Abstract
Key message
The qSCN10 locus with broad-spectrum SCN resistance was fine-mapped to a 379-kb region on chromosome 10 in soybean accession PI 567516C. Candidate genes and potential application benefits of this locus were discussed.
Abstract
Soybean cyst nematode (SCN, Heterodera glycines Ichinohe) is one of the most devastating pests of soybean, causing significant yield losses worldwide every year. Genetic resistance has been the major strategy to control this pest. However, the overuse of the same genetic resistance derived primarily from PI 88788 has led to the genetic shifts in nematode populations and resulted in the reduced effectiveness in soybean resistance to SCN. Therefore, novel genetic resistance resources, especially those with broad-spectrum resistance, are needed to develop new resistant cultivars to cope with the genetic shifts in nematode populations. In this study, a quantitative trait locus (QTL) qSCN10 previously identified from a soybean landrace PI 567516C was confirmed to confer resistance to multiple SCN HG Types. This QTL was further fine-mapped to a 379-kb region. There are 51 genes in this region. Four of them are defense-related and were regulated by SCN infection, suggesting their potential role in mediating resistance to SCN. The phylogenetic and haplotype analyses of qSCN10 as well as other information indicate that this locus is different from other reported resistance QTL or genes. There was no yield drag or other unfavorable traits associated with this QTL when near-isogenic lines with and without qSCN10 were tested in a SCN-free field. Therefore, our study not only provides further insight into the genetic basis of soybean resistance to SCN, but also identifies a novel genetic resistance resource for breeding soybean for durable, broad-spectrum resistance to this pest.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean cyst nematode (SCN, Heterodera glycines Ichinohe) is the most damaging pathogen of soybean, causing over $1 billion in yield losses annually in the USA alone (Allen et al. 2017). Furthermore, the nematode continues to spread to other soybean-growing areas in the North America (Tylka and Marett 2017). Therefore, more yield losses are possible. Non-host crop rotation, resistant cultivars, and nematicide applications are conventional strategies for controlling SCN. Among them, the deployment of cultivars with genetic resistance to SCN is the most cost-effective and environment-friendly method to combat this pest. Genetic resistance to SCN in soybean is a multi-genetic and quantitatively inherited trait (Arelli et al. 2009; Vuong et al. 2010; Liu et al. 2012; Yu and Diers 2017). Two major QTL, rhg1 (for resistance to Heterodera glycines 1) and Rhg4, responsible for SCN resistance in numerous soybean germplasm and cultivars, have been precisely localized and extensively characterized (Kim et al. 2010; Cook et al. 2012; Liu et al. 2012, 2017; Lakhssassi et al. 2019). For more than 2 decades, rhg1 from plant introduction (PI) PI 88,788 has been the major resistance introgressed into modern soybean varieties for resistance to SCN (Concibido et al. 2004). Approximately 95% of soybean resistant varieties carry this rhg1 resistance. Unfortunately, the overuse of the same resistance has led to the genetic shifts in nematode populations and resulted in the reduced effectiveness in soybean resistance to SCN (Niblack et al. 2008; Mulrooney et al. 2010; Lian et al. 2016, 2017; Howland et al. 2018). Therefore, other genetic resistance resources or genes are needed for sustainable soybean production for an ever-growing world population. For this purpose, in the past two decades, a large number of soybean accessions have been screened for novel resistance to SCN (Arelli et al. 1997, 2000; Han et al. 2015; Kadam et al. 2016; Zhang et al. 2016; Hua et al. 2018; Klepadlo et al. 2018). As a result, a number of exotic soybean germplasm, such as PI 90763, PI 437654, PI 438489B, PI 464925B, and PI 567516C, have been revealed to be resistant to multiple SCN races or HG types. Furthermore, QTL responsible for the observed resistance have been mapped in some of these accessions (Guo et al. 2005; Winter et al. 2006; Wu et al. 2009; Vuong et al. 2010, 2011).
Among these accessions, PI 567516C was shown to be resistant to multiple SCN HG Types, including 2.5.7 (race 1), 1.2.5.7 (race 2), 0 (race 3), 2.5.7 (race 5), 1.3.6.7 (race 14), and a synthetic nematode population LY1 (HG Type 1.2.3.4.5.6.7) (Young 1998; Vuong et al. 2010). The previous mapping study for this accession using F2:3 progeny derived from a Magellan × PI 567516C cross -identified 2 novel QTL on Chr. 10 (qSCN10) and Chr. 18 (qSCN18), respectively (Vuong et al. 2010). The qSCN10 locus was associated with resistance to 5 HG Types: 2.5.7 (race 1), 0, 2.5.7 (race 5), 1.3.6.7, and 1.2.3.4.5.6.7, with the likelihood of odds (LOD) scores ranging from 6.6 to 13.2 and explained 7.9% to 21.7% of the phenotypic variation. qSCN10 was mapped to a 15.3 cM interval (2,542 kb) between SSR markers Satt592 and Sat_038 on Chr. 10 (Vuong et al. 2010). This locus was also detected in a genome-wide association study (GWAS) using a diverse panel of 553 soybean germplasm (Vuong et al. 2015).
PI 567516C was reported to be genetically different from major resistant sources, such as PI 88788 (Chen et al. 2006; Arelli et al. 2009). According to whole-genome resequencing (WGRS) and haplotype analysis, PI 567516C carries three copies of rhg1, similar to Peking and PI 437654 (Jiao et al. 2015); however, PI 567516C does not contain the specific Rhg4 haplotype that is present in Peking-type resistant accessions (Lakhssassi et al. 2019). The initial mapping showed that qSCN10 was a different locus from the known resistance loci, rhg1 and Rhg4, since it is located on a different chromosome (on chr. 10) from rhg1 (on chr. 18) and Rhg4 (on chr. 8) (Vuong et al. 2010). Therefore, the qSCN10 locus very likely harbors gene(s) with a unique defense mechanism against SCN and shall be a good resistance source for breeding soybean varieties with durable, broad-spectrum resistance to SCN. To ensure the proper application of this unique QTL in breeding, qSCN10 needs to be further confirmed and fine-mapped to a reasonably small region, and to be shown that there are no adverse agronomic traits, especially yield drag, associated with it. For these purposes, our present study not only confirmed the effectiveness of the qSCN10 locus in conferring resistance to multiple SCN HG types, but also narrowed down the QTL interval to a 379-kb region for future breeding application. Furthermore, our study also demonstrated that there was no yield drag or other adverse agronomic traits associated with this QTL. Thus, qSCN10 is an excellent novel genetic resource for breeding soybean for durable, broad-spectrum resistance to SCN.
Materials and methods
Plant materials
An F6:7 recombinant inbred line (RIL) population was previously developed from a cross between the cultivar (cv.) Magellan (PI 595362, SCN susceptible) and PI 567516C (SCN resistant) for QTL mapping study (Vuong et al. 2010). A RIL line MPC2090 was selected as a donor parent based on the homozygous genotype at the target region and the multi-SCN resistance phenotype. In this study, MPC2090 was crossed with Magellan at the Bradford Research and Extension Center (BREC), the University of Missouri, Columbia, MO, USA. Heterozygous F1 plants were backcrossed with the recurrent parent, Magellan, for 4 generations (BC4F1) using marker-assisted backcrossing (MABC) method. Heterozygous BC4F1 plants were self-pollinated to advance the BC4F2 population. Over 900 BC4F2 seedlings planted in BREC and screened for recombinants containing breakpoints in the target qSCN10 region with 2 single nucleotide polymorphism (SNP) markers labeled SNP1 and SNP-O8 (Supplementary Table S1). Out of 972 BC4F2 plants, 159 were identified as recombinants and self-pollinated seeds (BC4F2:3) were harvested. Four pairs of independent segregating lines (BC4F2:3) derived from BC4F2 plants with different heterozygous segments for the target QTL region were selected to develop BC4F3:4 progeny lines of the recombinants (Q041, Q150, Q543, Q660) in a greenhouse, the University of Missouri. These progeny lines were utilized to conduct progeny test of SCN phenotyping for fine-mapping (Fig. 1). Another set of BC4F3:4 progeny lines, Q713-Mgl and Q713-516C, was also developed as described above but carry entire Magellan and PI 567516C segments without a breakpoint at the qSCN10 locus, respectively. Q713-Mgl and 516C were used as near-isogenic lines (NILs) to evaluate SCN resistance of the qSCN10 locus and to test yield drag effects.
A flowchart of qSCN10 fine-mapping and development of near-isogenic lines (NILs) in Magellan background. The construction of recombinant inbred line (RIL) MPC2090 was selected from the previously reported cross of Magellan × PI 567516C (Vuong et al. 2010)
Phenotypic evaluation of SCN resistance
Different SCN populations [HG Types 2.5.7 (race 1), 1.2.5.7, 0, 2.5.7 (race 5), 1.3.6.7, and 1.2.3.4.5.6.7] have been separately maintained in a greenhouse at the University of Missouri–Columbia by reproduction in small population sizes and believed to be near-homogeneous (Arelli et al. 2000). SCN resistance bioassays were conducted following an established method (Arelli et al. 2000; Niblack et al. 2009; Brown et al. 2010). Briefly, seeds were germinated at room temperature for 4 days in paper germination pouches. Seedlings were then transplanted into crocks containing micropots filled with sterilized sandy loam soil. Crocks were placed in water bath tanks with the temperature maintained at 27 °C. Plants were inoculated with about 2000 SCN eggs at 3 days after transplanting and watered daily. After 30 days, the root samples were harvested. During the harvesting procedure, seedling roots were gently swirled in water to remove soil, leaving roots undisturbed. The cysts from each root sample were then flushed with high-pressure water, transferred to a petri dish, scanned, and counted using a fluorescence-based imaging system (Brown et al. 2010). Indicator soybean lines (‘Peking’, PI 88788, PI 90763, PI 437654, PI 209332, PI 89772, PI 54831, and ‘Pickett’) and two standard susceptible controls ‘Hutcheson’ and ‘Lee 74′ were simultaneously evaluated for HG Types (Niblack et al. 2002) and race designation (Schmitt and Shannon 1992). Tested soybean plants were arranged in a randomized complete block design (RCBD) with over ten technical replicates per soybean line for each SCN population.
The female index (FI%) was calculated to evaluate the response of plants to SCN populations: FI (%) = (Number of female cyst nematodes on a given individual/Average number of female nematodes on the susceptible ‘Lee 74′) × 100. Indicator and test lines were scored according to their FI: FI < 10 = resistant (R); FI 10 to < 30 = moderately resistant (MR); FI 30 to 60 = moderately susceptible (MS); and FI > 60 = susceptible (S).
Observation of nematode development in Magellan and PI 567516C roots
To observe the nematode invasion and development in the two parental lines, their seedlings were inoculated with SCN HG Type 0 as described above. Roots were carefully removed from the soil at 2, 4, 6, and 8 days after inoculation (dai). Five root samples of each parent were harvested at each time point. Fresh roots were stained with acid-fuchsin (Byrd et al. 1983). Briefly, roots were washed with tap water and cleared by soaking roots in 5.25% sodium hypochlorite for 4 min. Roots were completely rinsed with running tap water and then boiled in the 37% acid-fuchsin staining solution for about 30 s in a microwave followed by rinsing with running tap water. The roots were then de-stained with boiling glycerol. After staining, the SCN juveniles from the J2–J4 stages inside the roots were morphologically examined with a dissecting microscope (Cook et al. 2012).
DNA extraction, SNP marker and Kompetitive allele-specific PCR (KASP) assay development
Genomic DNA (gDNA) was extracted from two parental lines, Magellan and PI 567516C, and from individual plants of NILs using the CTAB (cetyl trimethyl ammonium bromide) method (Murray and Thompson 1980).
KASP assays (LGC Genomics, UK) were utilized for genotyping of recombinants and NILs. Genome sequence information in the qSCN10 region was obtained from the WGRS data for Magellan (unpublished data) and PI 567516C (Valliyodan et al. 2016). The SNPs identified at this locus using Genome Analysis Toolkit (GATK) (McKenna et al. 2010; https://gatk.broadinstitute.org) software were reconfirmed by examining read alignments in the Integrative Genomics Viewer (IGV) tool (Robinson et al. 2011; https://igv.org/). A set of SNPs were selected, and corresponding primers (2 allele-specific forward primers, along with tail sequences and one common reverse primer) were designed for the development of KASP assays (Supplementary Table S1). KASP reactions were run in 10 µl reaction volume, which included 5 µl of 2 × premade KASP master mix, 5 µl of 10–25 ng/µl gDNA, and 0.14 µl of primers mix. The following cycling conditions were used: 15 min at 95 °C, followed by 10 touch-down cycles of 20 s at 94 °C, 1 min at 65–57 °C (dropping 0.8 °C per cycle) and then 23 cycles of 20 s at 94 °C, 1 min at 57 °C. The fluorescent end-point genotyping method was carried out using a Roche LightCycler 480 instrument (Roche Applied Sciences, Indianapolis, IN, USA).
Expression analysis from RNA-Seq and microarray profiles
Expression patterns of candidate genes in different tissues and under presence/absence of SCN infection in different soybean lines were derived from published RNA-Seq (Severin et al. 2010) and microarray profiles (Wan et al. 2015). In microarray assay by Wan et al. (2015), samples were collected from roots of Magellan and PI 567516C at 0, 3, and 8 days after inoculation with SCN HG Type 0. In RNA-Seq assay by Severin et al (2010), root samples were collected from SCN susceptible cultivar Williams 82 seedlings at 5 and 30 days after inoculation with SCN HG Type 0 or water (mock). Expression profiles were downloaded from the Gene Expression Omnibus (GEO) database in series GSE64492 and GSE125103. The average gene expression levels from three replicates were analyzed through Microsoft Excel and shown as normalized RPKM (reads/Kb/Million) data in RNA-Seq and unscaled expression levels in microarray.
Phylogenetic analysis
A panel of 106 diverse soybean genotypes, including 56 G. max elite lines, 43 G. max landraces, and 7 G. soja accessions (Valliyodan et al. 2016), were explored to understand the origin and evolution of the qSCN10 region. A total of 2,336 SNPs in the delimited 379-kb region were used to build the phylogenetic tree by MEGA X (Kumar et al. 2018). The statistical method was inferred by using the maximum likelihood method and the Tamura-Nei model (Tamura et al. 1993). The tree with the highest log likelihood (− 26,052.38) was shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site.
Yield test of the near-isogenic lines (NILs)
For yield drag test of the qSCN10 locus, abundant seeds were harvested from homozygous BC4F3:4 Q713 NIL plants grown at the BREC, the University of Missouri–Columbia, MO, USA, in 2017. Yield test was conducted under normal growth conditions without SCN stress at BREC in 2018. Five replicates for each line were planted using a randomized complete block design. Around 200 seeds of each replicate were planted in the 2-row plots with 0.76 m row spacing and 3 m length. Seeds were harvested at maturity for yield analysis.
Results
SCN growth and development was restricted in PI 567516C
To reveal any possible differences in SCN infection efficiency between PI 567516C and Magellan, nematode growth and development were monitored over time after inoculation. As shown in Fig. 2, no significant difference was observed between 2 parental lines in the number of nematodes (J2 stage) inside the roots at 2 days after inoculation (dai), indicating that the resistance in PI 567516C did not affect the penetration of nematodes in soybean roots. However, an obvious delay of nematode development was observed in PI 567516C at 4 dai: The nematodes were thinner (in diameter) in PI 567516C than in Magellan. The delay was even more obvious at 8 dai: Approximately half of the nematodes were at the J4 stage in Magellan, but majority (80%) of nematodes were at the J3 stage and only a few early J4 nematodes were found in PI 567516C. Clearly, the growth of nematodes in PI 567516C was restrained, similar to the phenomenon observed in other SCN resistant soybean lines, such as Peking and PI 88788 (Mitchum 2016; Chen et al. 2020).
Observation of SCN (HG Type 0) development in roots of Magellan and PI 567516C. a Representative images of nematode-infected roots at 2, 4, 6, and 8 days after inoculation (dai) were showed. Different developmental stages of SCN in the roots were indicated in the images. J2, second-stage juvenile; J3, third-stage juvenile; and J4, fourth-stage juvenile. b Statistics of constituent ratios of each nematode developmental stage at 2, 4, 6, and 8 dai. Data are represented by the means ± SE
qSCN10 was confirmed to confer resistance to multiple SCN HG types
To validate the qSCN10 locus, the BC4F3:4 Q713 NIL pair along with the 2 parental lines were examined for their response to six SCN HG Types [2.5.7 (race 1), 1.2.5.7, 0, 2.5.7 (race 5), 1.3.6.7, and 1.2.3.4.5.6.7] in a greenhouse. As shown in Fig. 3b, Magellan, as expected, showed susceptibility (S) to all 6 SCN HG Types; meanwhile, PI 567516C, as the source of the qSCN10 locus, showed different levels of resistance to different HG Types: resistant (R) to HG Type 2.5.7 (race 5); moderately resistant (MR) to HG Type 2.5.7 (race 1), 0, and 1.2.3.4.5.6.7; and moderately susceptible (MS) to HG Types 1.2.5.7 and 1.3.6.7. Although NIL Q713-516C did not show strong resistance to these SCN HG Types (MS to HG Type 0 and S to other populations) when compared with Magellan, it showed significant resistance to all these six HG types when compared with NIL Q713-Mgl, because significant reductions of the female index (FI) were observed in Q713-516C when compared with Q713-Mgl: 29, 26, 45, 14, 32, and 26% for HG Type 2.5.7 (race 1), 1.2.5.7, 0, 2.5.7 (race 5), 1.3.6.7, and 1.2.3.4.5.6.7, respectively. These results suggest that the qSCN10 locus confers broad-spectrum and partial resistance to SCN.
SCN resistance evaluation of the pair of near-isogenic lines (NILs) (Q713) and parents. a Schematic graph of chromosome 10 of a pair of Q713 NIL lines and the 2 parents. The name and genetic locus of the SSR markers and the position of qSCN10 are indicated as described previously (Vuong et al. 2010). b Responses of Q713 NILs and their parents to 6 SCN populations. Q713-Mgl and Q713-516C comprise homozygous Magellan and PI 567516C allele in the qSCN10 region, respectively. The female index (FI) is shown as mean ± SE (n > 10). According to the Student’s t test, * and ** indicate 5% and 1% significance level, respectively. Response scores of NILs and 2 parents to each HG Type based on FI scores were labeled on top of each bar. R, resistant (FI < 10); MR, moderately resistant (10 < FI < 30); MS, moderately susceptible (30 < FI < 60); and S, susceptible (FI > 60). c Yield drag test for NILs. The NILs were planted in the field under normal growth conditions (none-SCN stress field). Data shown are means ± SE of yield estimated from 3-m rows with 5 replications
No yield drag or other unfavorable traits associated with qSCN10
To examine whether there are any adverse agronomic traits, especially yield drag, associated with qSCN10, Q713-516C and Q713-Mgl, a pair of NILs, were tested in a field without the presence of SCN. As shown in Fig. 3c, no significant difference was observed in yield between Q713-516C (4.32 t ha−1) and Q713-Mgl (4.29 t ha−1), indicating there was no yield drag associated with the QTL. Additionally, no other obvious undesirable traits were found associated with the QTL. Therefore, the qSCN10 locus is suitable for application in breeding soybeans for resistance to SCN by introgression.
qSCN10 was fine-mapped to a 379-kb region
To further narrow down the qSCN10 region for future application in breeding and the cloning of the underlying gene(s), 159 individual BC4F2 plants were identified as cross-recombinants in the target qSCN10 region by two SNP markers (SNP1 and SNP-O8). Four more SNP markers (SNP1.1, SNP2, SNP3, and SNP4) were designed between these two SNP markers to further genotype these recombinants (Supplementary Table S1). Eventually, 4 pairs of NILs (Q041, Q150, Q543, and Q660) were selected for SCN phenotyping due to their different crossover sites within the QTL region (Fig. 4a).
Fine-mapping of qSCN10 for multiple SCN populations. a The qSCN10 locus was delimited into a 379-kb region by the progeny test of four recombinants in the qSCN10 region. b Zoomed-in physical map and recombinants at the delimited region. c Responses of the progeny lines of the recombinants to six SCN HG Types. The female index (FI) is shown as means ± SE. The numbers of homozygous plant individuals used for phenotyping analysis were indicated in the charts. According to the Student’s t test, * and ** indicate 5% and 1% significance level, respectively, and “ns” represents no significant difference
Progeny tests were performed for these four pairs of NILs in response to 6 SCN HG Types in the greenhouse. The progeny lines with the genomic fragment between SNP2 and SNP-O8 from PI 567516C showed significant reductions in female indexes (FIs): The Q041 line showed significant reductions of 32 and 38% in response to SCN HG Type 2.5.7 (race 1) and 1.3.6.7, respectively; the Q660 line showed significant reductions of 44, 47, 49, 35, and 51% in response to HG Type 2.5.7 (race 1), 1.2.5.7, 2.5.7 (race 5), 1.3.6.7, and 1.2.3.4.5.6.7, respectively. No significant difference was found between progeny lines of Q150 and Q543. Hence, the qSCN10 locus can be narrowed down to a 379-kb interval between markers SNP2 (Gm10:42,430,713) and SNP-O8 (Gm10:42,809,800) based on the above phenotypic as well as genotypic information (Fig. 4). Additional SNP markers (Fig. 4b, Supplementary Table S1) developed from the fine-mapped region were used to genotypes these NILs. Although these markers did not reduce the target interval any further, they can be used in the next round of fine-mapping.
Analysis of candidate genes in the fine-mapped QTL region
There are 51 genes in the delimited 379-kb interval according to the gene models Glyma. Wm82.a2.v1 (Table 1). DNA sequence variations within the QTL region were acquired from of our previous WGRS studies (Valliyodan et al. 2016; Xu et al. 2013). Sequence variations in annotated genes between Magellan and PI 567516C, including synonymous/non-synonymous changes in exons and nucleotide insertion/deletion/substitutions in non-coding regions (including introns, 5′UTR and 3′UTR), are shown in Supplementary Table S2. A total of 700 variations were detected in 38 genes, including 91 non-synonymous variations in the exons of 24 genes. A 5-bp deletion in Glyma.10G195000 encoding a cytosolic 5′-nucleotidase III-like protein was identified in the susceptible parent (Magellan). This deletion was found to cause open-reading frame shift and therefore a possible functional change.
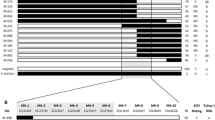
Expression profiles of the 51 annotated genes in response to SCN and in different tissues were obtained from the published microarray and RNA-Seq transcriptomes that have been deposited in the Gene Expression Omnibus (GEO) database and the Soybase website (GSE64492, Wan et al. 2015; GSE1251033, Neupane et al. 2019; www.soybase.org). Average expression levels of three replicates for each sample treatment were shown (Fig. 5). Over half of the genes were expressed in the soybean roots, where they may play a role in mediating resistance to SCN. Among them, Glyma.10G192500, Glyma.10G193200, Glyma.10G193500, Glyma.10G193600, Glyma.10G194800, and Glyma.10G195600 were up-regulated under SCN infection; Glyma.10G193400, Glyma.10G194500, and Glyma.10G195700 were down-regulated; and Glyma.10G191700 and Glyma.10G196700 were induced at 5 dai and suppressed at 30 dai. Notably, Glyma.10G194800, Glyma.10G195600, Glyma.10G195700, and Glyma.10G196700, encoding a bZIP transcription factor, a SNF7 family protein, a receptor-like kinase, and a CC-NBS-LRR protein, respectively, were annotated to be involved in defense responses. Therefore, they were considered to be potential candidate genes underlying the qSCN10.
Heat map of candidate gene expressions. Raw data from three sets of transcriptome profiles were downloaded from published database and analyzed in Excel: a Microarrays from Gene Expression Omnibus (GEO) database GSE64492 (Wan et al. 2015); b RNA-Seq from GEO database GSE1251033 (Neupane et al. 2019); c RNA-Seq from Soybase database (Severin et al. 2010). Average expression values of 3 replicates for datasets a and b were shown. Dots indicate no available probe-sets in microarray or no corresponding genes in RNA-Seq; red asterisks close to Gene IDs indicate genes that are regulated by SCN infection; two red asterisks indicate defense-related genes. Genes expressing in the roots (RPKM ≥ 1) are highlighted in blue font. Mgl, Magellan; 516C, PI567516C; W82, Williams 82; mock, control treatment with water; SCN, samples with SCN inoculation (color figure online)
Phylogenetic and haplotype analyses of the qSCN10 locus
To understand the evolution and distribution of the qSCN10 locus, a phylogenetic tree was constructed using 2,336 SNPs within the delimited 379-kb interval from 106 soybean lines, including seven wild soybean relatives (non-domesticated), 43 landraces (semi-domesticated), and 56 elite lines (Valliyodan et al. 2016; Lakhssassi et al. 2019) (Fig. 6). Based on the constructed tree, PI 567516C and 5 more landraces (PI 567,305, PI 567336B, PI 567,343, PI 567,357, PI 567,387) originating from China were clustered into a small separate group located at the end of the tree. Interestingly, neither do the elite breeding lines nor do the wild relatives in this diverse panel carry this locus. This result suggests the qSCN10 locus originated from an ancestor soybean in China. Due to its absence in the elite lines, this unique locus can be introgressed into elite breeding lines to increase genetic diversity for resistance to SCN.
Phylogenetic analysis for the qSCN10 locus. The dendrogram of 106 soybean lines was analyzed based on 2,336 SNPs derived from Valliyodan et al. (2016) within the delimited 379-kb qSCN10 region. Blue circle-shaped bullets show 56 elite lines; yellow triangle-shaped bullets show 43 landraces; and red diamond-shaped bullets show 7 wild races. Subtree containing PI 567516C is highlighted with green branches (color figure online)
Haplotypes and phenotypes of the six clustered landraces were further analyzed to investigate the relationship between the known rhg1 and Rhg4 loci and the qSCN10 locus (Supplementary Table S3). Based on a previous analysis (Kadam et al. 2016), 5 SNP markers identified for KASP assays were used to differentiate different alleles of rhg1, Rhg4 and qSCN10: Rhg1-2 and Rhg1-5 for rhg1; Rhg4-3 and Rhg4-5 for Rhg4; and SNP-O8 for qSCN10. The Rhg1-5 marker was used to select the rhg1 resistance allele, and the Rhg1-2 marker was employed to differentiate Peking and PI 88788-type resistance (Kadam et al. 2016). The WGRS data and the KASP assays revealed that PI 567305, PI 567516C, and PI 567336B carry the same genotype at five SNPs positions in the following loci or genes: Peking-type rhg1, PI 88,788/Williams 82-type Rhg4, and PI 567516C type qSCN10. The genotype is consistent with the closest clustering of these three PIs in the phylogenetic tree. Correspondingly, these three lines showed notable resistance to all the tested SCN HG Types. PI 567357 possesses susceptible haplotypes in all three genes, consistent with its susceptibility to SCN. PI 567343 showed opposite haplotypes from PI 567387 at the rhg1 and qSCN10 locus, and correspondingly the opposite response to all the tested HG Types from PI 567387. Based on this result, we speculated that the qSCN10 locus may need to work together with the Peking-type rhg1 to fulfill strong resistance to multiple SCN HG types, similar to Rhg4, which also requires rhg1 for resistance to SCN, because the qSCN10 locus alone only showed evident resistance to HG Type 0 and 1.3.6.7.
Discussion
qSCN10 confers broad-spectrum resistance to SCN
Previously, the qSCN10 locus was shown to confer resistance to SCN HG Types 2.5.7 (race 1), 0, 2.5.7 (race 5), 1.3.6.7, and 1.2.3.4.5.6.7 (Vuong et al. 2010). Our present study not only confirmed its resistance to these SCN HG types, but also identified additional resistance to HG Type 1.2.5.7. Thus, the qSCN10 locus is capable of conferring resistance to all the SCN HG types that have been tested. However, the resistance conferred by qSCN10 alone was moderate to low when compared with the resistance observed in the original resistant parent, PI 567516C. The reason that PI 567516C confers strong resistance to these HG types is that, in addition to qSCN10, it also contains qSCN18 as well as three copies of the Peking-type rhg1, although this rhg1 was not mapped as a resistance QTL in this accession in the previous study (Vuong et al. 2010). Therefore, qSCN10 very likely needs to interact with other QTL or genes, such as qSCN18 and rhg1, to fulfill strong broad-spectrum resistance to SCN.
qSCN10 is a unique genetic resource different from any known QTL or genes
Two pieces of evidence support that qSCN10 is a unique genetic resource different from any known QTL or genes. First, qSCN10 was mapped to chr. 10, different from any known QTL or genes, e.g., rhg1 (on chr. 18), Rhg4 (on chr. 8), and qSCN18 (on chr. 18). Second, phylogenetic analysis showed that qSCN10 locus existed only in 6 exotic landraces originating from China and was absent in both the current US elite lines and wild races. This data suggests that the qSCN10 locus may have originated from an ancestor soybean in China and evolved to become a resistance QTL due to nematode stress or domestication.
DNA sequence variations in the fine-mapped region
In the present study, the qSCN10 locus was narrowed down to a 379-kb region on chr. 10. Fifty-one genes are present in this region based on the Williams 82 reference genome (version 2). By analyzing the DNA sequence variations within the qSCN10 locus, no large DNA fragment insertion or deletion (> 50 bp) was detected, which may lead to introgression or deletion of any gene/genes other than the indicated 51 genes. Among these genes, 24 genes were found to have non-synonymous variations in the exons that may cause function changes of their coded proteins. Notably, a 5-bp deletion in Glyma.10G195000, which encodes a cytosolic 5′-nucleotidase III-like protein, caused a frameshift and therefore a possible functional change. In addition to these non-synonymous variations, sequence variations in the intergenic regions, introns, and 5′-and3′-UTRs may also affect gene functions by regulating their expressions.
To further narrow down the QTL region and eventually clone the underlying gene(s), another round of fine-mapping, which requires new progeny with recombinant haplotypes between marker SNP2 and SNP-O8, is needed. For this purpose, a large set of BC4F4 progeny will be generated and screened with KASP assays for SNPs that have been discovered. New pairs of NILs will be developed for fine-mapping. After the second round of fine-mapping, possible candidate genes will be selected and functionally validated using transgenic approaches for their possible role in conferring resistance to SCN.
Possible candidate genes and defense mechanism underlying the resistance of qSCN10 to SCN
Among the 51 annotated genes, none of them show significant similarities to rhg1 or Rhg4 genes, further supporting the genetic novelty of this locus. Of these genes, 11 were regulated by SCN infection. Among them are the following four defense-related genes. Glyma.10G194800 encodes a bZIP transcription factor. bZIP transcription factors have been indicated in defense against bacterial pathogens in some plants (Li et al. 2017; Lim et al. 2015). Glyma.10G195600 encodes a SNF7 family protein. A recent study showed that a SNF7 family protein, together with other proteins, was recruited to repair the membrane damage caused by pathogen infection in Dictyostelium discoideum (López-Jiménez et al. 2018). Glyma.10G195700 encodes a receptor-like kinase. Receptor-like kinases are well known to be important in both plant biotic and abiotic responses (Tang et al. 2017; Ye et al. 2017). Glyma.10G196700 encodes a CC-NBS-LRR protein. NBS-LRR proteins are typical plant disease resistance proteins important in defending plants against infection by various pathogens, such as viruses, bacteria, and fungi (Eitas et al. 2010; McHale et al. 2006). Multiple non-synonymous variations were found in Glyma.10G194800, Glyma.10G195700, and Glyma.10G196700. Except some sequence variations in the non-coding regions, there were no variations in the exons of Glyma.10g195600. Due to their potential role in defense, these four genes are good candidates for the qSCN10 QTL. However, other non-traditional defense-related genes cannot be completely overlooked. Since nematodes are different pathogens from bacteria and fungi, soybean plants may use a different mechanism to combat them. For example, the cloned Rhg4 gene is not a traditional defense-related gene: the gene encodes a serine hydroxy methyltransferase, responsible for interconversion of serine and glycine and essential for cellular one-carbon metabolism (Liu et al. 2012). Additionally, it is possible that more than one gene is involved in mediating the resistance to SCN by qSCN10, as in the case of rhg1, which requires the contributions from three neighboring genes to achieve resistance (Cook et al. 2014). Further work is needed to eventually identify the gene(s) and the corresponding mechanism that are responsible for the resistance to SCN conferred by qSCN10.
Our present work also suggests that defense against SCN in PI 567516C occurred after SCN entered the roots. The nematodes appeared to be able to enter the root normally to establish a feeding site (syncytium) in PI 567516C. However, their growth and development in PI 567516C were delayed or restricted compared to that in Magellan (Fig. 2). The previous transcriptome analysis in PI 567516C and Magellan with/without SCN infection showed that genes involved in ethylene and protein degradation were significantly enriched in PI 567516C over Magellan and regulated by SCN infection (Wan et al. 2015). Studies suggest that ethylene signaling can induce programmed cell death, and protein degradation is an important part of cell signaling and cell death (Bouchez et al. 2007; Steffens et al. 2009; Kidd et al. 1998). Therefore, the suppressed development of nematodes in PI 567516C may be due to their inability to properly maintain the feeding sites (syncytia) (syncytium degeneration) to provide sufficient nutrients for their growth and development. Although the growth of nematodes in PI 567516C was slowed down, a small portion of nematodes were still able to fully develop, unlike the complete resistance that observed in Peking and PI 88788 (Mitchum 2016; Chen et al. 2020). The previous transcriptomic studies also support this suggestion, as different gene expression profiles were observed in PI 567516C and PI 437654 (Peking-type rhg1) during SCN infection. Therefore, the cloning of the gene(s) underlying the qSCN10 QTL in PI 567516C will likely discover a novel SCN defense mechanism.
Application of qSCN10 in breeding soybean for broad-spectrum resistance to SCN
Our present study not only confirmed that the qSCN10 QTL is a novel source conferring resistance to multiple SCN HG types, but also narrowed the QTL down to a 379-kb region. Furthermore, there was no yield drag or other unfavorable traits associated with this QTL. Therefore, qSCN10 shall be an excellent resistance source to be introgressed into elite soybean varieties for broad-spectrum resistance to SCN via marker-assisted selection (MAS). However, qSCN10 alone only conferred moderate-to-low resistance to different HG types, in comparison with the original resistant parent, PI 567516C, which confers much stronger resistance to different SCN HG types. As discussed afore, PI 567516C contains not only qSCN10, but also qSCN18 as well as three copies of the Peking-type rhg1 (Vuong et al. 2010; Patil et al. 2019), suggesting that the strong broad-spectrum resistance observed in PI 567516C is due to the synergistic interactions among these QTLs or genes. Therefore, to achieve strong broad-spectrum resistance to different HG types, qSCN10 needs to be introgressed into elite soybean varieties with other resistance QTL or genes, such as qSCN18 and rhg1.
References
Allen TW, Bradley CA, Sisson AJ, ByamukamaE Chilvers MI, Coker CM et al (2017) Soybean yield loss estimates due to diseases in the United States, and Ontario, Canada, from 2010 to 2014. Plant Health Prog 18:19–27
Arelli PR, Sleper DA, Yue P, Wilcox JA (2000) Soybean reactions to races 1 and 2 of Heterodera glycines. Crop Sci 40:824–826
Arelli PR, Young LD, Concibido VC (2009) Inheritance of resistance in soybean PI 567516C to LY1 nematode population infecting cv Hartwig. Euphytica 165:1–4
Arelli PR, Wilcox JA, Myers J, Gibson PT (1997) Soybean germplasm resistant to races 1 and 2 of Heterodera glycines. Crop Sci 37:1367–1369
Bayless AM, Zapotocny RW, Grunwald DJ, Amundson KK, Diers BW, Bent AF (2018) An atypical N-ethylmaleimide sensitive factor enables the viability of nematode-resistant Rhg1 soybeans. Proc Natl Acad Sci USA 115:E4512–E4521. https://doi.org/10.1073/pnas.1717070115
Bouchez O, Huard C, Lorrain S, Roby D, Balague C (2007) Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1. Plant Physiol 145:465–477
Brown S, Yeckel G, Heinz R, Clark K, Sleper D, Mitchum MG (2010) A high-throughput automated technique for counting females of Heterodera glycines using a fluorescence-based imaging system. J Nematol 42:201–206
Byrd DW Jr, Kirkpatrick T, Barker KR (1983) An improved technique for clearing and staining plant tissue for detection of nematodes. J Nematol 15:142–143
Chen X, Li S, Zhao X, Zhu X, Wang Y, Xuan Y, Liu X, Fan H, Chen L, Duan Y (2020) Modulation of (homo)glutathione metabolism and H2O2 accumulation during soybean cyst nematode infections in susceptible and resistant soybean cultivars. Int J Mol Sci 21:388
Chen Y, Wang D, Arelli P, Ebrahimi M, Nelson RL (2006) Molecular marker diversity of SCN-resistant sources in soybean. Genome 49:938–949
Concibido VC, Diers BW, Arelli PR (2004) A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci 44:1121–1131
Cook DE, Bayless AM, Wang K, Guo X, Song Q, Jiang J, Bent AF (2014) Distinct copy number, coding sequence, and locus methylation patterns underlie Rhg1-mediated soybean resistance to soybean cyst nematode. Plant Physiol 165:630–647
Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, Diers BW, Jiang J, Hudson ME, Bent AF (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338:1206–1209. https://doi.org/10.1126/science.1228746
Davis EL, Meyers DM, Burton JW, Barker KR (1998) Resistance to root-knot, reniform, and soybean cyst nematodes in selected soybean breeding lines. J Nematol 30:530–541
Eitas TK, Dangl JL (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13:472–477
Guo B, Sleper DA, Arelli PR, Shannon JG, Nguyen HT (2005) Identification of QTLs associated with resistance to soybean cyst nematode races 2, 3 and 5 in soybean PI 90763. Theor Appl Genet 111:965–971
Han Y, Zhao X, Cao G, Wang Y, Li Y, Liu D, Teng W, Zhang Z, Li D, Qiu L, Zheng H, Li W (2015) Genetic characteristics of soybean resistance to HG type 0 and HG type 1.2.3.5.7 of the cyst nematode analyzed by genome-wide association mapping. BMC Genom 16:598
Howland A, Monnig N, Mathesius J, Nathan M, Mitchum MG (2018) Survey of Heterodera glycines population densities and virulence phenotypes during 2015–2016 in Missouri. Plant Dis 102(12):2407–2410
Hua C, Li C, Hu Y, Mao Y, You J, Wang M, Chen J, Tian Z, Wang C (2018) Identification of HG types of soybean cyst nematode Heterodera glycines and resistance screening on soybean genotypes in northeast China. J Nematol 50:41–50
Jiao Y, Vuong TD, Liu Y, Li Z, Noe J, Robbins RT, Joshi T, Xu D, Shannon JG, Nguyen HT (2015) Identification of quantitative trait loci underlying resistance to southern root-knot and reniform nematodes in soybean accession PI 567516C. Mol Breed 35:131. https://doi.org/10.1007/s11032-015-0330-5
Kadam S, Vuong TD, Qiu D, Meinhardt CG, Song L, Deshmukh R, Patil G, Wan J, Valliyodan B, Scaboo AM (2016) Genomic-assisted phylogenetic analysis and marker development for next generation soybean cyst nematode resistance breeding. Plant Sci 242:342–350. https://doi.org/10.1016/j.plantsci.2015.08.015
Kidd VJ (1998) Proteolytic activities that mediate apoptosis. Annu Rev Physiol 60:533–573. https://doi.org/10.1146/annurev.physiol.60.1.533
Kim M, Hyten DL, Bent AF, Diers BW (2010) Fine mapping of the SCN resistance locus rhg1-b from PI 88788. Plant Genom 3(2):81–89. https://doi.org/10.3835/plantgenome2010.02.0001
Klepadlo M, Meinhardt CG, Vuong TD, Patil G, Bachleda N, Ye H, Robbins RT, Li Z, Shannon JG, Chen P, Meksem K (2018) Evaluation of soybean germplasm for resistance to multiple nematode species: Heterodera glycines, Meloidogyne incognita, and Rotylenchulus reniformis. Crop Sci 58(6):2511–2522
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lakhssassi N, Patil G, Piya S, Zhou Z, Baharlouei A, Kassem MA, Lightfoot DA, Hewezi T, Barakat A, Nguyen HT, Meksem K (2019) Genome reorganization of the GmSHMT gene family in soybean showed a lack of functional redundancy in resistance to soybean cyst nematode. Sci Rep 9:1506. https://doi.org/10.1038/s41598-018-37815-w
Li YH, Chen SY, Young ND (2004a) Effect of the Rhg1 gene on penetration, development and reproduction of Heterodera glycines race 3. Nematology 6:729–736
Li Y, Chen S, Young ND (2004b) Effect of rhg1 gene on penetration, development, and reproduction of Heterodera glycines race 3. Nematology 6:727–734
Li X, Fan S, Hu W, Liu G, Wei Y, He C, Shi H (2017) Two cassava basic leucine zipper (bZIP) transcription factors (MebZIP3 and MebZIP5) confer disease resistance against cassava bacterial blight. Front Plant Sci 8:2110. https://doi.org/10.3389/fpls.2017.02110
Lian Y, Guo JQ, Li HC, Wu YK, Wei H, Wang JS, Li JY, Lu WG (2017) A new race (X12) of soybean cyst nematode in China. J Nematol 49(3):321–326
Lian Y, Wang JS, Li HC, Wei H, Li JY, Wu YK, Lei CF, Zhang H, Wang SF, Guo JQ, Li YX, Li ZH, Jin QL, Xu SX, Zhang ZM, Yang CY, Yu HY, Geng Z, Shu WT, Lu WG (2016) Race distribution of soybean cyst nematode in the main soybean producing area of Huang-Huai Rivers Valley. Acta Agron Sin 42(10):1479–1486
Lim CW, Baek W, Lim S, Han SW, Lee SC (2015) Expression and functional roles of the pepper pathogen-induced bZIP transcription factor, CabZIP2, in enhanced disease resistance to bacterial pathogen infection. Mol Plant Microbe Interact 28:825–833
Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, Hill J, Baum TJ, Cianzio S, Whitham SA, Korkin D, Mitchum MG, Meksem K (2012) A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492:256–260
Liu S, Kandoth PK, Lakhssassi N, Kang J, Colantonio V, Heinz R, Yechel G, Zhou Z, Bekal S, Dapprich J, Rotter B, Cianzio S, Mitchum MG, Meksem K (2017) The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat Commun 8:14822
López-Jiménez AT, Cardenal-Muñoz E, Leuba F (2018) The ESCRT and autophagy machineries cooperate to repair ESX-1-dependent damage at the Mycobacterium-containing vacuole but have opposite impact on containing the infection. PLoS Pathog 14(12):e1007501
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7(4):212. https://doi.org/10.1186/gb-2006-7-4-212
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Meksem K, Pantazopoulos P, Njiti VN, Hyten LD, Arelli PR, Lightfoot DA (2001) ‘Forrest’ resistance to soybean cyst nematode is bigenic: saturation mapping of the Rhg1 and Rhg4 loci. Theor Appl Genet 103:710–717
Mitchum MG (2016) Soybean resistance to the soybean cyst nematode Heterodera glycines: an update. Phytopathology 106:1444–1450
Mulrooney RP, Gregory NF, Heinz RD (2010) Soybean cyst nematode populations in Delaware are shifting in response to widespread planting of soybean cultivars with resistance from PI88788. J Nematol 42(3):259–260
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Neupane S, Mathew FM, VarenhorstAJ, et al (2019) Transcriptome profiling of interaction effects of soybean cyst nematodes and soybean aphids on soybean. Sci Data 6:133. https://doi.org/10.1038/s41597-019-0140-4
Niblack TL, Arelli PR, Noel GR, Opperman CH, Orf JH, Schmitt DP, Shannon JG, Tylka GL (2002) A revised classification scheme for genetically diverse populations of Heterodera glycines. J Nematol 34:279–288
Niblack TL, Colgrove AL, Colgrove K, Bond JP (2008) Shift in virulence of soybean cyst nematode is associated with use of resistance from PI 88788. Plant Health Prog 9:29
Niblack T, Tylka GL, Arelli P, Bond J, Diers B, Donald P, Faghihi J, Ferris VR, Gallo K, Heinz RD, Lopez-Nicora H, Von Qualen R, Welacky T, Wilcox J (2009) A standard greenhouse method for assessing soybean cyst nematode resistance in soybean: sCE08 (standardized cyst evaluation 2008). Plant Health Prog. https://doi.org/10.1094/PHP-2009-0513-01-RV
Patil GB, Lakhssassi N, Wan J, Song L, Zhou Z, Klepadlo M, Vuong TD, Stec AO, Kahil SS, Colantonio V, Valliyodan B, Rice JH, Piya S, Hewezi T, Stupar RM, Meksem K, Nguyen HT (2019) Whole-genome re-sequencing reveals the impact of the interaction of copy number variants of the rhg1 and Rhg4 genes on broad-based resistance to soybean cyst nematode. Plant Biotechnol J 17:1595–1611. https://doi.org/10.1111/pbi.13086
Robinson J, Thorvaldsdóttir H, Winckler W et al (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. https://doi.org/10.1038/nbt.1754
Schmitt DP, Shannon JG (1992) Differentiating soybean responses to Heterodera glycines races. Crop Sci 32:275–277
Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, Graham MA, Cannon SB, May GD, Vance CP, Shoemaker RC (2010) RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. https://doi.org/10.1186/1471-2229-10-160
Steffens B, Sauter M (2009) Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 21:184–196
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29:618–637
Tylka GL, Marett CC (2017) Known distribution of the soybean cyst nematode, Heterodera glycines, in the United States and Canada: 1954 to 2017. Plant Health Prog 18:167–168
Valliyodan B, Qiu D, Patil G, Zeng P, Huang J, Dai L, Chen C, Li Y, Joshi T, Song L, Vuong TD, Musket TA, Xu D, Shannon JG, Shifeng C, Liu X, Nguyen HT (2016) Landscape of genomic diversity and trait discovery in soybean. Sci Rep. https://doi.org/10.1038/srep23598
Vuong TD, Sleper DA, Shannon JG et al (2011) Confirmation of quantitative trait loci for resistance to multiple-HG types of soybean cyst nematode (Heterodera glycines Ichinohe). Euphytica 181:101
Vuong TD, Sleper DA, Shannon JG, Nguyen HT (2010) Novel quantitative trait loci for broad-based resistance to soybean cyst nematode (Heterodera glycines Ichinohe) in soybean PI 567516C. Theor Appl Genet 121:1253–1266
Vuong TD, Sonah H, Meinhardt CG, Deshmukh R, Kadam S, Nelson RL, Shannon JG, Nguyen HT (2015) Genetic architecture of cyst nematode resistance revealed by genome-wide association study in soybean. BMC Genom. https://doi.org/10.1186/s12864-015-1811-y
Wan J, Vuong T, Jiao Y, Joshi T, Zhang H, Dong Xu, Nguyen HT (2015) Whole-genome gene expression profiling revealed genes and pathways potentially involved in regulating interactions of soybean with cyst nematode (Heterodera glycines Ichinohe). BMC Genom 16:1–48
Whitham SA, Qi M, Innes RW, Ma W, Lopes-Caitar V, Hewezi T (2016) Molecular soybean–pathogen interactions. Ann Rev Phytopathol 54:443–468
Winter SMJ, Shelp BJ, Anderson TR, Welacky TW, Rajcan I (2006) QTL associated with horizontal resistance to soybean cyst nematode in Glycine soja PI 464925B. Theor Appl Genet 114:461–472
Wu X, Blake S, Sleper DA, Shannon JG, Cregan P, Nguyen HT (2009) QTL, additive and epistatic effects for SCN resistance in PI 437654. Theor Appl Genet 118:1093–1105
Xu X, Zeng L, Tao Y, Vuong T, Wan J, Boerma R, Noe J, Li Z, Finnerty S, Pathan SM, Shannon JG, Nguyen HT (2013) Pinpointing genes underlying the quantitative trait loci for root-knot nematode resistance in palaeopolyploid soybean by whole genome resequencing. Proc Natl Acad Sci USA 110(33):13469–74
Young LD (1998) Heterodera glycines populations selected for reproduction on Hartwig soybean. J Nematol 30:523
Young LD (1999) Soybeans resistant to Heterodera glycines populations attacking Hartwig cultivar. J Nematol 31:583
Yu N, Diers BW (2017) Fine mapping of the SCN resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Euphytica 213:54
Zhang H, Li C, Davis EL, Wang J, Griffin JD, Kofsky J, Song BH (2016) Genome-wide association study of resistance to soybean cyst nematode (Heterodera glycines) HG type 2.5.7 in wild soybean (Glycine soja). Front Plant Sci. https://doi.org/10.3389/fpls.2016.01214
Acknowledgements
The authors gratefully acknowledge the financial support for this study provided by the United States Department of Agriculture—National Institute of Food and Agriculture (USDA-NIFA) Grant 2019-67013-29370 and the USDA multiple state/hatch project MO-MSPS0002.
Author information
Authors and Affiliations
Contributions
LZ, LS, HY, and HTN conceived and designed the project; LS and TV contributed to the development of the mapping populations; LZ, LS, YL, and MK collected plant materials and performed genotyping and phenotyping experiments. LZ and LS contributed to the data analysis; LZ wrote the manuscript; HY, TV, and JW contributed to discussion of the manuscript; and all authors critically revised the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Communicated by Istvan Rajcan.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, L., Song, L., Lian, Y. et al. Genetic characterization of qSCN10 from an exotic soybean accession PI 567516C reveals a novel source conferring broad-spectrum resistance to soybean cyst nematode. Theor Appl Genet 134, 859–874 (2021). https://doi.org/10.1007/s00122-020-03736-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03736-4