Abstract
Key message
A major QTL for oviposition deterrence to orange wheat blossom midge was detected on chromosome 1A in the Canadian breeding line BW278 that was inherited from the Chinese variety Sumai-3.
Abstract
Orange wheat blossom midge (OWBM, Sitodiplosis mosellana Géhin, Diptera: Cecidomyiidae) is an important insect pest of wheat (Triticum aestivum L.) that reduces both grain yield and quality. Oviposition deterrence results in a reduction of eggs deposited on spikes relative to that observed on a wheat line preferred by OWBM. Quantification of oviposition deterrence is labor-intensive, so wheat breeders require efficient DNA markers for the selection of this trait. The objective of this study was to identify quantitative trait loci (QTL) for oviposition deterrence in a doubled haploid (DH) population developed from the spring wheat cross Superb/BW278. The DH population and check varieties were evaluated for OWBM kernel damage from five field nurseries over three growing seasons. QTL analysis identified major effect loci on chromosomes 1A (QSm.mrc-1A) and 5A (QSm.mrc-5A). Reduced kernel damage was contributed by BW278 at QSm.mrc-1A and Superb at QSm.mrc-5A. QSm.mrc-1A mapped to the approximate location of the oviposition deterrence QTL previously found in the American variety Reeder. However, haplotype analysis revealed that BW278 inherited this oviposition deterrence allele from the Chinese spring wheat variety Sumai-3. QSm.mrc-5A mapped to the location of awn inhibitor gene B1, suggesting that awns hinder OWBM oviposition. Single-nucleotide polymorphisms (SNPs) were identified for predicting the presence or absence of QSm.mrc-1A based upon haplotype. Functional annotation of candidate genes in 1A QTL intervals revealed eleven potential candidate genes, including a gene involved in terpenoid biosynthesis. SNPs for QSm.mrc-1A and fully awned spikes provide a basis for the selection of oviposition deterrence to OWBM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The orange wheat blossom midge (OWBM) (Sitodiplosis mosellana Géhin; Diptera: Cecidomyiidae) is among the most damaging insect pests of spring wheat, Triticum aestivum L., in western Canada, northern USA and many countries of Europe and Asia (Smith et al. 2014; Wise et al. 2015; Kassa et al. 2016). Recent outbreaks of OWBM have been reported in many wheat-growing European countries including UK, France, Belgium and Germany, causing significant yield losses (Thomas et al. 2005; Kassa et al. 2016). In China, two serious outbreaks caused by OWBM had been reported in the 1950s and 1980s, resulting in nearly 50% loss in wheat production (Zhang et al. 2020). In Canada, the first major outbreak of wheat midge occurred in northeast Saskatchewan in 1983 and caused over $30 million in yield losses (Olfert et al. 2012). Since that time, OWBM has become a significant pest of spring wheat throughout all major wheat-growing areas of western Canada, causing seed damage, grade reduction and extensive yield losses during population outbreaks (Lamb et al. 1999; Olfert et al. 2009; Elliott et al. 2011). In Canada, the estimated annual wheat losses caused by OWBM exceeded $60 million CAD, prior to the introduction of resistant wheat varieties carrying the OWBM resistance gene Sm1 (Kassa et al. 2016).
The OWBM adults emerge from the soil at the time of wheat heading, mate and lay eggs (oviposit) on the emerging wheat spikes in response to characteristic wheat odor components (Barnes 1956). Upon hatching, larvae feed on developing kernels causing shriveled, cracked or distorted grains, contributing to poor seed quality and reduction in end-use suitability (Dexter et al. 1987; Lamb et al. 2000). Canadian spring and durum wheat varieties differ in their susceptibility to wheat midge damage. The severity of kernel damage by OWBM depends on several factors including larval population density (number of larvae present per spikelet), their spatial distribution and timing of oviposition relative to heading and anthesis (Wright and Doane 1987; Elliott and Mann 1996). Wheat spikes are most susceptible to damage when oviposition occurs during the heading stage and damage declines dramatically after the anthers are visible (Elliott and Mann 1996).
The development of wheat varieties with antibiotic resistance (antibiosis) and/or oviposition deterrence (antixenosis) to OWBM is an important component of integrated pest management (Lamb et al. 2002; Thomas et al. 2005). Sm1 is the only described antibiosis gene for OWBM resistance (Berzonsky et al. 2002; McKenzie et al. 2002) and is located on wheat chromosome arm 2BS (Thomas et al. 2005; Kassa et al. 2016). The resistance mechanism of Sm1 depends on antibiosis (inhibition of larval growth) and mediated through the production of phenolic acids in the seed coat in response to larval feeding on the developing kernel. Phenolic acids such as ferulic acid and/or p-coumaric acid cause larvae to leave the kernel they are feeding on and die of starvation (Ding et al. 2000).
Oviposition deterrence is a resistance mechanism employed by plants to deter or reduce colonization by insects (Morando et al. 2015). Oviposition deterrence has been identified in both common and durum wheat (Lamb et al. 2001, 2002). Wheat varieties with oviposition deterrence to OWBM have fewer eggs laid on their spikes and are often associated with a higher proportion of eggs deposited on the rachis rather than the florets. The tiny, newly hatched larvae have to travel longer distances to feed on developing kernels and have a greater risk of desiccation en route (Lamb et al. 2002). Although the underlying mechanism that affects oviposition deterrence behavior in wheat is not fully understood, the deterrent wheat may have ability to repel females, provide an insufficient oviposition stimulus or physically block a female's access through chemical or morphological characteristics of wheat spikes during oviposition (Gharalari et al. 2009). Previous studies suggest that more than one gene controls oviposition deterrence to OWBM (Lamb et al. 2002; Gharalari et al. 2009). Blake et al. (2011) identified a major oviposition deterrence QTL on chromosome 1A (QSm.mst-1A) from spring wheat variety Reeder.
In this study, the genetic basis of oviposition deterrence to OWBM was explored using a DH population developed from a cross between Canadian hard red spring wheat variety Superb and oviposition deterrent breeding line BW278. The main objectives of this study were to: 1) identify QTL for oviposition deterrence and 2) identify SNP markers suitable for high-throughput marker-assisted selection (MAS).
Materials and methods
Plant materials and field trials
A DH population of 142 lines derived from the cross Superb/BW278 was used to identify QTL for oviposition deterrence to OWBM. Superb (pedigree: Grandin*2/AC Domain) is an awned, semi-dwarf spring wheat variety registered in the Canada Western Red Spring (CWRS) market class (Townley-Smith et al. 2010). BW278 (pedigree: AC Domain*2/Sumai-3) is a spring wheat breeding line with oviposition deterrence to OWBM that was developed to introgress Fusarium head blight (FHB) resistance from the Chinese variety Sumai-3 into Canadian germplasm. Sumai-3 is a well-known source of FHB resistance used in wheat breeding programs around the world (Dhokane et al. 2016). Field nurseries were grown from 2012 to 2014 at four locations in western Canada (Winnipeg, Manitoba (Wpg); Glenlea, Manitoba (Gln); Brandon, Manitoba (Bdn); Saskatoon, Saskatchewan (Stn)). The spring wheat varieties Reeder, Waskada, 5602HR, Andrew, Harvest, Thatcher, Infinity, Lillian and AC Barrie were grown as control genotypes/checks in field tests. Field experiments were arranged in an alpha lattice design with 12 incomplete blocks and three replicates per environment. The experimental plot was a 10 cm row in which 15 seeds were sown. The Superb/BW278 population and checks were grown in field nurseries and exposed to natural populations of OWBM.
Phenotyping and statistical analysis
The Superb/BW278 DH population and checks were evaluated for OWBM kernel damage in five environments over three growing seasons (Wpg 2012, Gln 2013, Wpg 2013, Bdn 2014, Stn 2014). In this study, midge-damaged kernels (MDK) were quantified rather than counting eggs or larvae within spikes. Egg counting is extremely laborious and time-consuming, and must be done on fresh green spike samples, which was not possible based on the number of samples needed for QTL analysis. Counting larvae requires collection of green spikes and would also be time-consuming and difficult. Both methods are time-constrained as refrigerated material must be analyzed before eggs or larvae die. Therefore, MDK were used a proxy for egg counts and were quantified according to the protocol described previously (Wise et al. 2015). Briefly, wheat spikes were harvested at maturity from each plot, and 10 spikes per plot were examined for seed damage. Each spike was dissected with forceps under a dissection microscope and the number of damaged and total seeds found in each spike were recorded. MDK were separated by weight as being harvestable (> 8 mg; HMDK) or unharvestable (< 8 mg; UMDK) (Fig. 1). MDK, HMDK and UMDK were expressed as a percentage of all kernels from the dissected spikes. Heading date (HD) and plant height (HT) data were also collected. HD was recorded as the Julian date at which 50% of spikes had emerged from flag leaf sheaths. HD data were collected in Winnipeg 2012, Brandon 2014 and Saskatoon 2014. HT was measured as the distance from the soil surface to the top of the wheat spikes (excluding awns). HT data were collected in Brandon and Saskatoon 2014. The presence and absence of awns on wheat spikes were also recorded.
The software Multi Environment Trial Analysis with R for Windows (META-R) Version 6.0 (Alvarado et al. 2017) was used for analysis of variance (ANOVA) and to calculate best linear unbiased predictors (BLUPs) for HD, HT, MDK, HMDK and UMDK in each individual environment and across all environments. Genotype (Gen), environment (Env), replicate (Rep) and incomplete block (Block) were considered random effects. Broad-sense heritability (H2), least significant difference (LSD) and coefficient of variation (CV) were also calculated using META-R. META-R software uses the lme4 package in R software version 3.3.1. The linear model for lattice designs is:
where i is the ith environment, j is the jth replicate, k is the kth incomplete block and l is the lth genotype.
Genotyping
Genomic DNA was extracted from lyophilized leaf tissue with the DNeasy 96 Plant Kit (Qiagen, Toronto, Canada) and quantified using PicoGreen stain (Molecular Probes, Inc., Eugene, Oregon, USA). SNP markers were genotyped on the DH population and parents using the Illumina Infinium 90 K wheat SNP beadchip (Illumina, San Diego, CA) (Cavanagh et al. 2013; Wang et al. 2014). The raw data were analyzed with genotyping module of GenomeStudio version 2011.1 software (Illumina, San Diego, USA) using default clustering parameters.
Construction of genetic maps
A total of 142 DHs were used to construct a genetic linkage map of the Superb/BW278 population for mapping QTL for OWBM resistance and disease-related morphological traits. All monomorphic SNPs and those with > 10% missing data were excluded from linkage analysis. Each marker was tested for deviation from the expected 1:1 ratio using Chi-squared test. Markers showing significant (P < 0.001) segregation distortions were also discarded. Markers were placed into preliminary linkage bins using the BIN module in QTL IciMapping version 4.0.6.0 (Li et al. 2007). A single marker with the least missing data was selected from each linkage bin and used for linkage analysis with MapDisto version 1.7.7 (Lorieux 2012). A minimum logarithm of the odds (LOD) score of 3.0 and maximum recombination fraction of 0.2 was used to identify linkage groups. Recombination fractions were converted into map distances using the Kosambi mapping function (Kosambi 1943). Linkage groups were assigned to chromosomes based on existing high-density consensus SNP maps of wheat (Maccaferri et al. 2014; Wang et al. 2014).
DNA markers were located on IWGSC (International Wheat Genome Sequencing Consortium) Chinese Spring RefSeq v1.0 (Appels et al. 2018) by Basic Local Alignment Search Tool (BLAST). The dwarfing gene Rht-B1 (Peng et al. 1999), DNA markers for the earliness per se (Eps) QTL QEet.fcu-5AL (Liu et al. 2005) and the B1 awn inhibitor (Huang et al. 2019) were also located on RefSeq version 1.0 with BLAST.
QTL analysis
QTL analysis was conducted with interval mapping (IM) and inclusive composite interval mapping (ICIM) using QTL IciMapping version 4.1.0.0 (Li et al. 2007, 2008). For all traits, QTL analyses were carried out using BLUPs for test entries in each individual environment and pooled over all environments. Analysis for additive effect QTL was conducted with 0.1 cM steps, and the 5% LOD significance threshold was estimated with 10,000 permutations. The LOD significance threshold was 2.96 for both IM and ICIM additive effect QTL analysis. Additive effect QTL were declared when the LOD score exceeded the LOD significance threshold in two or more environments, or one or more environments plus the pooled dataset, based upon IM or ICIM. QTL analysis statistics for these declared additive effect QTL were also reported for additional environments in which the LOD score exceeded 2.5. The proportion of phenotypic variance explained by each QTL was determined by the square of the partial correlation coefficient (R2). Analysis for epistatic QTL was conducted with 2.0 cM steps and a default LOD significance threshold of 5.0. Epistatic QTL were reported when the LOD exceeded 5.0 in three or more environments, based upon QIME (i.e., interval mapping) and QICE (i.e., inclusive composite interval mapping) epistasis modules.
Haplotyping wheat lines with SNP markers linked to 1A QTL interval
Haplotype analysis was performed on a diverse collection of wheat germplasm, consisting of 75 wheat lines with varying levels of wheat midge resistance (Supplementary Table S6). Most wheat lines in the haplotype panel were susceptible to wheat midge (i.e., do not carry the OWBM resistance gene Sm1). The wheat lines BW278, Parshall, Reeder, Vesper and Waskada are known carriers of oviposition deterrence. A total of 159 Infinium SNP markers from the 1A QTL region were used for haplotype analysis to identify SNP markers for MAS.
Identification of the putative candidate genes (CGs) within the 1A QTL interval
SNP markers defining the boundaries of 1A QTL confidence interval were aligned to the IWGSC Chinese Spring RefSeq v1.0 (Appels et al. 2018) to define the physical location of the interval and identify putative CGs. Gene models found within the physical QTL intervals were retrieved using BioMart tool found in Ensembl Plants database (https://plants.ensembl.org/index.html). All genes in the candidate region were subjected to gene function annotation using GO analysis, and biological functions of a gene product were described with GO Term. GO annotations can be found in the IWGSC database (Appels et al. 2018). The metabolic pathway annotation was carried out using Blast2GO (https://www.blast2go.com/) against KEGG database (https://www.genome.jp/kegg/pathway.html) for genes in the 1A QTL interval.
Results
Phenotypic data analyses
Histograms of MDK, HD and HT in Superb/BW278 DH population are illustrated in Fig. 2. BW278 and the oviposition deterrent check lines Reeder and Waskada had fewer MDK relative to the mean of the Superb/BW278 DH population (Table 1). Superb, Andrew, Harvest, Thatcher, Infinity, Lillian, AC Barrie and Intrepid were more susceptible to OWBM than mean of the Superb/BW278 DH population. Superb flowered almost a day earlier and was 14 cm shorter than BW278. The average plant height among the 142 DHs was 94 cm, and average heading date was Julian date 201. There was some transgressive segregation for plant height, but most DHs were within the means of the parents. Heading date was approximately normally distributed in the Superb/BW278 populations (Fig. 2). Analysis of variance showed significant (P < 0.001) differences among genotypes for all traits (HD, HT, MDK, HMDK and UMDK) (Table 2 and Supplementary Table S1). Broad-sense heritability estimates were 0.75 for heading date, 0.88 for plant height, 0.73 for MDK, 0.74 for HMDK and 0.69 for UMDK in the Superb/BW278 DH population (Table 2). Broad-sense heritability estimates for individual environments are presented in Supplementary Table S2. For each individual trait, the data were positively correlated in all pairwise comparison between the environments (Supplementary Table S3). Correlation analysis revealed a significant positive correlation between midge damage (MDK, HMDK, UMDK) and HD, and a negative correlation between midge damage (MDK, HMDK, UMDK) and HT (Table 3).
Frequency distribution of best linear unbiased predictors (BLUPs) for orange wheat blossom midge (OWBM, Sitodiplosis mosellana)-related phenotypic traits: heading date, plant height and midge-damaged kernel (MDK) in the Superb/BW278 DH population in five environments over 2012, 2013 and 2014. Means of the parents are indicated
Linkage map
A high-density whole genome linkage map was developed for the Superb/BW278 DH population using the 90 K wheat Infinium SNP chip. The Superb/BW278 linkage map consisted of 7,158 SNPs. The total map length across the 32 linkage groups was 1882.3 cM (Supplementary Table S4). In some instances, a chromosome consisted of two or three linkage groups (e.g., chromosomes 2A, 2D, 3B, 4D, 5A, 5B, 6A, 6D and 7D), which was not unexpected since some regions of the genome will be identical by descent given the relatedness of the parents. Of the 7,158 polymorphic markers, 2,803 SNPs (39.2%) mapped to the A genome spanning 618 cM, 3,993 SNPs (55.8%) mapped to the B genome covering 619 cM, and 362 SNPs (5.1%) mapped to the D genome spanning 645 cM. The average spacing between neighboring markers was 0.3 cM (Supplementary Table S5). The B1 awn inhibitor gene mapped to chromosome arm 5AL (linkage group 5A.2 at position 27.1 cM) (Supplementary Table S4).
Heading date QTL
A major QTL for heading date was identified on the chromosome 5A in the Superb/BW278 DH population (linkage group 5A.2 at approximately 12 cM) (Table 4). Superb carried the early allele at QHd.mrc-5A. These results were consistent with Superb being 0.9 day earlier heading than BW278. QHd.mrc-5A mapped to the expected location of earliness per se (Eps) QTL QEet.fcu-5AL from American hard red spring wheat variety Grandin (Liu et al. 2005).
Plant height QTL
A major additive effect QTL for plant height was identified on chromosome 4B at approximately 32 cM and was named QHt.mrc-4B (Table 4). The BW278 allele increased plant height at this QTL, consistent with BW278 being 14 cm taller than Superb in the field experiments. QHt.mrc-4B explained 58.3% to 73.8% of the phenotypic variation and is consistent with the location of Rht-B1 locus.
Oviposition deterrence QTL
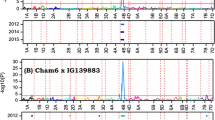
Six additive effect QTL for oviposition deterrence (MDK, HMDK and UMDK) were identified using IM and ICIM analysis with the additive effect module of QTL IciMapping (Table 4). BW278 contributed midge resistance at four of these QTL, which is consistent with BW278 showing fewer MDK than Superb (Table 1 and unpublished data, Marjorie Smith). BW278 contributed resistance at QSm.mrc-1A (chromosome 1A at 105.4 cM), which was consistent with oviposition deterrence QTL QSm.mst-1A reported previously from the American oviposition deterrent spring wheat variety Reeder (Blake et al. 2011). The QSm.mrc-1A was detected under all five environments with the LOD scores ranging from 4.6 to 31.6 and explained 10.9% to 58.3% of the phenotypic variation (Fig. 3 and Table 4). A second major oviposition deterrence QTL QSm.mrc-4B mapped on chromosome 4B (31.9 cM) coincided with HT QTL QHt.mrc-4B. At the plant height QTL QHt.mrc-4B (i.e., Rht-B1), the reduced height allele was consistently associated with increased OWBM damage.
The most prominent oviposition deterrence QTL from Superb mapped on chromosome 5A (QSm.mrc-5A), explaining up to 21% of the phenotypic variation. This 5A QTL was statistically significant in three of the five individual environments and the pooled dataset. QSm.mrc-5A mapped approximately 15 cM distal of the HD QTL QHd.mrc-5A (Table 4 and Supplementary Table S4), which explains the positive correlation between HD and OWBM damage in the Superb/BW278 population. QSm.mrc-5A coincided with the B1 awn inhibitor gene (Huang et al. 2019), which mapped to chromosome arm 5AL (linkage group 5A.2) at position 27.1 cM (Table 4 and Supplementary Table S4). Figure 4 illustrates the combined effects of the two major QTL QSm.mrc-1A and QSm.mrc-5A on MDK.
Three minor QTL for oviposition deterrence were identified on chromosomes 3B and 7A (QSm.mrc-3B, QSm.mrc-7A.1 and QSm.mrc-7A.2) based on ICIM, which were not detected by IM. Their effects were lower and less consistent than the QTL on chromosomes 1A, 4B and 5A (Table 4). In addition, these QTL were detected based upon HMDK or UMDK data, but not detected based upon total MDK.
Epistatic QTL were not detected for oviposition deterrence or other phenotypic traits (heading date and plant height) with the QIME (i.e., interval mapping) and QICE epistatic module (i.e., inclusive composite interval mapping) at the LOD significance threshold of 5.
Haplotype analysis
Haplotype analysis of a panel of wheat lines revealed that the QSm.mrc-1A region had considerable haplotype diversity among the spring wheat varieties and breeding lines. The parent BW278 (pedigree: AC Domain*2/Sumai-3) had the same haplotype as Chinese wheat variety Sumai-3, but not AC Domain, which indicates that BW278 inherited QSm.mrc-1A from Sumai-3 (BW278 haplotype). The Canada Western Red Spring (CWRS) variety Waskada which has a pedigree of BW278/2*Superb inherited this chromosome region from BW278 (Supplementary Table S6). Interestingly, BW278 had a different haplotype at QSm.mrc-1A than the American spring wheat variety Reeder, carrier of the QSm.mst-1A allele located at the same genomic region (Blake et al. 2011). Known OWBM oviposition deterrent varieties Reeder and Parshall had similar but slightly different haplotypes at QSm.mrc-1A. Vesper, another source of oviposition deterrence had the same haplotype as OWBM susceptible wheat Roblin (Roblin haplotype). Spring wheat AC Splendor had the same haplotype as the susceptible parent Superb (Superb haplotype), suggesting it also lacks the oviposition QTL QSm.mrc-1A. A majority of wheat lines in the diversity wheat panel did not have the haplotype of either BW278 or Superb (Supplementary Table S6).
Accurate MAS of QSm.mrc-1A can be accomplished with combinations of the following SNPs: wsnp_Ex_c28900_37982485, BS00023935_51, RAC875_c41993_582, wsnp_CAP8_c4785_2322876, wsnp_BE443588A_Ta_2_2, TA005289-1104, BS00070560_51, Excalibur_c23598_1632, BobWhite_c44164_151, Tdurum_contig81011_244, wsnp_CAP11_c146_160903, CAP12_c6629_301 and Tdurum_contig62584_770. These SNPs differentiate the BW278 haplotype from other haplotypes (Supplementary Table S6). The spring wheat varieties Frontana, Nyubai and Wangshuibai had similar haplotypes to BW278 from 94.1 to 103.3 cM, but differed from 105.4 to 111.3 cM (Supplementary Table S6). Different combinations of SNPs may be needed depending on the germplasm present in specific breeding programs. Presently, BW278 and Reeder carry the only two haplotypes known to confer OWBM oviposition deterrence on chromosome 1A.
Identification of putative candidate genes (CGs) at QSm.mrc-1A
QSm.mrc-1A mapped to a 3.6 cM interval (102.5–106.1 cM) on chromosome 1A linkage map which corresponded to a 42.7 Mb (536,613,453–579,299,659 bp) physical region in the IWGSC Chinese Spring reference genome RefSeq v1.0 (Appels et al. 2018) based upon BLAST locations of SNP markers defining the boundaries of QSm.mrc-1A. The 42.6 Mb candidate region on chromosome 1A contained 815 protein coding genes. Based on gene function annotation, 11 CGs may be related to OWBM resistance (Supplementary Table S7). In the candidate region, three genes (TraesCS1A02G355300, TraesCS1A02G398100 and TraesCS1A02G398200) had predicted biological function in defense responses. The CG TraesCS1A02G384300 was predicted to be involved in the biosynthesis of terpenoids. The gene TraesCS1A02G382900 was annotated as a serine/threonine protein phosphatase 2A regulatory subunit, and genes TraesCS1A02G382900 and TraesCS1A02G389500 were involved in signal transduction pathways. Several other annotated genes carrying domains that may have roles in plant defense against insects such as hydrolases superfamily protein and protein kinase were also identified (Supplementary Table S7).
Discussion
OWBM-resistant wheat varieties carrying the antibiosis gene Sm1 have been successfully used to manage OWBM in Canada, USA and Europe (https://midgetolerantwheat.ca/; https://ahdb.org.uk/wheat-blossom-midges; https://www.usda.gov/). However, the heavy reliance on Sm1 leaves wheat crops vulnerable if Sm1-virulent OWBM populations evolve. Therefore, alternate genetic resources to control OWBM are needed. To date, very few alternate genetic resistances to Sm1 have been identified. For example, Blake et al. (2011) identified a QTL (QSm.mst-1A) that reduces midge damage in the American Spring wheat variety Reeder and Zhang et al. (2020) detected two QTL on chromosome 4A conferring resistance to OWBM from Chinese wheats Henong215 and Jimai24.
In this study, the genetic basis of oviposition deterrence was characterized in a DH population derived from the cross between a Canadian spring wheat variety Superb and breeding line BW278. QTL analysis identified six QTL controlling oviposition deterrence in the Superb/BW278 population on chromosomes 1A, 3B, 4B, 5A and 7A, which are different from either the location of Sm1 (Thomas et al. 2005) or two recently reported resistance QTL on chromosome 4A (Zhang et al. 2020). BW278 contributed resistant alleles at four these loci, which is consistent with BW278 being more deterrent to oviposition in the field nurseries (Table 1). A major QTL QSm.mrc-1A, on chromosome 1A that was associated with reduced midge-damaged kernels was identified in BW278. This QTL mapped to the same region of chromosome 1A as the previously reported oviposition deterrence QTL QSm.mst-1A in the American spring wheat variety Reeder (Blake et al. 2011). However, haplotype analysis using 159 Infinium SNP markers in this region of chromosome 1A in a panel of wheat lines revealed that BW278 (AC Domain*2/Sumai-3) inherited this region from the Chinese spring wheat Sumai-3, which has been used as a major source of FHB resistance in Canadian spring wheat breeding programs (Dhokane et al. 2016). The OWBM oviposition deterrent variety Waskada (BW278/2*Superb) (Fox et al. 2009) had the same haplotype as BW278, as did the Canadian soft white spring variety Sadash. Interestingly, BW278 had a different haplotype in the 1A QTL region than the oviposition deterrent variety Reeder, carrier of QSm.mst-1A (Blake et al. 2011).
Functional annotation of genes within the QSm.mrc-1A confidence interval of 42.7 Mb (536,613,453–579,299,659 bp) revealed 11 OWBM resistance-related candidate genes (CGs). Several different classes of insect resistance genes have been reported in plants, including genes encoding inhibitors of proteases (serine and cysteine) and alpha-amylase, plant lectins and enzymes such as chitinases and lipoxygenases (Gatehouse and Gatehouse 1998; Malone et al. 2008). However, few genes have been implicated in OWBM resistance (Hao et al. 2019). TraesCS1A02G384300 has been annotated as a gene involved in chemical reactions and pathways resulting in the biosynthesis of terpenoids. As reported previously, female midge uses fine-scale features and surface chemicals, including volatiles, to select oviposition sites on the wheat spike (Gharalari et al. 2009, 2012). Volatile organic compound(s) produced by deterrent genotypes may reduce oviposition on preferred genotypes and may cause the females to lay their eggs further from potential larval feeding sites (Lamb et al. 2003; Gharalari et al. 2009). No morphological trait has been identified that accounts for oviposition preferences, apart from a small effect of inter-spikelet distance (Lamb et al. 2001; Gharalari et al. 2009).
The oviposition deterrence QTL on chromosome 4B from BW278 is colocated with a major gene affecting plant height in the Superb/BW278 DH population. There was a strong association between the dwarfing alleles at Rht-B1 and increased kernel damage (QSm.mrc-4B) in the Superb/BW278 population (Table 4). Superb has the dwarfing allele Rht-B1b, which was associated with increased kernel damage at QSm.mrc-4B. Despite the widespread use of Rht-B1b and Rht-D1b in modern wheat varieties for increasing wheat grain yields and providing lodging resistance, their associations with increased susceptibility to some plant pathogens have been reported several times in past studies (Srinivasachary et al. 2009; Saville et al. 2011). Previously, a QTL mapping study of yield and yield components in a spring wheat cross between a high-yielding variety, Superb (Grandin*2/AC Domain) and BW278 revealed that Superb carried a dwarfing allele at Rht-B1 from Grandin and associated with increased yield (https://mspace.lib.umanitoba.ca/bitstream/handle/1993/21231/Cuthbert_Molecular_Mapping.pdf), but also provides a microclimate more favorable for pathogen establishment due to reduced height (Scott et al. 1985). However, this association between the Rht-B1b and increased OWBM damage may not translate into increased OWBM damage in commercial fields. OWBM may preferentially oviposit on short genotypes in small plot experiments, possibly to avoid the wind. This would explain the correlation observed between plant height and OWBM damage in this study. Such an effect may not occur in commercial fields planted to monoculture.
The awned parent Superb contributed oviposition deterrence at the QTL QSm.mrc-5A, which colocated with the position of the B1 awn inhibitor locus (Huang et al. 2019) in the Superb/BW278 population. QSm.mrc-5A and B1 mapped approximately 15 cM distal of the HD QTL QHd.mrc-5A on chromosome 5A (Table 4 and Supplementary Table S4). Since QHd.mrc-5A does not colocate with QSm.mrc-5A, the significant correlation between HD and OWBM kernel damage in the Superb/BW278 DH population is most likely due to linkage rather than pleiotropy (Tables 3, 4). The earlier maturing parent Superb contributed earliness allele at the HD QTL QHd.mrc-5A in the Superb/BW278 DH population. Earliness per se genes (Eps) are known to regulate flowering time independently of vernalization genes (Vrn) and photoperiod genes (Ppd), and are important for the fine-tuning of flowering time (Lewis et al. 2008). Eps genes have been reported to induce earlier flowering, even in the presence of Vrn and Ppd genes (van Beem et al. 2005), and they have been mapped as QTL for heading time on different chromosomes on wheat (Kamran et al. 2013; Zanke et al. 2014). Liu et al. (2005) identified a major Eps QTL designated as QEet.fcu-5AL on chromosome 5A for earliness contributed by Grandin in the Grandin/BR34 RIL population. Grandin is a parent of Superb (pedigree: Grandin*2/AC Domain).
Additional QTL for oviposition deterrence were identified on chromosomes 3B and 7A, and their effects were lower and less consistent than the major oviposition deterrence QTL on chromosomes 1A, 4B and 5A. Two minor effect QTL, QSm.mrc-3B and QSm.mrc-7A.2 detected based on HMDK from BW278, and one minor QTL, QSm.mrc-7A.1 detected based on UMDK from Superb, were also responsible for reducing OWBM damage in the Superb/BW278 DH population (Table 4).
Gharalari et al. (2009) suggested that oviposition deterrence is controlled by multiple genes, with complementary interaction among genes. Difficult phenotyping and complex genetics would make it difficult to incorporate this trait into breeding programs. The present genetic study revealed that oviposition deterrence was controlled by several genes. However, QSm.mrc-1A had a major impact on MDK such that MAS of this QTL combined with phenotypic selection of awned genotypes would provide a basis for efficient selection of oviposition deterrence to OWBM. The 11 OWBM candidate genes for QSm.mrc-1A reported in this study would be an appropriate set of genes for follow-up genetic research on oviposition deterrence. In conclusion, the findings of this study provide insight into the inheritance of oviposition deterrence to OWBM and provide information for pyramiding the OWBM resistance gene Sm1 with oviposition deterrence.
Availability of data and material
Data supporting the current study can be obtained by contacting the corresponding author (curt.mccartney@canada.ca).
Abbreviations
- ANOVA:
-
Analysis of variance
- BLAST:
-
Basic local alignment search tool
- BLUPs:
-
Best linear unbiased predictors
- DH:
-
Doubled haploid
- HMDK:
-
Harvestable midge-damaged kernels
- HD:
-
Heading date
- ICIM:
-
Inclusive composite interval mapping
- IM:
-
Interval mapping
- LOD:
-
Logarithm of odds
- MAS:
-
Marker-assisted selection
- MDK:
-
Midge-damaged kernels
- META-R:
-
Multi Environment Trial Analysis with R
- OWBM:
-
Orange wheat blossom midge
- HT:
-
Plant height
- PVE:
-
Phenotypic variation explained
- QTL:
-
Quantitative trait loci
- SNP:
-
Single-nucleotide polymorphism
- UMDK:
-
Unharvestable midge-damaged kernels
References
Alvarado G, López M, Vargas M, Pacheco Á, Rodríguez F, Burgueño J, Crossa J (2017) META-R (Multi Environment Trail Analysis with R for Windows) Version 6.01. CIMMYT research data & software repository network. https://hdl.handle.net/11529/10201.
Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, Poland J et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:7191. https://doi.org/10.1126/science.aar7191
Barnes HF (1956) Gall Midges of Economic Importance. Gall midges of cereal crops. Crosby Lockwood & Son Ltd., London
Berzonsky WA, Ding H, Haley SD, Harris MO, Lamb RJ, McKenzie RI, Ohm HW, Patterson FL, Peairs FB, Porter DR, Ratcliffe RH, Shanower TG (2002) Breeding Wheat for Resistance to Insects. In: Janick J (ed) Plant Breeding Reviews. vol 22, pp 221–296. https://doi.org/10.1002/9780470650202.ch5
Blake NK, Stougaard RN, Weaver DK, Sherman JD, Lanning SP, Naruoka Y, Xue Q, Martin JM, Talbert LE (2011) Identification of a quantitative trait locus for resistance to Sitodiplosis mosellana (Géhin), the orange wheat blossom midge, in spring wheat. Plant Breed 130:25–30. https://doi.org/10.1111/j.1439-0523.2010.01809.x
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, See D, Bai G, Pumphrey M, Tomar L, Wong D, Kong S, Reynolds M, da Silva ML, Bockelman H, Talbert L, Anderson JA, Dreisigacker S, Baenziger S, Carter A, Korzun V, Morrell PL, Dubcovsky J, Morell MK, Sorrells ME, Hayden MJ, Akhunov E (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Nat Acad Sci USA 110:8057–8062. https://doi.org/10.1073/pnas.1217133110
Dexter JE, Preston KR, Cooke LA, Morgan BC, Kruger JE, Kilborn RH, Elliott RH (1987) The influence of orange wheat blossom midge (Sitodiplosis mosellana Géhin) damage on hard red spring wheat quality and the effectiveness of insecticide treatments. Can J Plant Sci 67:697–712. https://doi.org/10.4141/cjps87-097
Dhokane D, Karre S, Kushalappa AC, McCartney C (2016) Integrated metabolo-transcriptomics reveals Fusarium head blight candidate resistance genes in wheat QTL-Fhb2. PLoS ONE 11:e0155851. https://doi.org/10.1371/journal.pone.0155851
Ding H, Lamb RJ, Ames N (2000) Inducible production of phenolic acids in wheat and antibiotic resistance to Sitodiplosis mosellana. J Chem Ecol 26:969–985. https://doi.org/10.1023/a:1005412309735
Elliott B, Olfert O, Hartley S (2011) Management practices for wheat midge, Sitodiplosis mosellana (Géhin). Prairie Soils Crops 4:8–13
Elliott RH, Mann LW (1996) Susceptibility of red spring wheat, Triticum aestivum L. cv. Katepwa, during heading and anthesis to damage by wheat midge, Sitodiplosis mosellana (Géhin) (Diptera: Cecidomyiidae). Can Entomol 128:367–375. https://doi.org/10.4039/Ent128367-3
Fox SL, Thomas JB, Wise IL, Smith MAH, Humphreys DG, Brown PD, Townley-Smith TF, McCallum BD, Fetch TG, Menzies JG, Gilbert JA, Fernandez MR, Despins T, Niziol D (2009) Waskada hard red spring wheat. Can J Plant Sci 89:929–936. https://doi.org/10.4141/CJPS08222
Gatehouse AMR, Gatehouse JA (1998) Identifying proteins with insecticidal activity: use of encoding genes to produce insect-resistant transgenic crops. Pestic Sci 52:165–175. https://doi.org/10.1002/(SICI)1096-9063(199802)52:2%3c165::AID-PS679%3e3.0.CO;2-7
Gharalari AH, Fox SL, Smith MAH, Lamb RJ (2009) Oviposition deterrence in spring wheat, Triticum aestivum, against orange wheat blossom midge, Sitodiplosis mosellana: implications for inheritance of deterrence. Entomol Exp Appl 133:74–83. https://doi.org/10.1111/j.1570-7458.2009.00906.x
Gharalari AH, Smith MAH, Fox SL, Lamb RJ (2012) Behaviour of Sitodiplosis mosellana (Diptera: Cecidomyiidae) on spring wheat spikes with and without oviposition deterrence. Can Entomol 142:574–583. https://doi.org/10.4039/n10-030
Hao Z, Geng M, Hao Y, Zhang Y, Zhang L, Wen S, Wang R, Liu G (2019) Screening for differential expression of genes for resistance to Sitodiplosis mosellana in bread wheat via BSR-seq analysis. Theor Appl Genet 132:3201–3221. https://doi.org/10.1007/s00122-019-03419-9
Huang D, Zheng Q, Melchkart T, Bekkaoui Y, Konkin DJF, Kagale S, Martucci M, You FM, Clarke M, Adamski NM, Chinoy C, Steed A, McCartney CA, Cutler AJ, Nicholson P, Feurtado JA (2019) Dominant inhibition of awn development by a putative zinc-finger transcriptional repressor expressed at the B1 locus in wheat. New Phytol 225:340–355. https://doi.org/10.1111/nph.16154
Kamran A, Iqbal M, Navabi A, Randhawa H, Pozniak C, Spaner D (2013) Earliness per se QTLs and their interaction with the photoperiod insensitive allele Ppd-D1a in the Cutler x AC Barrie spring wheat population. Theor Appl Genet 126:1965–1976. https://doi.org/10.1007/s00122-013-2110-0
Kassa MT, Haas S, Schliephake E, Lewis C, You FM, Pozniak CJ, Kramer I, Perovic D, Sharpe AG, Fobert PR, Koch M, Wise IL, Fenwick P, Berry S, Simmonds J, Hourcade D, Senellart P, Duchalais L, Robert O, Forster J, Thomas JB, Friedt W, Ordon F, Uauy C, McCartney CA (2016) A saturated SNP linkage map for the orange wheat blossom midge resistance gene Sm1. Theor Appl Genet 129:1507–1517. https://doi.org/10.1007/s00122-016-2720-4
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175. https://doi.org/10.1111/j.1469-1809.1943.tb02321.x
Lamb RJ, McKenzie RIH, Wise IL, Barker PS, Smith MAH, Olfert OO (2000) Resistance to Sitodiplosis mosellana (Diptera: Cecidomyiidae) in spring wheat (Gramineae). Can Entomol 132:591–605. https://doi.org/10.4039/Ent132591-5
Lamb RJ, Smith MAH, Wise IL, Clarke P, Clarke J (2001) Oviposition deterrence to Sitodiplosis mosellana (Diptera: Cecidomyiidae): a source of resistance for durum wheat (Gramineae). Can Entomol 133:579–591. https://doi.org/10.4039/Ent133579-4
Lamb RJ, Sridhar P, Smith MAH, Wise IL (2003) Oviposition preference and offspring performance of a wheat midge Sitodiplosis mosellana (Géhin) (Diptera: Cecidomyiidae) on defended and less defended wheat plants. Environ Entomol 32:414–420
Lamb RJ, Wise IL, Olfert OO, Gavloski J, Barker PS (1999) Distribution and seasonal abundance of Sitodiplosis mosellana (Diptera: Cecidomyiidae) in spring wheat. Can Entomol 131:387–397. https://doi.org/10.4039/Ent131387-3
Lamb RJ, Wise IL, Smith MAH, McKenzie RIH, Thomas J, Olfert OO (2002) Oviposition deterrence against Sitodiplosis mosellana (Diptera: Cecidomyiidae) in spring wheat (Gramineae). Can Entomol 134:85–96. https://doi.org/10.4039/Ent13485-1
Lewis S, Faricelli ME, Appendino ML, Valarik M, Dubcovsky J (2008) The chromosome region including the earliness per se locus Eps-Am 1 affects the duration of early developmental phases and spikelet number in diploid wheat. J Exp Bot 59:3595–3607. https://doi.org/10.1093/jxb/ern209
Li H, Ribaut JM, Li Z, Wang J (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116:243–260. https://doi.org/10.1007/s00122-007-0663-5
Li H, Ye G, Wang J (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374. https://doi.org/10.1534/genetics.106.066811
Liu ZH, Anderson JA, Hu J, Friesen TL, Rasmussen JB, Faris JD (2005) A wheat intervarietal genetic linkage map based on microsatellite and target region amplified polymorphism markers and its utility for detecting quantitative trait loci. Theor Appl Genet 111:782–794. https://doi.org/10.1007/s00122-005-2064-y
Lorieux M (2012) MapDisto: fast and efficient computation of genetic linkage maps. Mol Breed 30:1231–1235. https://doi.org/10.1007/s11032-012-9706-y
Maccaferri M, Cane MA, Sanguineti MC, Salvi S, Colalongo MC, Massi A, Clarke F, Knox R, Pozniak CJ, Clarke JM, Fahima T, Dubcovsky J, Xu S, Ammar K, Karsai I, Vida G, Tuberosa R (2014) A consensus framework map of durum wheat (Triticum durum Desf) suitable for linkage disequilibrium analysis and genome-wide association mapping. BMC Genomics 15:873. https://doi.org/10.1186/1471-2164-15-873
Malone LA, Gatehouse AMR, Barratt BIP (2008) Beyond Bt: Alternative Strategies for Insect-Resistant Genetically Modified Crops. In: Romeis J, Shelton AM, Kennedy GG (eds) Integration of Insect-Resistant Genetically Modified Crops within IPM Programs. Springer, Netherlands. https://doi.org/10.1007/978-1-4020-8373-0_13
McKenzie RIH, Lamb RJ, Aung T, Wise IL, Barker P, Olfert OO, McIntosh RI (2002) Inheritance of resistance to wheat midge, Sitodiplosis mosellana, in spring wheat. Plant Breed 121:383–388. https://doi.org/10.1046/j.1439-0523.2002.745267.x
Morando R, Baldin ELL, Cruz PL, Lourenção AL, Chiorato AF (2015) Antixenosis of bean genotypes to Chrysodeixis includens (Lepidoptera: Noctuidae). Pesqui Agropecu Bras 50:450–458. https://doi.org/10.1590/s0100-204x2015000600003
Olfert O, Elliott RH, Hartley S (2009) Non-native insects in agriculture: Strategies to manage the economic and environmental impact of wheat midge, Sitodiplosis mosellana, in Saskatchewan. Biol Invasions 11:127–133. https://doi.org/10.1007/s10530-008-9324-0
Olfert OO, Mukerji MK, Doane JF (2012) Relationship between infestation levels and yield loss caused by wheat midge, Sitodiplosis mosellana (Géhin) (Diptera: Cecidomyiidae), in spring wheat in Saskatchewan. Can Entomol 117:593–598. https://doi.org/10.4039/Ent117593-5
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400:256–261. https://doi.org/10.1038/22307
Saville RJ, Gosman N, Burt CJ, Makepeace J, Steed A, Corbitt M, Chandler E, Brown JKM, Boulton MI, Nicholson P (2011) The ‘Green Revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. J Exp Bot 63:1271–1283. https://doi.org/10.1093/jxb/err350
Scott PR, Benedikz PW, Zones HG, Ford MA (1985) Some effects of canopy structure and microclimate on infection of tall and short wheats by Septoria nodorum. Plant Pathol 34:578–593. https://doi.org/10.1111/j.1365-3059.1985.tb01410.x
Smith MAH, Wise IL, Fox SL, Vera CL, DePauw RM, Lukow OM (2014) Seed damage and sources of yield loss by Sitodiplosis mosellana (Diptera: Cecidomyiidae) in resistant wheat varietal blends relative to susceptible wheat cultivars in western Canada. Can Entomol 146:335–346. https://doi.org/10.4039/tce.2013.77
Srinivasachary GN, Steed A, Simmonds J, Leverington-Waite M, Wang Y, Snape J, Nicholson P (2009) Susceptibility to Fusarium head blight is associated with the Rht-D1b semi-dwarfing allele in wheat. Theor Appl Genet 116:1145–1153. https://doi.org/10.1007/s00122-008-0742-2
Thomas J, Fineberg N, Penner G, McCartney C, Aung T, Wise I, McCallum B (2005) Chromosome location and markers of Sm1: a gene of wheat that conditions antibiotic resistance to orange wheat blossom midge. Mol Breed 15:183–192. https://doi.org/10.1007/s11032-004-5041-2
Townley-Smith TF, Humphreys DG, Czarnecki E, Lukow OM, McCallum BM, Fetch TG, Gilbert JA, Menzies JG, Brown PD (2010) Superb hard red spring wheat. Can J Plant Sci 90:347–352. https://doi.org/10.4141/CJPS09087
van Beem J, Mohler V, Lukman R, van Ginkel M, William M, Crossa J, Worland AJ (2005) Analysis of genetic factors influencing the developmental rate of globally important CIMMYT wheat cultivars. Crop Sci 45:2113–2119. https://doi.org/10.2135/cropsci2004.0665
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E, International Wheat Genome Sequencing C (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796. https://doi.org/10.1111/pbi.12183
Wise IL, Fox SL, Smith MAH (2015) Seed damage by Sitodiplosis mosellana (Diptera: Cecidomyiidae) to spring wheat cultivars with the Sm1 gene. Can Entomol 147:754–765. https://doi.org/10.4039/tce.2014.75
Wright AT, Doane J (1987) Wheat midge infestation of spring cereals in northeastern Saskatchewan. Can J Plant Sci 67:117–120. https://doi.org/10.4141/cjps87-013
Zanke C, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Hinze M, Beier S, Ganal MW, Roder MS (2014) Genetic architecture of main effect QTL for heading date in European winter wheat. Front Plant Sci 5:217. https://doi.org/10.3389/fpls.2014.00217
Zhang L, Geng M, Zhang Z, Zhang Y, Yan G, Wen S, Liu G, Wang R (2020) Molecular mapping of major QTL conferring resistance to orange wheat blossom midge (Sitodiplosis mosellana) in Chinese wheat varieties with selective populations. Theor Appl Genet 133:491–502. https://doi.org/10.1007/s00122-019-03480-4
Acknowledgements
The authors thank Sheila Wolfe, Leslie Bezte, Suzanne Enns, Alain Ngantcha and other technical staff from the participating labs for their contributions to this research. The authors also wish to thank Zhen Yao for bioinformatics assistance. This project was funded by the Saskatchewan Agriculture Development Fund (grant 20140250), the Western Grains Research Foundation and the Saskatchewan Wheat Development Commission.
Author information
Authors and Affiliations
Contributions
All authors planned and organized the study. CM, IW, MS and SF designed the study. CM, CP, SK and AB grew field nurseries. CM, IW, MS and AC assessed OWBM damage. DT and CM developed the linkage maps, conducted QTL analyses, interpreted results and wrote the first draft of the manuscript. All authors contributed to and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and animals participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by Aimin Zhang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thambugala, D., Pozniak, C.J., Kumar, S. et al. Genetic analysis of oviposition deterrence to orange wheat blossom midge in spring wheat. Theor Appl Genet 134, 647–660 (2021). https://doi.org/10.1007/s00122-020-03720-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03720-y