mitochondrial genes, respectively. Protein derived from S-orfs responsible for male sterility. a S-orf is located independently of mitochondrial gene. They are co-transcribed, and then translated into two independent proteins. b S-orf fused with mitochondrial genes is co-transcribed, and then translated into a fusion protein. The fusion protein is proteolytic cleavage into two proteins. c S-orf fused with mitochondrial genes is co-transcribed, and then translated into a fusion protein. d S-orfs are solitary
CMS-associated ORFs vary in their primary structure, but several groups may be possible at biochemical level
The above-mentioned studies indicate that the protein product of S-orf likely alters mitochondrial function, but the application of sequence-based approaches to infer S-orf function has been difficult because of the great nucleotide-sequence diversity among S-orfs. However, it has been shown that the translation products predicted through S-orfs have highly hydrophobic domains (Dewey et al. 1987; Köhler et al. 1991; Yamamoto et al. 2005; Kazama et al. 2008; Okazaki et al. 2013). Note that this is not always consistent with their strict membrane localization because petunia PCF protein and wild beet ORF129 protein (derived from I-12CMS(3)/E-type cytoplasm) were detected from both the mitochondrial membrane and matrix (Nivison et al. 1994; Yamamoto et al. 2008), suggesting that they are loosely associated with the mitochondrial membrane.
S-orfs can be classified according to the biochemical properties of their translation products. One is the oligomer-forming group, to which maize urf13-T (from T-type CMS), radish orf138 (Ogura-type CMS), and sugar beet preSatp6 (Owen-type CMS) belong (Rhoads et al. 1998; Duroc et al. 2005; Yamamoto et al. 2005). The translation products of these ORFs are highly hydrophobic and they are detected only in a fraction of the membrane, indicating that they are likely integrated into the mitochondrial membrane to form oligomers, whereas the specific association of another protein was not supported (Duroc et al. 2005, 2009; Yamamoto et al. 2005). Rhoads et al. (1998) proposed a model of molecular pore made from the translation products of urf13-T. This molecular pore is formed under the presence of specific chemicals such as T-toxin produced by Cochliobolus heterostrophus race T (the causal fungus of Southern corn leaf blight) and the pore acts as an uncoupler. A similar molecular pore may be formed in radish and sugar beet CMS plants, although this remains to be examined. The amount of oligomer-formed preSATP6 protein exhibits a good correlation with the severity of the CMS phenotype (Arakawa et al. 2018, 2019b, 2020a), suggesting that oligomer-formed preSATP6 protein is quantitatively associated with the CMS phenotype.
Another group consists of the protein having the ability to bind specific proteins other than itself. This group includes rice WA352c (WA-type CMS) and rice orfH79 (HL-type CMS). WA352c protein was shown to bind to COX11 protein, which is a copper metallochaperone involved in the cytochrome c oxidase assembly (Luo et al. 2013; Wang et al. 2018). ORFH79 seems to be able to bind a protein(s) but its partner may not be a single protein (Liu et al. 2012; Wang et al. 2013). The question of whether an additional group or alternative classification of S-orf is possible remains to be answered.
Functional aspects of CMS-associated ORF
The fact that there are trans-specific similarity among S-orf gene products at biochemical level, such as those seen in the oligomer-forming group, points to the presence of a shared mechanism that underlies the expression male sterility. The translation product of S-orf is believed to impair mitochondrial function, thereby causing pollen sterility (Horn et al. 2014). Given that many S-orfs are constitutively expressed (Touzet and Meyer 2014), the degree of impairment should be small or largely compensated in other organs than anthers, otherwise large deleterious effects would incur a fitness penalty, which will reduce the yield of hybrid crops. Three working hypotheses have been discussed about the expression of CMS: one evokes an anther-specific factor to activate the protein. This hypothesis was proposed based on the fact that URF13-T protein (maize T-type CMS) becomes an uncoupler under the presence of certain chemicals (Flavell 1974; Levings 1993). The second hypothesis postulates a difference in the relative importance of mitochondria among organs, of which the anther is highly dependent on mitochondria (Levings 1993). According to this hypothesis, a subtle effect of S-orf in non-anther organs is expected; however, it may be undetectable. Maize with T-type CMS shows slight morphological differences such as in its height, leaf number, and grain yield (Duvick 1965). Rice plants expressing orfH79 (HL-type CMS) exhibited shorter roots, altered mitochondrial function, and increased reactive oxygen species (ROS) compared to plants with N-cytoplasm and these alterations disappeared when orfH79 is suppressed (Yu et al. 2015), apparently consistent with this hypothesis. Some S-orf proteins are reported to be cytotoxic in non-anther cells (e.g., Wang et al. 2006; Kojima et al. 2010; Nakai et al. 1995). Note that, although some S-cytoplasm causes vegetative abnormalities such as chlorosis, these are attributed to the plastid genome and can be genetically separated from the male sterility phenotype (Yamagishi and Bhat 2014). Presence/absence of S-orf is not the only factor that differentiates the N- and S-cytoplasm. Additionally, differences have been found between N- and S mitochondrial DNA sequences (e.g., Satoh et al. 2004; Wang 2020). Detailed analyses are necessary to ascertain whether other phenotypes, other than male sterility, are caused by S-orf. In the third hypothesis, S-orf is translated only in the anther. Translation products of common bean orf239 and rice WA352c were detected from the anther but not from other organs (Abad et al. 1995; Luo et al. 2013). In this case, the effect of the S-orf protein may not be necessarily weak.
The way how the S-orf proteins impair mitochondrial function is unclear. If the S-orf protein binds to a specific mitochondrial protein, this binding may be deleterious to the protein’s function, thereby mitochondria are damaged (Luo et al. 2013). Sabar et al. (2003) investigated sunflower PET-type CMS and detected a decrease in the enzymatic capacity of the Complex V (F1F0-ATP synthase), for which translation product of orf522, an S-orf of this CMS, is suggested to compete with atp8 due to shared homology in their 5' coding regions. This notion may be applicable for S-orfs having homology to genuine mitochondrial genes. These data imply that some S-orfs directly impair function of mitochondrial proteins. For the oligomer-forming-type S-orfs, it has been postulated that the oligomer may act as a mild uncoupler (Duroc et al. 2009), but this notion needs further physiological support. While the enzymatic capacity of respiratory complexes is unchanged in the plant with orf138 (Duroc et al. 2009), a decrease in the Complex V was found in the plant with preSatp6 (Wesołowski et al. 2015). It was reported that not all preSATP6 proteins form oligomers under certain extraction conditions (Wesołowski et al. 2015). Given the high hydrophobicity of these oligomer-forming proteins, it may be possible that their non-specific binding with other mitochondrial proteins has a small harmful effect, but its effect accumulates eventually and crosses the threshold in the anther. This explanation is congruent with the above-mentioned notion that protein–protein interactions play a key role in the impairment of mitochondrial function, and may be a common mechanism that unifies the expression of CMS. On the other hand, mitochondrial function can be regulated by other mechanisms that have been extensively investigated within the context of CMS. For example, there is intimate associated between mitochondrial shape and function, and this link has been shown to drive changes in the respiratory supercomplex (Cogliati et al. 2013). Interestingly, Meyer et al. (2018) showed an alteration in the mitochondrial supercomplex in wild beet with G-type CMS. We believe that not enough evidence has been brought forth to conclude that the protein–protein interaction model explains all the dynamics of CMS. Post-translational action of oligomer-forming type S-orf should be investigated further.
Mitochondrial alteration by CMS-associated ORF
Mitochondria are involved in the production of energetic substances such as ATP through the respiratory chain in the inner membrane (Logan 2006). Considering the hypothesis that anther development highly depends on mitochondria, anthers may consume more ATP than other organs, and ATP shortage by mitochondrial impairment could impact on its development. However, Touzet and Meyer (2014) argue against this notion based on the phenotypes of mutants with defects in the subunits of the respiratory complexes. A detailed study is necessary on whether changes in ATP/ADP ratio observed in some CMS plants are the cause of CMS or by-products. Mitochondria not only produce ATP, but they also involved in the catabolism of amino acids and the provision of carbon skeletons for the biosynthesis of an array of compounds (Sweetlove et al. 2007). Whether and how S-orf affects these biosynthetic pathways remains to be examined.
Because mitochondria are a site of ROS production (Scheffler 1999), their impairment sometimes leads to the overproduction of ROS. An increase in ROS accumulation is frequently detected in CMS plants (Horn et al. 2014). Whereas ROS can directly damage biological macromolecules (Scheffler 1999), ROS is also associated with biological processes such as programmed cell death (PCD) (Gechev et al. 2006). In the developmental process of pollen, the anther tapetum tissue undergoes PCD at a certain stage of microspore development (Tariq et al. 2022). Anther tapetum is a sporophytic tissue surrounding microspore mother cells/microspores and plays important roles in pollen development, primarily by providing components for pollen production (Tariq et al. 2022). The PCD of tapetum is crucial for pollen development because mutants with defects in this process fail to produce functional pollen (Tariq et al. 2022). In sunflowers with PET-type CMS and rice with WA-type CMS, the onset of PCD is earlier than in the N-cytoplasm plant, which may be caused by altered ROS accumulation (Balk and Leaver 2001; Luo et al. 2013). On the other hand, Arakawa et al. (2019b) observed the correlation between the persistence of tapetal debris and the severity of sugar beet Owen-type CMS, suggesting that the onset of PCD is delayed or that PCD proceeds slowly. These observations point to mitochondria intervening in or modulating pollen development to induce male sterility. This viewpoint has been investigated by Geddy et al. (2005); they found that rapeseed with nap CMS exhibited altered expression pattern of apetala 3, a floral homeotic gene, pointing to the anther being impacted by a developmental abnormality. The link of mitochondrial function to the expression of developmental abnormalities is apparent, as homeotic conversion of the stamen into other floral organs such as the carpel and petal is seen in carrot and wheat CMS (Linke et al. 2003; Murai et al. 2002). In these CMS plants, the expression of the so-called B-class MADS box genes is disrupted (Linke et al. 2003; Murai et al. 2002). Overall, mitochondria are likely involved in anther development; however, the details underlying this involvement remain unknown. Hence, our understanding of CMS should be rooted in clearly elucidating the role of mitochondria in floral development; specifically, there should be investigation delving into how mitochondria affect nuclear gene expression within the context of floral development (i.e., retrograde signaling).
Genetic and molecular aspects of restorer-of-fertility
It is not always the case that one Rf is identified in an S-cytoplasm. There have been cases in which multiple Rfs have been identified (Schnable and Wise 1998; Kaul 1988). In general, an Rf specifically counteracts the cognate S-cytoplasm, a genetic rule used for discriminating S-cytoplasms (Duvick 1965). Test cross is a simple procedure to know whether a plant has a specific Rf, but it is time-consuming and laborious and a major obstacle for hybrid breeding programs in some crops. To save the cost, alternative methods such as DNA marker-assisted selection are necessary. Molecular organization and polymorphism of Rf is crucial information to develop DNA markers of Rf.
Table 1 summarizes Rf-gene products based on cloned Rf or its strong candidates. Note that the gene product of maize Rf4 is not a mitochondrial protein (Jaqueth et al. 2020). In many cases, the Rf-gene product is reported to be proteins with a pentatricopeptide repeat (PPR) motif (Gaborieau et al. 2016) (PPR-Rf). PPR genes are involved in post-transcriptional processes in mitochondria and plastids, and their translation products are generally considered to be able to bind to RNA in a sequence-specific manner (Barkan and Small 2014). Genes encoding PPR proteins occur 400 to more than 1000 times in land-plant genomes, and they are classified according to the difference in the PPR motif and the presence or absence of auxiliary domains (Barkan and Small 2014). Among the classes of PPR proteins, the P-class protein is the gene product of PPR-Rf, except for two cases (Table 1). Within the P-class, Fujii et al. (2011) identified a distinctive subgroup to which PPR-Rfs belong (termed restorer-to-fertility-like, RFL). Because it had been frequently observed that the expression of S-orfs is altered to reduce S-orf protein upon fertility restoration (Brown et al. 2003; Wang et al. 2006; Tang et al. 2014), how PPR-Rf alters S-orf expression is the principal question. Direct or indirect association between PPR-Rf protein and S-orf mRNA has been shown (Kazama et al. 2008; Qin et al. 2014; Jiang et al. 2022). A large protein complex including PPR-Rf protein is reported in several cases (Gillman et al. 2007; Hu et al. 2012), suggesting that an additional protein is involved in fertility restoration. Radish Rfo is a well-characterized PPR-Rf (Brown et al. 2003; Qin et al. 2014; Yamagishi et al. 2021). In the Rfo locus, multiple RFLs are clustered, of which PPR-B (or orf687) has been identified as the gene responsible for fertility restoration (Koizuka et al. 2003; Desloire et al. 2003; Uyttewaal et al. 2008). The translation product of PPR-B is shown to bind to the coding region of orf138 mRNA to inhibit translation (Wang et al. 2021), a clear instance of post-transcriptional suppression by PPR-Rf. Yamagishi et al. (2021) showed that a single nucleotide substitution in orf138 that coincides with the binding site is enough to prevent fertility restoration by Rfo, indicating that interaction between PPR-B and orf138 mRNA occurs in a highly sequence-specific manner. On the other hand, Sorghum Rf1 and barley Rfm1 belong to another class of PPR protein (PLS-DYW), but the function of these genes is unknown partly due to the absence of knowledge about their S-orf.
The post-transcriptional mechanism is also involved in a restoration of fertility that is governed by other types of Rf. Rice Rf2 (for Lead Rice-type CMS) encodes a glycine-rich protein that can reduce the translation product of orf79 (the S-orf of this CMS) by mRNA degradation (Itabashi et al. 2011; Kazama et al. 2016). The recessive rf2 allele (i.e., male sterility-inducing allele) has a missense mutation. Barley Rfm3 and rye Rfp1 likely encode mitochondrial transcription termination factor family (mTERF) proteins, which are known to be involved in transcription, splicing, or tRNA maturation (Quesada 2016). Therefore, they may restore pollen fertility via post-transcriptional mechanism. Small et al. (2019) pointed out that RNA processing enzymes make up about 15% of plant mitochondrial proteome, suggesting the importance of post-transcriptional mechanisms in plant mitochondria. This can be associated with the prevalence of post-transcriptional mechanisms in fertility restoration.
The post-translational mechanisms associated with the inactivation of S-orf protein are found in sugar beet Rf1 (for Owen-type CMS) (Kitazaki et al. 2015). The molecular action of sugar beet Rf1 is not obvious in the amount of preSatp6 (S-orf of this CMS) mRNA or its protein product; however, although the total amount of preSATP6 protein is almost unchanged, its oligomer form is highly reduced in anther where Rf1 is expressed, suggesting that Rf1 can alter higher-order structure of preSATP6 protein (Kitazaki et al. 2015). The nucleotide sequence of sugar beet Rf1 resembles Oma1, known to be involved in the quality control of mitochondria (Matsuhira et al. 2012). Sugar beet Rf1 is not the orthologue of Oma1, but a paralogue evolved through segmental duplication (Arakawa et al. 2020a). Unlike authentic Oma1, sugar beet Rf1 has a defect in zinc-binding domain that is crucial for peptidase activity (Matsuhira et al. 2012). On the other hand, the translation product of sugar beet Rf1 (dominant allele) can bind preSATP6 protein but neither that of recessive rf1 nor authentic Oma1 does (Arakawa et al. 2019a). It is inferred that the translation product of Rf1 acts as a molecular chaperone on fertility restoration.
It should be noted that the relationship between S-orf and Rf-gene products is unclear in some cases. Rice Rf17 (for CW-type CMS) encodes a protein resembling acyl carrier protein (named RMS) (Fujii and Toriyama 2009). Among the alleles of Rf17, those with reduced expression restore pollen fertility but neither wild type- nor knockout allele does (Suketomo et al. 2020). Perhaps RMS may play an important role in pollen development (hence plant with a knockout allele is male sterile) but CW-mitochondria use RMS to express male sterility (i.e., RMS transmits a signal from CW-mitochondria for the CMS expression). Although RMS encodes a mitochondrial protein, its relationship with orf307 (S-orf of CW-CMS) is unknown (Fujii and Toriyama 2009). Maize Rf4 (for C-type CMS) encodes a basic helix-loop-helix protein, whose null allele had previously been identified as ms23, which is responsible for genic male sterility (Jaqueth et al. 2020). For the recessive rf4 allele for C-type CMS to allow male sterility but not for N-cytoplasm, it must have a missense mutation that inhibits hetero dimer formation of this protein with a partner protein (Jaqueth et al. 2020). It is unknown how this allele interacts with C-type mitochondria, its functioning points to the association between flower development and mitochondria. Maize Rf2a (for T-type CMS) is the first cloned Rf in the plant (Cui et al. 1996), which encode mitochondrial aldehyde dehydrogenase. The enzymatic activity of RF2a indicated that it has a broad range of substrate specificity (Liu and Schnable 2002), but its substrate in the anther is unknown. Both null allele and missense alleles are known for rf2a. A long-standing and unanswered question is how Rf2a interacts with urf13-T.
Rfs belonging to unique gene families
RFL is a subgroup of the P-class PPR gene family and is nearly ubiquitous in plants irrespective of the presence of the CMS/Rf system (Fujii et al. 2011). PPR-type Rf is frequently embedded within the RFL gene cluster (Gaborieau et al. 2016), which shows copy number variation polymorphism of clustered genes. Although sugar beet Rf1 (for Owen-type CMS) is a paralogue of Oma1 but not PPR-type Rf, it was identified from a gene cluster consisting of the Oma1 paralogues resembling the Rf1. This Oma1-paralogue cluster has evolved after the establishment of Amaranthaceae, perhaps in the genus Beta (Arakawa et al. 2020b). The Oma1-paralogue cluster seems to be ubiquitous in sugar beet and leaf beet irrespective of the Rf1/rf1 genotype (Ohgami et al. 2016; Arakawa et al. 2018). Like RFL, copy number variation is seen between sugar beet lines (Moritani et al. 2013; Arakawa et al. 2020b). Another Rf belonging to a unique gene family is the barley Rfm3 and rye Rfp1(Bernhard et al. 2019; Hackauf et al. 2017), which both encode or are associated with mTERF proteins. Melonek and Small (2022) found that plant mTERF genes form a gene family but a subgroup of this family greatly expands in cereals, to which rye Rfp1 belongs. These data suggest that some Rfs have evolved from gene families that formed and expanded in certain lineages.
Evolutionary patterns of RFL and the Oma1-paralogues are similar; they tend to form gene cluster that shows copy number variation, and they have traces of positive selection (Fujii et al. 2011; Arakawa et al. 2020b). This may be the case for a subgroup of the cereal mTERF gene family to which rye Rfp1 belongs (Melonek and Small 2022). Such evolutionary patterns are reminiscence of resistance (R) gene cluster (Young 2000), suggesting that the evolutionary forces underlying these gene families are common despite the difference in their gene products. It has been proposed that both the R gene and Rf have evolved to cope with ever-rising pathogens and S-orfs, respectively. The mitochondrial incentive to evolve S-orfs stems from the maternal inheritance of mitochondria because pollen is useless for them and male sterility is beneficial to maximize their transmission to progeny if it allows resources reallocation (Touzet 2012). In contrast, the nuclear genome incurs fitness penalty from S-orfs and favors the deployment of its suppressor (Touzet 2012). This situation evokes co-evolution between opposing parties, the so-called arms race (Fujii et al. 2011). However, it needs more study before considering all the Rf and Rf-like genes (i.e., RFLs and the Oma1 paralogues) as weapons. For example, maize Rf2a, Rf4, and rice Rf17 appear to be out of an arms race. Among the Arabidopsis RFLs, RFL8 is shown to be involved in the expression of a genuine mitochondrial gene ccmFN2, which needs Brassicaceae-specific compensation because of its truncated gene organization in this lineage (Nguyen et al. 2021).
We note that less attention is paid to the gene cluster to which Rf belongs, with regard to its function. In Arabidopsis, RPP1 locus contains an R gene cluster whose constituents encode nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins conferring resistance to downy mildew (Botella et al. 1998). The clustered genes were dissected and examined for their specificity to the races of the pathogen. To our interest, multiple NBS-LRR genes in the cluster exhibited resistance to a race, indicating that RPP1 is a complex locus (Botella et al. 1998). In the case of sugar beet Rf1, multiple Oma1 paralogues in a haplotype are likely to participate in fertility restoration; hence, we consider Rf1 as a complex locus (Arakawa et al. 2020a, b). Wang et al. (2006) described rice Rf-1 locus contains two PPR genes that can restore Boro II-type CMS. Intraspecific polymorphism of the Rf locus has been reported in several crops (Yamagishi and Bhat 2014; Aarakawa et al. 2018; Melonek et al. 2019), but the functional analysis of haplotypes has been scarce. Is it only a single ORF or multiple ORFs in a haplotype that participate in the restoration of fertility? Moreover, Arakawa et al. (2018, 2019b, 2020a) found that haplotypes of sugar beet Rf1 reflect the alleles of different strengths in the level of fertility restoration (i.e., multiple allelism) (Fig. 2). Haplotype polymorphism should be considered for DNA marker development to discriminate Rf alleles.
Allelo morphs and organization of restorer-of-fertility 1 (Rf1) alleles in sugar beet. Sugar beet Rf1 is a complex locus consisting of ORFs encoding OMA1-like protein. Boxes and Wedges represent exons and introns, respectively. Gene directions are from right to left. Names of individual ORF copies are labeled above. A vertical arrow indicates the position of a premature stop codon. Protein products from ORFs indicated by striped boxes have ability to bind preSATP6 protein (the protein product S-orf of Owen-type CMS), whereas translation products from ORFs indicated by open boxes do not. This figure is based on Arakawa et al. (2020a)
Unstable CMS: its cause and potential application
Penetrance of CMS is an important factor for a CMS to be implemented into a breeding program. Low penetrance (i.e., phenotypically unstable CMS) means occasional pollen production in the CMS line, leading to contamination of self-pollinated plants in the hybrids. Xiao et al. (2022) found a variant of maize S-type CMS that exhibits a more stable male sterility phenotype. Although both the original and the variant mitochondria expressed orf355 (the S-orf of maize S-type CMS) at a similar level, the gene dosage of nad1 was different (Xiao et al. 2022), suggesting that mitochondrial genetic architecture affects CMS expression. It may be possible to genetically modify S-mitochondria to be more stable by mitochondrial genome editing.
Factors that impact CMS penetrance include low- and high temperature, air humidity, day length, and soil moisture (Kim et al. 2013; Bernhard et al. 2017; Downes and Marshall 1971; Reddy and Reddi 1970; Van der Veken et al. 2018; Duvick 1965; Bueckmann et al. 2016; Murai and Tsunewaki 1993; Bueckmann et al. 2016; Elkonin and Tsvetova 2012). In soybean, CMS-cytoplasm derived from the NJCMS1A line was combined with two different nuclear backgrounds and the two lines were grown at high temperatures during the flowering period. The CMS stability was different between the two lines (Ding et al. 2020, 2021). This is an instance that genetic background plays an important role in CMS stability.
We propose that there might be cases where environment-sensitive CMS is caused by environment-sensitive Rf. For example, when dominant Rf and environment-sensitive Rf are prevalent but true rf is scarce in a gene pool, the cognate CMS seems to be environmentally sensitive until it is combined with the true rf. In some cases, known Rf has been associated with environmental sensitivity. Rice Rf6 (for HL-type cytoplasm) can restore male fertility more stable than Rf5 under high temperatures (Zhang et al. 2017). Maize Rf9 expression is influenced by temperature (Gabay-Laughnan et al. 2009). Rf1 locus of a Japanese leaf beet accession is occupied by very weak Rf1 that conditions male sterility but occasionally restores partial male fertility under low temperatures (Arakawa et al. 2018; T. Arakawa, unpublished observation). In sugar beet, Owen-type mitochondria were combined with various genetic backgrounds, and the plants were grown under two different environments (normal temperature and high temperature). The plants’ phenotypes are classified into three groups; constant male sterile, constant male fertile, and male fertile under normal temperature but male sterile under high temperature (i.e., temperature-sensitive male sterility) (Matsuhira et al. 2022). In this experiment, the examined genetic background included different Rf1 haplotypes; the association of these elements with the three phenotypic groups was investigated (Matsuhira et al. 2022). Each of the three phenotypic groups has unique Rf1 haplotypes, suggesting an association between temperature-sensitive male sterility and the Rf1 allele (Matsuhira et al. 2022).
Even though genetic background has been known to affect the stability of CMS, the identification of genes involved in such a genetic background is challenging because of their subtle effects. The expression of environment-sensitive Rfs may be masked depending on the conditions under which the plant is growth. Additionally, the segregation of fertility restoration by such Rfs may deviate from Mendelian fashion; hence, such Rf should be identified as QTL. This strategy has been adopted and succeeded in the identification of the QTL in maize (Tie et al. 2006; Kohls et al. 2011; Feng et al. 2015; Su et al. 2016). A detailed study on the environment-sensitive Rf would provide clues to obtain stable CMS.
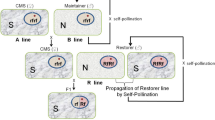
A breeding line equipped with stable CMS requires a maintainer line that has the same nuclear genotype as the CMS line but its cytoplasm is N for the propagation of CMS line (Fig. 3a). The cost to prepare the maintainer line for CMS line propagation could be saved if the male sterility phenotype is conditional, that is, the male sterile line can self-pollinate under permissive conditions (Fig. 3b). A similar approach is adopted in the case of hybrid breeding of rice using genic male sterility that is sensitive to photoperiod (Ding et al. 2012). This indicates that environmental sensitivity can be regarded as environment inducibility if a permissive- and nonpermissive condition is defined. Wheat photoperiod-sensitive CMS (D2-type) has been applied to maintainer-less hybrid breeding in which a line is self-pollinated under short-day conditions, whereas this line is converted into seed parents for hybrid production (Murai et al. 2008). CMS sugar beet (Owen-type) with the genotype expressing male sterility under high temperature, whereas male fertile under normal temperature was tested for its applicability to the maintainer-less system (Matsuhira et al 2022). The results showed that it is a good seed parent under nonpermissive conditions but self-pollinates to set seeds under permissive conditions (Fig. 3c). Genes associated with environment inducibility should be identified to establish a novel breeding system.
Genotypes and phenotypes of plants with different types of male sterility-inducing cytoplasms CMS. Outer circles and inner circles indicate cytoplasm and genotypes, respectively. a Breeding system using stable CMS (shown as S) requires maintainer line, containing N-cytoplasm and recessive rf alleles in homozygous, to propagate the cognate CMS line. CMS line is crossed with restorer line, harboring dominant Rf alleles in homozygous, to produce male fertile hybrids. b Plant with environment-sensitive CMS (the cytoplasm is shown as SES) is male fertile under permissive condition but male sterile under nonpermissive condition. The male fertile plant can self-pollinate to set seeds; hence, maintainer line is not necessary. The male sterile plant can be used for seed parent in hybrid seed production. c Plant with stable CMS (shown as S) is male fertile under permissive condition but male sterile under nonpermissive condition when the plant has environment-sensitive Rf genes (RfES)
Conclusions and perspectives
The implementation of CMS into the breeding programs of a variety of crop species has been a desirable outcome, but one that has been difficult to achieve. CMS expression may involve intricate interactions with known and unknown genes. Fine tuning of CMS expression would be possible if these genes and interactions are unveiled. The mitochondrial genome had been untouchable in terms of genetic manipulation for a long time, but genome editing technology is changing the situation. As novel technology emerges in this field (c.f., Nakazato et al. 2022), approaches from the mitochondrial genome will be increased in research and practical application. CMS expression has quantitative nature, part of which is associated with the expression level of CMS-associated ORF (Kazama et al. 2016; Onodera et al. 2015), the amount of the oligomer form of CMS-associated protein (Arakawa et al. 2018, 2019b, 2020a), and the amount of tapetal debris persisting in the later developmental stages (Arakawa et al. 2019b). This may imply that S-orf emits a substance that the anther issue responds to. A possible candidate for such a signal may be ROS, but it needs to be investigated whether the behavior and biological role of ROS is consistent with this notion. The involvement of mitochondria in the development of flowers should be studied in detail, with the aim of elucidating its association with CMS expression. It is possible that S-orf does not play a significant, but only slightly modifies flower development through a currently unknown mechanism.
PPR-Rf is prevalent in the crop but instances of non-PPR-Rf are increasing. The molecular basis of S-cytoplasm specificity of Rf can be associated with the ability of PPR-Rf protein to recognize S-orf mRNA in sequence-specific manner. But many Rfs lack such molecular bases. Some Rfs belong to unique gene families to specific lineages (e.g., sugar beet Oma1 paralogues and cereal mTERF genes). Evolutionary patterns of such Rfs show a striking similarity to those of RFL and R genes. It reflects arms race evolution, but a detailed study is necessary to find other possibilities. The Rf locus may be a complex locus in which multiple constituent genes can counteract single S-cytoplasm. Moreover, differences in their haplotypes may reflect functionally different allelomorphs (e.g., sugar beet Rf1 alleles differ in their strength of fertility restoration).
CMS expression is more or less affected by environmental factors. Perhaps CMS expression depends on the delicate balance between the action of S-orf and (both nuclear and mitochondrial) genes. Expression of S-orf and these genes may be affected by environmental factors. The instability of CMS expression can be corrected by nuclear background or, in the future, mitochondrial genome editing. Control of CMS by the environment has the potential to be a tool for breeding if the appropriate nuclear background and the permissive and nonpermissive condition is defined. The appropriate genetic background may include alleles of known Rf, but further study is necessary to clarify the genes involved in this genetic background.
References
Abad AR, Mehrtens BJ, Mackenzie SA (1995) Specific expression in reproductive tissues and fate of a mitochondrial sterility-associated protein in cytoplasmic male-sterile bean. Plant Cell 7:271–285. https://doi.org/10.1105/tpc.7.3.271
Arakawa T, Uchiyama D, Ohgami T, Ohgami R, Murata T, Honma Y, Hamada H, Kuroda Y, Taguchi K, Kitazaki K, Kubo T (2018) A fertility-restoring genotype of beet (Beta vulgaris L.) is composed of a weak restorer-of-fertility gene and a modifier gene tightly linked to the Rf1 locus. PLoS ONE 13:e0198409. https://doi.org/10.1371/journal.pone.0198409
Arakawa T, Sugaya H, Katsuyama T, Honma Y, Matsui K, Matsuhira H, Kuroda Y, Kitazaki K, Kubo T (2019a) How did a duplicated gene copy evolve into a restorer-of-fertility gene in a plant? The case of Oma1. R Soc Open Sci 6:190853. https://doi.org/10.1098/rsos.190853
Arakawa T, Ue S, Sano C, Matsunaga M, Kagami H, Yoshida Y, Kuroda Y, Taguchi K, Kitazaki K, Kubo T (2019b) Identification and characterization of a semi-dominant restorer-of-fertility 1 allele in sugar beet (Beta vulgaris). Theor Appl Genet 132:227–240. https://doi.org/10.1007/s00122-018-3211-6
Arakawa T, Kagami H, Katsuyama T, Kitazaki K, Kubo T (2020a) A lineage-specific paralog of Oma1 evolved into a gene family from which a suppressor of male sterility-inducing mitochondria emerged in plants. Genome Biol Evol 12:2314–2327. https://doi.org/10.1093/gbe/evaa186
Arakawa T, Matsunaga M, Matsui K, Itoh K, Kuroda Y, Matsuhira H, Kitazaki K, Kubo T (2020b) The molecular basis for allelic differences suggests Restorer-of-fertility 1 is a complex locus in sugar beet (Beta vulgaris L.). BMC Plant Biol 20:503. https://doi.org/10.1186/s12870-020-02721-9
Bailey-Serres J, Hanson DK, Fox TD, Leaver CJ (1986) Mitochondrial genome rearrangement leads to extension and relocation of the cytochrome c oxidase subunit I gene in sorghum. Cell 47:567–576. https://doi.org/10.1016/0092-8674(86)90621-5
Balk J, Leaver CJ (2001) The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13:1803–1818. https://doi.org/10.1105/tpc.010116
Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65:415–442. https://doi.org/10.1146/annurev-arplant-050213-040159
Bernhard T, Friedt W, Snowdon RJ, Wittkop B (2017) New insights into genotypic thermodependency of cytoplasmic male sterility for hybrid barley breeding. Plant Breed 136:8–17. https://doi.org/10.1111/pbr.12435
Bernhard T, Koch M, Snowdon RJ, Friedt W, Wittkop B (2019) Undesired fertility restoration in msm1 barley associates with two mTERF genes. Theor Appl Genet 132:1335–1350. https://doi.org/10.1007/s00122-019-03281-9
Bohra A, Jha UC, Adhimoolam P, Bisht D, Singh NP (2016) Cytoplasmic male sterility (CMS) in hybrid breeding in field crops. Plant Cell Rep 35:967–993. https://doi.org/10.1007/s00299-016-1949-3
Bosemark NO (1998) Genetic diversity for male sterility in wild and cultivated beets. In: Frese L, Panella L, Strivastava HM, Lange W (eds) International Beta genetic resources network A report on the 4th international Beta genetic resources workshop and world beta network conference held at the Aegean Agricultural Research Institute. Izmir, Turkey, International Plant Genetic Resources Institute, Rome, pp 44–56
Botella MA, Parker JE, Frost LN, Bittner-Eddy PD, Beynon JL, Daniels MJ, Holub EB, Jones JDG (1998) Three genes of the arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10:1847–1860. https://doi.org/10.1105/tpc.10.11.1847
Brown GG, Formanová N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS (2003) The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J 35:262–272. https://doi.org/10.1046/j.1365-313x.2003.01799.x
Bueckmann H, Thiele K, Schiemann J (2016) Influence of soil moisture and air temperature on the stability of cytoplasmic male sterility (CMS) in maize (Zea mays L.). Agric Sci 7:70. https://doi.org/10.4236/as.2016.72007
Burns D, Scarth R, McVetty P (1991) Temperature and genotypic effects on the expression of pol cytoplasmic male sterility in summer rape. Can J Plant Sci 71:655–661. https://doi.org/10.4141/cjps91-097
Chaumont F, Bernier B, Buxant R, Williams ME, Levings C 3rd, Boutry M (1995) Targeting the maize T-urf13 product into tobacco mitochondria confers methomyl sensitivity to mitochondrial respiration. Proc Natl Acad Sci U S A 92:1167–1171. https://doi.org/10.1073/pnas.92.4.1167
Chen L, Liu Y-G (2014) Male sterility and fertility restoration in crops. Annu Rev Plant Biol 65:579–606. https://doi.org/10.1146/annurev-arplant-050213-040119
Chen Z, Zhao N, Li S, Grover CE, Nie H, Wendel JF, Hua J (2017) Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit Rev Plant Sci 36:55–69. https://doi.org/10.1080/07352689.2017.1327762
Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC (2013) Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155:160–171. https://doi.org/10.1016/j.cell.2013.08.032
Cui X, Wise RP, Schnable PS (1996) The rf2 Nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272:1334–1336. https://doi.org/10.1126/science.272.5266.1334
Dahan J, Mireau H (2013) The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol 10:1469–1476. https://doi.org/10.4161/rna.25568
Darracq A, Varré J-S, Maréchal-Drouard L, Courseaux A, Castric V, Saumitou-Laprade P, Oztas S, Lenoble P, Vacherie B, Barbe V (2011) Structural and content diversity of mitochondrial genome in beet: a comparative genomic analysis. Genome Biol Evol 3:723–736. https://doi.org/10.1093/gbe/evr042
Desloire S, Gherbi H, Laloui W, Marhadour S, Clouet V, Cattolico L, Falentin C, Giancola S, Renard M, Budar F (2003) Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep 4:588–594. https://doi.org/10.1038/sj.embor.embor848
Dewey R, Timothy D, Levings C III (1987) A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proc Natl Acad Sci U S A 84:5374–5378. https://doi.org/10.1073/pnas.84.15.5374
Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q (2012) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci U S A 109:2654–2659. https://doi.org/10.1073/pnas.1121374109
Ding X, Guo Q, Li Q, Gai J, Yang S (2020) Comparative transcriptomics analysis and functional study reveal important role of high-temperature stress response gene GmHSFA2 during flower bud development of CMS-based F1 in soybean. Front Plant Sci 11:600217. https://doi.org/10.3389/fpls.2020.600217
Ding X, Guo J, Zhang Q, Yu L, Zhao T, Yang S (2021) Heat-responsive miRNAs participate in the regulation of male fertility stability in soybean CMS-Based F1 under high temperature stress. Int J Mol Sci 22:2446. https://doi.org/10.3390/ijms22052446
Downes R, Marshall D (1971) Low temperature induced male sterility in Sorghum bicolor. Aust J Exp Agric 11:352–356. https://doi.org/10.1071/EA9710352
Ducos E, Touzet P, Boutry M (2001) The male sterile G cytoplasm of wild beet displays modified mitochondrial respiratory complexes. Plant J 26:171–180. https://doi.org/10.1046/j.1365-313x.2001.01017.x
Dufay M, Champelovier P, Käfer J, Henry J-P, Mousset S, Marais G (2014) An angiosperm-wide analysis of the gynodioecy–dioecy pathway. Ann Bot 114:539–548. https://doi.org/10.1093/aob/mcu134
Duroc Y, Gaillard C, Hiard S, Defrance M-C, Pelletier G, Budar F (2005) Biochemical and functional characterization of ORF138, a mitochondrial protein responsible for Ogura cytoplasmic male sterility in Brassiceae. Biochimie 87:1089–1100. https://doi.org/10.1016/j.biochi.2005.05.009
Duroc Y, Hiard S, Vrielynck N, Ragu S, Budar F (2009) The Ogura sterility-inducing protein forms a large complex without interfering with the oxidative phosphorylation components in rapeseed mitochondria. Plant Mol Biol 70:123–137. https://doi.org/10.1007/s11103-009-9461-6
Duvick DN (1965) Cytoplasmic pollen sterility in corn. Adv Genet 13:1–56
Duvick DN (2001) Biotechnology in the 1930s: the development of hybrid maize. Nat Rev Genet 2:69–74. https://doi.org/10.1038/35047587
Elkonin LA, Tsvetova MI (2012) Heritable effect of plant water availability conditions on restoration of male fertility in the “9E” CMS-inducing cytoplasm of sorghum. Front Plant Sci 3:91. https://doi.org/10.3389/fpls.2012.00091
Feng Y, Zheng Q, Song H, Wang Y, Wang H, Jiang L, Yan J, Zheng Y, Yue B (2015) Multiple loci not only Rf3 involved in the restoration ability of pollen fertility, anther exsertion and pollen shedding to S type cytoplasmic male sterile in maize. Theor Appl Genet 128:2341–2350. https://doi.org/10.1007/s00122-015-2589-7
Flavell R (1974) A model for the mechanism of cytoplasmic male sterility in plants, with special reference to maize. Plant Sci Lett 3:259–263. https://doi.org/10.1016/0304-4211(74)90096-0
Fujii S, Toriyama K (2009) Suppressed expression of RETROGRADE-REGULATED MALE STERILITY restores pollen fertility in cytoplasmic male sterile rice plants. Proc Natl Acad Sci U S A 106:9513–9518. https://doi.org/10.1073/pnas.0901860106
Fujii S, Bond CS, Small ID (2011) Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc Natl Acad Sci U S A 108:1723–1728. https://doi.org/10.1073/pnas.1007667108
Gabay-Laughnan S, Kuzmin EV, Monroe J, Roark L, Newton KJ (2009) Characterization of a novel thermosensitive restorer of fertility for cytoplasmic male sterility in maize. Genetics 182:91–103. https://doi.org/10.1534/genetics.108.099895
Gaborieau L, Brown GG, Mireau H (2016) The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Front Plant Sci 7:1816. https://doi.org/10.3389/fpls.2016.01816
Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28:1091–1101. https://doi.org/10.1002/bies.20493
Geddy R, Mahé L, Brown GG (2005) Cell-specific regulation of a Brassica napus CMS-associated gene by a nuclear restorer with related effects on a floral homeotic gene promoter. Plant J 41:333–345. https://doi.org/10.1111/j.1365-313X.2004.02305.x
Gillman JD, Bentolila S, Hanson MR (2007) The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J 49:217–227. https://doi.org/10.1111/j.1365-313X.2006.02953.x
Greiner S, Sobanski J, Bock R (2015) Why are most organelle genomes transmitted maternally? BioEssays 37:80–94. https://doi.org/10.1002/bies.201400110
Hackauf B, Bauer E, Korzun V, Miedaner T (2017) Fine mapping of the restorer gene Rfp3 from an Iranian primitive rye (Secale cereale L.). Theor Appl Genet 130:1179–1189. https://doi.org/10.1007/s00122-017-2879-3
Handa H, Gualberto JM, Grienenberger J-M (1995) Characterization of the mitochondrial orfB gene and its derivative, orf224, a chimeric open reading frame specific to one mitochondrial genome of the “Polima” male-sterile cytoplasm in rapeseed (Brassica napus L.). Curr Genet 28:546–552. https://doi.org/10.1007/BF00518167
He S, Abad AR, Gelvin SB, Mackenzie SA (1996) A cytoplasmic male sterility-associated mitochondrial protein causes pollen disruption in transgenic tobacco. Proc Natl Acad Sci U S A 93:11763–11768. https://doi.org/10.1073/pnas.93.21.11763
Horn R, Gupta KJ, Colombo N (2014) Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 19:198–205. https://doi.org/10.1016/j.mito.2014.04.004
Hu J, Wang K, Huang W, Liu G, Gao Y, Wang J, Huang Q, Ji Y, Qin X, Wan L (2012) The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24:109–122. https://doi.org/10.1105/tpc.111.093211
Huang J-Z, Zhang H-L, Shu Q-Y (2014) Workable male sterility systems for hybrid rice: genetics, biochemistry, molecular biology, and utilization. Rice 7:1–14. https://doi.org/10.1186/s12284-014-0013-6
Itabashi E, Iwata N, Fujii S, Kazama T, Toriyama K (2011) The fertility restorer gene, Rf2, for Lead Rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. Plant J 65:359–367. https://doi.org/10.1111/j.1365-313X.2010.04427.x
Janska H, Sarria R, Woloszynska M, Arrieta-Montiel M, Mackenzie SA (1998) Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10:1163–1180. https://doi.org/10.1105/tpc.10.7.1163
Jaqueth JS, Hou Z, Zheng P, Ren R, Nagel BA, Cutter G, Niu X, Vollbrecht E, Greene TW, Kumpatla SP (2020) Fertility restoration of maize CMS-C altered by a single amino acid substitution within the Rf4 bHLH transcription factor. Plant J 101:101–111. https://doi.org/10.1111/tpj.14521
Jiang H, Lu Q, Qiu S, Yu H, Wang Z, Yu Z, Lu Y, Wang L, Xia F, Wu Y (2022) Fujian cytoplasmic male sterility and the fertility restorer gene OsRf19 provide a promising breeding system for hybrid rice. Proc Natl Acad Sci U S A 119:e2208759119. https://doi.org/10.1073/pnas.2208759119
Jing B, Heng S, Tong D, Wan Z, Fu T, Tu J, Ma C, Yi B, Wen J, Shen J (2012) A male sterility-associated cytotoxic protein ORF288 in Brassica juncea causes aborted pollen development. J Exp Bot 63:1285–1295. https://doi.org/10.1093/jxb/err355
Jones H (1936) A male sterile onion. Proc Am Soc Hortic Sc 34:582–585
Jones H (1943) Inheritance of male sterility in the onion and the production of hybrid seed. Proc Am Soc Hortic Sc 43:189–194
Kaul MLH (1988) Male sterility in higher plants. Springer, New York, pp 3–14
Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K (2008) Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J 55:619–628. https://doi.org/10.1111/j.1365-313X.2008.03529.x
Kazama T, Itabashi E, Fujii S, Nakamura T, Toriyama K (2016) Mitochondrial ORF79 levels determine pollen abortion in cytoplasmic male sterile rice. Plant J 85:707–716. https://doi.org/10.1111/tpj.13135
Kazama T, Okuno M, Watari Y, Yanase S, Koizuka C, Tsuruta Y, Sugaya H, Toyoda A, Itoh T, Tsutsumi N (2019) Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat Plants 5:722–730. https://doi.org/10.1038/s41477-019-0459-z
Kim YJ, Zhang D (2018) Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci 23:53–65. https://doi.org/10.1016/j.tplants.2017.10.001
Kim DH, Kang JG, Kim B-D (2007) Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.). Plant Mol Biol 63:519–532. https://doi.org/10.1007/s11103-006-9106-y
Kim YM, Jo YD, Kwon J-K, Kim IT, Kang B-C (2013) Characterization and inheritance of a novel thermo-sensitive restoration of cytoplasmic male sterility in Capsicum annuum. Sci Hortic 164:512–520. https://doi.org/10.1016/j.scienta.2013.10.014
Kitazaki K, Arakawa T, Matsunaga M, Yui-Kurino R, Matsuhira H, Mikami T, Kubo T (2015) Post-translational mechanisms are associated with fertility restoration of cytoplasmic male sterility in sugar beet (Beta vulgaris). Plant J 83:290–299. https://doi.org/10.1111/tpj.12888
Klein RR, Klein PE, Mullet JE, Minx P, Rooney WL, Schertz KF (2005) Fertility restorer locus Rf1of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor Appl Genet 111:994–1012. https://doi.org/10.1007/s00122-005-2011-y
Köhler RH, Horn R, Lössl A, Zetsche K (1991) Cytoplasmic male sterility in sunflower is correlated with the co-transcription of a new open reading frame with the atpA gene. Mol Gen Genet 227:369–376. https://doi.org/10.1007/BF00273925
Kohls S, Stamp P, Knaak C, Messmer R (2011) QTL involved in the partial restoration of male fertility of C-type cytoplasmic male sterility in maize. Theor Appl Genet 123:327–338. https://doi.org/10.1007/s00122-011-1586-8
Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J (2003) Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J 34:407–415. https://doi.org/10.1046/j.1365-313x.2003.01735.x
Kojima H, Kazama T, Fujii S, Toriyama K (2010) Cytoplasmic male sterility-associated ORF79 is toxic to plant regeneration when expressed with mitochondrial targeting sequence of ATPase γ subunit. Plant Biotechnol 27:111–114. https://doi.org/10.5511/plantbiotechnology.27.111
Kubo T, Arakawa T, Honma Y, Kitazaki K (2020) What does the molecular genetics of different types of restorer-of-fertility genes imply? Plants 9:361. https://doi.org/10.3390/plants9030361
Kuwabara K, Arimura S-i, Shirasawa K, Ariizumi T (2022) orf137 triggers cytoplasmic male sterility in tomato. Plant Phys 189:465–468. https://doi.org/10.1093/plphys/kiac082
Laser KD, Lersten NR (1972) Anatomy and cytology of microsporogenesis in cytoplasmic male sterile angiosperms. Bot Rev 38:425–454. https://doi.org/10.1007/BF02860010
Levings C 3rd (1993) Thoughts on cytoplasmic male sterility in cms-T maize. Plant Cell 5:1285. https://doi.org/10.2307/3869781
Linke B, Nothnagel T, Börner T (2003) Flower development in carrot CMS plants: mitochondria affect the expression of MADS box genes homologous to GLOBOSA and DEFICIENS. Plant J 34:27–37. https://doi.org/10.1046/j.1365-313X.2003.01703.x
Liu F, Schnable PS (2002) Functional specialization of maize mitochondrial aldehyde dehydrogenases. Plant Physiol 130:1657–1674. https://doi.org/10.1104/pp.012336
Liu F, Cui X, Horner HT, Weiner H, Schnable PS (2001) Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. Plant Cell 13:1063–1078. https://doi.org/10.1105/tpc.13.5.1063
Liu G, Tian H, Huang Y-Q, Hu J, Ji Y-X, Li S-Q, Feng Y-Q, Guo L, Zhu Y-G (2012) Alterations of mitochondrial protein assembly and jasmonic acid biosynthesis pathway in Honglian (HL)-type cytoplasmic male sterility rice. J Biol Chem 287:40051–40060. https://doi.org/10.1074/jbc.M112.382549
Logan DC (2006) The mitochondrial compartment. J Exp Bot 57:1225–1243. https://doi.org/10.1093/jxb/erj151
Luo D, Xu H, Liu Z, Guo J, Li H, Chen L, Fang C, Zhang Q, Bai M, Yao N, Wu H, Wu H, Ji C, Zheng H, Chen Y, Ye S, Li X, Zhao X, Li R, Liu Y-G (2013) A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet 45:573–577. https://doi.org/10.1038/ng.2570
Makarenko MS, Usatov AV, Tatarinova TV, Azarin KV, Logacheva MD, Gavrilova VA, Horn R (2019) Characterization of the mitochondrial genome of the MAX1 type of cytoplasmic male-sterile sunflower. BMC Plant Biol 19:41–47. https://doi.org/10.1186/s12870-019-1637-x
Marienfeld JR, Unseld M, Brandt P, Brennicke A (1997) Mosaic open reading frames in the Arabidopsis thaliana mitochondrial genome. Biol Chem 378:859–892. https://doi.org/10.1515/bchm.1997.378.8.859
Marinković R, Miller JF (1995) A new cytoplasmic male sterility source from wild Helianthus annuus. Euphytica 82:39–42. https://doi.org/10.1007/BF00028708
Matsuhira H, Kagami H, Kurata M, Kitazaki K, Matsunaga M, Hamaguchi Y, Hagihara E, Ueda M, Harada M, Muramatsu A, Yui-Kurino R, Taguchi K, Tamagake H, Mikami T, Kubo T (2012) Unusual and typical features of a novel Restorer-of-Fertility Gene of Sugar Beet (Beta vulgaris L.). Genetics 192:1347–1358. https://doi.org/10.1534/genetics.112.145409
Matsuhira H, Kitazaki K, Matsui K, Kubota K, Kuroda Y, Kubo T (2022) Selection of nuclear genotypes associated with the thermo-sensitivity of Owen-type cytoplasmic male sterility in sugar beet (Beta vulgaris L.). Theor Appl Genet 135:1457–1466. https://doi.org/10.1007/s00122-022-04046-7
Melonek J, Small I (2022) Triticeae genome sequences reveal huge expansions of gene families implicated in fertility restoration. Curr Opin Plant Biol 66:102166. https://doi.org/10.1016/j.pbi.2021.102166
Melonek J, Zhou R, Bayer PE, Edwards D, Stein N, Small I (2019) High intraspecific diversity of Restorer-of-fertility-like genes in barley. Plant J 97:281–295. https://doi.org/10.1111/tpj.14115
Meyer EH, Lehmann C, Boivin S, Brings L, De Cauwer I, Bock R, Kühn K, Touzet P (2018) CMS-G from Beta vulgaris ssp. maritima is maintained in natural populations despite containing an atypical cytochrome c oxidase. Biochem J 475:759–773. https://doi.org/10.1042/BCJ20170655
Mikami T, Kishima Y, Sugiura M, Kinoshita T (1985) Organelle genome diversity in sugar beet with normal and different sources of male sterile cytoplasms. Theor Appl Genet 71:166–171. https://doi.org/10.1007/BF00252051
Mogensen HL (1996) Invited special paper: the hows and whys of cytoplasmic inheritance in seed plants. Am J Bot 83:383–404. https://doi.org/10.2307/2446172
Møller IM, Rasmusson AG, Van Aken O (2021) Plant mitochondria–past, present and future. Plant J 108:912–959. https://doi.org/10.1111/tpj.15495
Moritani M, Taguchi K, Kitazaki K, Matsuhira H, Katsuyama T, Mikami T, Kubo T (2013) Identification of the predominant nonrestoring allele for Owen-type cytoplasmic male sterility in sugar beet (Beta vulgaris L.): development of molecular markers for the maintainer genotype. Mol Breeding 32:91–100. https://doi.org/10.1007/s11032-013-9854-8
Murai K, Tsunewaki K (1993) Photoperiod-sensitive cytoplasmic male sterility in wheat with Aegilops crassa cytoplasm. Euphytica 67:41–48. https://doi.org/10.1007/BF00022723
Murai K, Takumi S, Koga H, Ogihara Y (2002) Pistillody, homeotic transformation of stamens into pistil-like structures, caused by nuclear-cytoplasm interaction in wheat. Plant J 29:168–181. https://doi.org/10.1046/j.0960-7412.2001.01203.x
Murai K, Tsutui I, Kawanishi Y, Ikeguchi S, Yanaka M, Ishikawa N (2008) Development of photoperiod-sensitive cytoplasmic male sterile (PCMS) wheat lines showing high male sterility under long-day conditions and high seed fertility under short-day conditions. Euphytica 159:315–323. https://doi.org/10.1007/s10681-007-9510-7
Nakai S, Noda D, Kondo M, Terachi T (1995) High-level expression of a mitochondrial orf 522 gene from the male-sterile sunflower is lethal to E. Coli Jpn J Breed 45:233–236. https://doi.org/10.1270/jsbbs1951.45.233
Nakazato I, Okuno M, Zhou C, Itoh T, Tsutsumi N, Takenaka M, Arimura S (2022) Targeted base editing in the mitochondrial genome of Arabidopsis thaliana. Proc Natl Acad Sci U S A 119:e2121177119. https://doi.org/10.1073/pnas.2121177119
Nguyen T-T, Planchard N, Dahan J, Arnal N, Balzergue S, Benamar A, Bertin P, Brunaud V, Dargel-Graffin C, Macherel D, Martin-Magniette M-L, Quadrado M, Namy O, Mireau H (2021) A case of gene fragmentation in plant mitochondria fixed by the selection of a compensatory restorer of fertility-like PPR gene. Mol Biol Evol 38:3445–3458. https://doi.org/10.1093/molbev/msab115
Nivison HT, Sutton CA, Wilson RK, Hanson MR (1994) Sequencing, processing, and localization of the petunia CMS-associated mitochondrial protein. Plant J 5:613–623. https://doi.org/10.1111/j.1365-313x.1994.00613.x
Ohgami T, Uchiyama D, Ue S, Yui-Kurino R, Yoshida Y, Kamei Y, Kuroda Y, Taguchi K, Kubo T (2016) Identification of molecular variants of the nonrestoring restorer-of-fertility 1 allele in sugar beet (Beta vulgaris L.). Theor Appl Genet 129:675–688. https://doi.org/10.1007/s00122-015-2656-0
Okazaki M, Kazama T, Murata H, Motomura K, Toriyama K (2013) Whole mitochondrial genome sequencing and transcriptional analysis to uncover an RT102-type cytoplasmic male sterility-associated candidate gene derived from Oryza rufipogon. Plant Cell Physiol 54:1560–1568. https://doi.org/10.1093/pcp/pct102
Omukai S, Arimura S-i, Toriyama K, Kazama T (2021) Disruption of mitochondrial open reading frame 352 partially restores pollen development in cytoplasmic male sterile rice. Plant Physiol 187:236–246. https://doi.org/10.1093/plphys/kiab236
Onodera Y, Arakawa T, Yui-Kurino R, Yamamoto MP, Kitazaki K, Ebe S, Matsunaga M, Taguchi K, Kuroda Y, Yamashita S, Sakai T, Kinoshita T, Mikami T, Kubo T (2015) Two male sterility-inducing cytoplasms of beet (Beta vulgaris) are genetically distinct but have closely related mitochondrial genomes: implication of a substoichiometric mitochondrial DNA molecule in their evolution. Euphytica 206:365–379. https://doi.org/10.1007/s10681-015-1484-2
Owen F (1945) Cytoplasmically inherited male-sterility in sugar beets. J Agric Res 71:423–440
Park JY, Lee YP, Lee J, Choi BS, Kim S, Yang TJ (2013) Complete mitochondrial genome sequence and identification of a candidate gene responsible for cytoplasmic male sterility in radish (Raphanus sativus L.) containing DCGMS cytoplasm. Theor Appl Genet 126:1763–1774. https://doi.org/10.1007/s00122-013-2090-0
Pelletier G, Budar F (2015) Brassica Ogu-INRA cytoplasmic male sterility: an example of successful plant somatic fusion for hybrid seed production. In: Li XQ, Donnelly D, Jensen T (eds) Somatic genome manipulation. Springer, New York, pp 199–216. https://doi.org/10.1007/978-1-4939-2389-2_9
Pfeiffer P (1998) Nucleotide sequence, genetic organization and expression strategy of the double-stranded RNA associated with the “447” cytoplasmic male sterility trait in Vicia faba. J Gen Virol 79:2349–2358. https://doi.org/10.1099/0022-1317-79-10-2349
Qin X, Warguchuk R, Arnal N, Gaborieau L, Mireau H, Brown GG (2014) In vivo functional analysis of a nuclear restorer PPR protein. BMC Plant Biol 14:1–12. https://doi.org/10.1186/s12870-014-0313-4
Quesada V (2016) The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol Plant 157:389–399. https://doi.org/10.1111/ppl.12416
Rathburn HB, Hedgcoth C (1991) A chimeric open reading frame in the 5′ flanking region of coxI mitochondrial DNA from cytoplasmic male-sterile wheat. Plant Mol Biol 16:909–912. https://doi.org/10.1007/BF00015083
Reddy BB, Reddi M (1970) Studies on the breakdown of male sterility and other related aspects in certain cytoplasmic male-sterile lines of pearl millet (Pennisetum typhoides Stapf and Hubb). Andhra Agric J 17:173–180
Reddy C, Sinha B, Reddy A, Reddy Y (2008) Maintenance of male sterility and fertility restoration in different CMS sources of sunflower (Helianthus annuus L.). Asian J Plant Sci 7:762–766. https://doi.org/10.3923/ajps.2008.762.766
Rhoads DM, Brunner-Neuenschwander B, Levings CS, Siedow JN (1998) Cross-linking and disulfide bond formation of introduced cysteine residues suggest a modified model for the tertiary structure of URF13 in the pore-forming oligomers. Arch Biochem Biophys 354:158–164. https://doi.org/10.1006/abbi.1998.0671
Rizzolatti C, Bury P, Tatara E, Pin PA, Rodde N, Bergès H, Budar F, Mireau H, Gielen JJL (2017) Map-based cloning of the fertility restoration locus Rfm1 in cultivated barley (Hordeum vulgare). Euphytica 213:276. https://doi.org/10.1007/s10681-017-2056-4
Rogers JS, Edwardson JR (1952) The utilization of cytoplasmic male-sterile inbreds in the production of corn hybrids 1. Agron J 44:8–13. https://doi.org/10.2134/agronj1952.00021962004400010004x
Sabar M, Gagliardi D, Balk J, Leaver CJ (2003) ORFB is a subunit of F1 FO-ATP synthase: insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep 4:381–386. https://doi.org/10.1038/sj.embor.embor800
Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, Mikami T (2004) The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Genet Genomics 272:247–256. https://doi.org/10.1007/s00438-004-1058-9
Sattari M, Kathiresan A, Gregorio GB, Hernandez JE, Nas TM, Virmani SS (2007) Development and use of a two-gene marker-aided selection system for fertility restorer genes in rice. Euphytica 153:35–42. https://doi.org/10.1007/s10681-006-9213-5
Saxena KB, Choudhary AK, Saxena RK, Varshney RK (2018) Breeding pigeonpea cultivars for intercropping: synthesis and strategies. Breed Sci 68:159–167. https://doi.org/10.1270/jsbbs.17105
Scheffler IE (1999) Mitochondria. Wiley, New York. https://doi.org/10.1002/0471223891
Schnable PS, Wise RP (1998) The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci 3:175–180. https://doi.org/10.1016/S1360-1385(98)01235-7
Small ID, Schallenberg-Rüdinger M, Takenaka M, Mireau H, Ostersetzer-Biran O (2019) Plant organellar RNA editing: what 30 years of research has revealed. Plant J 101:1040–1056. https://doi.org/10.1111/tpj.14578
Song J, Hedgcoth C (1994) Influence of nuclear background on transcription of a chimeric gene (orf256) and coxI in fertile and cytoplasmic male sterile wheats. Genome 37:203–209. https://doi.org/10.1139/g94-028
Stone JD, Koloušková P, Sloan DB, Štorchová H (2017) Non-coding RNA may be associated with cytoplasmic male sterility in Silene vulgaris. J Exp Bot 68:1599–1612. https://doi.org/10.1093/jxb/erx057
Su A, Song W, Xing J, Zhao Y, Zhang R, Li C, Duan M, Luo M, Shi Z, Zhao J (2016) Identification of genes potentially associated with the fertility instability of S-type cytoplasmic male sterility in maize via bulked segregant RNA-Seq. PLoS ONE 11:e0163489. https://doi.org/10.1371/journal.pone.0163489
Suketomo C, Kazama T, Toriyama K (2020) Fertility restoration of Chinese wild rice-type cytoplasmic male sterility by CRISPR/Cas9-mediated genome editing of nuclear-encoded RETROGRADE-REGULATED MALE STERILITY. Plant Biotechnol 20:0326b. https://doi.org/10.5511/plantbiotechnology.20.0326b
Sweetlove LJ, Fait A, Nunes-Nesi A, Williams T, Fernie AR (2007) The mitochondrion: an integration point of cellular metabolism and signalling. Crit Rev Plant Sci 26:17–43. https://doi.org/10.1080/07352680601147919
Takatsuka A, Kazama T, Si A, Toriyama K (2022) TALEN-mediated depletion of the mitochondrial gene orf312 proves that it is a Tadukan-type cytoplasmic male sterility-causative gene in rice. Plant J 110:994–1004. https://doi.org/10.1111/tpj.15715
Tang HV, Pring DR, Shaw LC, Salazar RA, Muza FR, Yan B, Schertz KF (1996) Transcript processing internal to a mitochondrial open reading frame is correlated with fertility restoration in male-sterile sorghum. Plant J 10:123–133. https://doi.org/10.1046/j.1365-313X.1996.10010123.x
Tang H, Luo D, Zhou D, Zhang Q, Tian D, Zheng X, Chen L, Liu Y-G (2014) The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol Plant 7:1497–1500. https://doi.org/10.1093/mp/ssu047
Tang H, Zheng X, Li C, Xie X, Chen Y, Chen L, Zhao X, Zheng H, Zhou J, Ye S (2017) Multi-step formation, evolution, and functionalization of new cytoplasmic male sterility genes in the plant mitochondrial genomes. Cell Res 27:130–146. https://doi.org/10.1038/cr.2016.115
Tariq N, Yaseen M, Dawei X, Rehman HM, Bibi M, Uzair M (2022) Rice anther tapetum; a vital reproductive cell layer for sporopollenin biosynthesis and pollen exine patterning. Plant Biol. https://doi.org/10.1111/plb.13485
Tie S, Xia J, Qiu F, Zheng Y (2006) Genome-wide analysis of maize cytoplasmic male sterility-S based on QTL mapping. Plant Mol Biol Rep 24:71–80. https://doi.org/10.1007/BF02914047
Toriyama K (2021) Molecular basis of cytoplasmic male sterility and fertility restoration in rice. Plant Biotechnol 38:285–295. https://doi.org/10.5511/plantbiotechnology.21.0607a
Touzet P (2012) Mitochondrial genome evolution and gynodioecy. In: Marechal-Drouard L (ed) Mitochondrial genome evolution. Academic Press, Oxford, pp 71–98. https://doi.org/10.1016/B978-0-12-394279-1.00004-1
Touzet P, Meyer EH (2014) Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion 19:166–171. https://doi.org/10.1016/j.mito.2014.04.009
Uyttewaal M, Arnal N, Quadrado M, Martin-Canadell A, Vrielynck N, Hiard S, Gherbi H, Bendahmane A, Budar F, Mireau H (2008) Characterization of Raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for Ogura cytoplasmic male sterility. Plant Cell 20:3331–3345. https://doi.org/10.1105/tpc.107.057208
Van der Veken J, Vandermoere S, Maudoux O, Notté C, Eeckhaut T, Baert J, Werbrouck S, Van Huylenbroeck J (2018) Temperature related instability of a naturally induced cytoplasmic male sterility in industrial chicory (Cichorium intybus L.). Plant Breed 137:423–432. https://doi.org/10.1111/pbr.12582
Vong M, Manny AR, Smith KL, Gao W, Nibert ML (2019) Beta vulgaris mitovirus 1 in diverse cultivars of beet and chard. Virus Res 265:80–87. https://doi.org/10.1016/j.virusres.2019.02.008
Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D (2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18:676–687. https://doi.org/10.1105/tpc.105.038240
Wang K, Gao F, Ji Y, Liu Y, Dan Z, Yang P, Zhu Y, Li S (2013) ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice. New Phytol 198:408–418. https://doi.org/10.1111/nph.12180
Wang X, Guan Z, Gong Z, Yan J, Yang G, Liu Y-G, Yin P (2018) Crystal structure of WA352 provides insight into cytoplasmic male sterility in rice. Biochem Biophys Res Commun 501:898–904. https://doi.org/10.1016/j.bbrc.2018.05.079
Wang Y, Wang Q, Hao W, Li J, Qi M, Zhang L (2020) Mitochondrial genome sequencing reveals orf463a may induce male sterility in NWB cytoplasm of radish. Genes 11:74. https://doi.org/10.3390/genes11010074
Wang C, Lezhneva L, Arnal N, Quadrado M, Mireau H (2021) The radish Ogura fertility restorer impedes translation elongation along its cognate CMS-causing mRNA. Proc Natl Acad Sci U S A 118:e2105274118. https://doi.org/10.1073/pnas.2105274118
Wesołowski W, Szklarczyk M, Szalonek M, Słowińska J (2015) Analysis of the mitochondrial proteome in cytoplasmic male-sterile and male-fertile beets. J Proteomics 119:61–74. https://doi.org/10.1016/j.jprot.2014.12.013
Wintz H, Chen H-C, Sutton CA, Conley CA, Cobb A, Ruth D, Hanson MR (1995) Expression of the CMS-associated urfS sequence in transgenic petunia and tobacco. Plant Mol Biol 28:83–92. https://doi.org/10.1007/BF00042040
Wise RP, Pring DR, Gengenbach BG (1987) Mutation to male fertility and toxin insensitivity in Texas (T)-cytoplasm maize is associated with a frameshift in a mitochondrial open reading frame. Proc Natl Acad Sci U S A 84:2858–2862. https://doi.org/10.1073/pnas.84.9.2858
Xiao S, Xing J, Nie T, Su A, Zhang R, Zhao Y, Song W, Zhao J (2022) Comparative analysis of mitochondrial genomes of maize CMS-S subtypes provides new insights into male sterility stability. BMC Plant Biol 22:469. https://doi.org/10.1186/s12870-022-03849-6
Xu F, Yang X, Zhao N, Hu Z, Mackenzie SA, Zhang M, Yang J (2022) Exploiting sterility and fertility variation in cytoplasmic male sterile vegetable crops. Hortic Res. https://doi.org/10.1093/hr/uhab039
Yamagishi H, Bhat SR (2014) Cytoplasmic male sterility in Brassicaceae crops. Breed Sci 64:38–47. https://doi.org/10.1270/jsbbs.64.38
Yamagishi H, Jikuya M, Okushiro K, Hashimoto A, Fukunaga A, Takenaka M, Terachi T (2021) A single nucleotide substitution in the coding region of Ogura male sterile gene, orf138, determines the effectiveness of a fertility restorer gene, Rfo, in radish. Mol Genet Genomics 296:705–717. https://doi.org/10.1007/s00438-021-01777-y
Yamamoto MP, Kubo T, Mikami T (2005) The 5′-leader sequence of sugar beet mitochondrial atp6 encodes a novel polypeptide that is characteristic of Owen cytoplasmic male sterility. Mol Gen Genet 273:342–349. https://doi.org/10.1007/s00438-005-1140-y
Yamamoto MP, Shinada H, Onodera Y, Komaki C, Mikami T, Kubo T (2008) A male sterility-associated mitochondrial protein in wild beets causes pollen disruption in transgenic plants. Plant J 54:1027–1036. https://doi.org/10.1111/j.1365-313X.2008.03473.x
Yang J, Liu X, Yang X, Zhang M (2010) Mitochondrially-targeted expression of a cytoplasmic male sterility-associated orf220 gene causes male sterility in Brassica juncea. BMC Plant Biol 10:1–10. https://doi.org/10.1186/1471-2229-10-231
Young ND (2000) The genetic architecture of resistance. Curr Opin Plant Biol 3:285–290. https://doi.org/10.1016/s1369-5266(00)00081-9
Yu C, Wang L, Xu S, Zeng Y, He C, Chen C, Huang W, Zhu Y, Hu J (2015) Mitochondrial ORFH79 is essential for drought and salt tolerance in rice. Plant Cell Physiol 56:2248–2258. https://doi.org/10.1093/pcp/pcv137
Zhang H, Che J, Ge Y, Pei Y, Zhang L, Liu Q, Gu M, Tang S (2017) Ability of Rf5 and Rf6 to Restore Fertility of Chinsurah Boro II-type Cytoplasmic Male Sterile Oryza sativa (ssp Japonica) Lines. Rice 10:2. https://doi.org/10.1186/s12284-017-0142-9
Zhong X, Chen D, Cui J, Li H, Huang Y, Kang J (2021) Comparative analysis of the complete mitochondrial genome sequences and anther development cytology between maintainer and Ogura-type cytoplasm male-sterile cabbage (B. oleracea Var. capitata). BMC Genomics 22:1–15. https://doi.org/10.1186/s12864-021-07963-x
Acknowledgements
The authors thank Prof. Dr. Tomohiko Kubo for his support.
Funding
This work was supported in part by NARO Bio-oriented Technology Research Advancement Institution (BRAIN) (Research program on development of innovative technology, Grant Number 30001A) to KK and the Japan Society for the Promotion of Science, Grant-in-Aid for Scientific Research, Grant Numbers 21H02159 (to KK), 22K05569 (to KK), and 22H02267 (to KK).
Author information
Authors and Affiliations
Contributions
KK conceived the idea and took the lead in writing the manuscript. KO, AA and RI collected and listed relevantarticles as references. All authors reviewed the manuscript draft and revised it. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Rajeev K. Varshney.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kitazaki, K., Oda, K., Akazawa, A. et al. Molecular genetics of cytoplasmic male sterility and restorer-of-fertility for the fine tuning of pollen production in crops. Theor Appl Genet 136, 156 (2023). https://doi.org/10.1007/s00122-023-04398-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04398-8