Abstract
Key message
Silage quality traits of maize hybrids between the Dent and Flint heterotic groups mostly involved QTL specific of each parental group, some of them showing unfavorable pleiotropic effects on yield.
Abstract
Maize (Zea mays L.) is commonly used as silage for cattle feeding in Northern Europe. In addition to biomass production, improving whole-plant digestibility is a major breeding objective. To identify loci involved in the general (GCA, parental values) and specific combining ability (SCA, cross-specific value) components of hybrid value, we analyzed an incomplete factorial design of 951 hybrids obtained by crossing inbred lines issued from two multiparental connected populations, each specific to one of the heterotic groups used for silage in Europe (“Dent” and “Flint”). Inbred lines were genotyped for approximately 20K single nucleotide polymorphisms, and hybrids were phenotyped in eight environments for seven silage quality traits measured by near-infrared spectroscopy, biomass yield and precocity (partly analyzed in a previous study). We estimated variance components for GCA and SCA and their interaction with environment. We performed QTL detection using different models adapted to this hybrid population. Strong family effects and a predominance of GCA components compared to SCA were found for all traits. In total, 230 QTL were detected, with only two showing SCA effects significant at the whole-genome level. More than 80% of GCA QTL were specific of one heterotic group. QTL explained individually less than 5% of the phenotypic variance. QTL co-localizations and correlation between QTL effects of quality and productivity traits suggest at least partial pleiotropic effects. This work opens new prospects for improving maize hybrid performances for both biomass productivity and quality accounting for complementarities between heterotic groups.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Silage maize is an important crop for cattle feeding, especially in Northern Europe. One important challenge is to improve maize varieties for their silage productivity (biomass production) as well as for their feeding value that corresponds to the energy available for cattle.
Silage maize energy is determined by (1) the proportion of grains and cellular content of stalks and leaves, which are assumed to be fully digestible, and (2) the digestibility of cell walls, which is highly variable. Cell wall is mainly composed of polysaccharidic compounds (cellulose and hemicelluloses) and of phenolic compounds (lignin and p-hydroxycinnamic acids). Cell wall digestibility is the most limiting factor of silage value (Barrière et al. 2003) and was found to mainly depend on lignin content and cross-linkages between lignin and the other cell wall components (Hartley 1972; Ralph et al. 1994; Grabber et al. 1996; Méchin et al. 2000; Fontaine et al. 2003; Zhang et al. 2011; El Hage et al. 2018). In Northern Europe, maize varieties commonly used as silage for cattle feeding are hybrids resulting from crosses between the “Dent” and the “Flint” heterotic groups. In a first step, lines are selected within each heterotic group for their general combining ability (GCA), i.e., the average performance of their hybrid progeny with lines from the other group. In a second step, selected lines are crossed in order to identify intergroup hybrids showing the best specific combining abilities (SCA). As SCA is only evaluated in the second step for pairs of lines already selected based on their GCA value, the observed variation for this component may be biased, potentially hindering the identification of the best hybrid combinations. One important issue for the optimization of hybrid mating designs is therefore to evaluate the magnitude of SCA compared to GCA and also to evaluate the sensitivity of these components to environmental conditions. The Dent group, mostly from recent US origin, generally confers productivity, whereas the Flint group confers adaptation to European conditions. In the 1980s, most of successful hybrids in Europe were derived from the cross between Dent lines from the Iodent subgroup, and Flint lines related to the Lacaune population, which produced, in particular, the key founder line F2. These two origins showed good combining ability for yield-related traits, but their use in Europe resulted in a decrease in cell wall digestibility due to a negative correlation between these traits (Barrière et al. 2003). To stop this decrease, a new trait, the Milk Fodder Unit (MFU), which represents the energy available for milk production per kg of silage dry matter was added in 1998 in the official tests performed in France for the registration of silage maize varieties. The correlation between silage yield and cell wall digestibility depends on the genetic material considered. For instance, no correlation was found by Argillier et al. (2000) on a set of hybrids between different heterotic groups, whereas Barrière and Emile (2000) and Surault et al. (2005) found a correlation of − 0.5 between these traits. These contrasting results call for a better understanding of the genetic architecture of digestibility-related traits and their relationship with biomass yield.
Since the later 1990s, several studies have been carried out in order to detect loci (or QTL) involved in the architecture of silage quality traits (Lübberstedt et al. 1997a, b, 1998; Méchin et al. 2001; Roussel et al. 2002; Krakowsky et al. 2003; Cardinal et al. 2003; Krakowsky et al. 2005, 2006; Barrière et al. 2007; Riboulet et al. 2008; Barrière et al. 2008, 2010; Courtial et al. 2013, 2014; Torres et al. 2014; Penning et al. 2014; Leng et al. 2018) and QTL related to cell wall digestibility and cell wall composition were found all over the maize genome. In a meta-analysis of QTL associated with maize silage quality traits based on eleven experiments, Truntzler et al. (2010) identified numerous meta-QTL (MQTL) for digestibility and cell wall composition traits (26 and 42 MQTL, respectively). These studies emphasize the genetic complexity of silage quality traits, which mainly involve QTL with small effects. QTL detections conducted so far for biomass quality traits were mostly carried out on recombinant inbred line (RIL) populations evaluated per se or on test-cross hybrids obtained by crossing RILs to a single inbred line from a complementary group (called tester). These populations cannot be used to detect dominance effects. Consequently, little is known on the additive/dominance or GCA/SCA QTL effects. Significant correlations between per se and test-cross values were found in some studies, suggesting a small impact of the dominance on silage quality traits (Barrière et al. 2003, 2010). Recently, Leng et al. (2018) found only moderate correlations between per se and test-cross performances for silage quality traits in a Dent and in a Flint population, resulting in a small consistency between QTL detected in both types of populations, therefore leaving the question of the role of dominance open. Another limit of studies conducted so far is that they mostly involved biparental populations analyzed separately, each representing only a small part of the variability available for breeding. Compared to biparental populations, multiparental populations enable to estimate allelic series at QTL, but they have never been used so far to study traits related to silage digestibility.
This work aimed at getting more insight into the genetic architecture of silage quality traits in Dent–Flint hybrids. We used an original hybrid population issued from a factorial design obtained by crossing lines issued from a Dent and a Flint multiparental populations, composed each of six connected unselected biparental families. This population was first analyzed for silage productivity traits and precocity (Giraud et al. 2017a, b). The family structure of the population enabled performing QTL detection using a combination of linkage and association mapping approaches that were found complementary. However, Giraud et al. (2017a, b) did not consider environmental variation. Our objectives in the present study were to (1) evaluate GCA/SCA variance components for silage quality traits and compare them with those found for productivity traits, with a focus on their interaction with the environment, (2) measure correlations between traits and (3) implement QTL detection for silage biomass quality traits considering GCA and SCA components or additive/dominance effects depending on the model. This enabled us to evaluate the group specificity of the QTL involved in hybrid variation and the importance of dominance/SCA for silage quality traits. We then compared QTL results with those obtained for biomass yield and precocity by Giraud et al. (2017a, b) to better decipher potential correlations between biomass production and quality traits. Finally, we evaluated prospects of these results for breeding.
Materials and methods
Biological material
We used the same hybrid population as Giraud et al. (2017a, b). This population was obtained by crossing segregating biparental families from the Dent and Flint group. In each group, four founder inbred lines (F373, F03802, F02803 and F7088 for the Flint group; F98902, F1808, F04401 and F7082 for the Dent group, Table S1) were crossed following a half-diallel mating design to derive six Dent biparental families (called D1 to D6) obtained by haplodiploidization and six Flint biparental families (called F1 to F6) obtained by four generations of single seed descent (SSD) (Table S2). Among the founders of each group, one inbred line (F7088 for the Flint and F7082 for the Dent) was selected for its good biomass digestibility, and the three others for their good agronomical potential for yield and precocity. In the Flint group, founder lines except F7088 mostly originated from material issued from the French Lacaune open-pollinated population crossed with other sources of early Flint material. F7088 derived from the cross between the F4 line and a commercial hybrid, both known for their good digestibility (Barrière et al. 2017, and Bauland, personal communication). In the Dent group, founder lines except F7082 derived from synthetics of commercial hybrids mostly related to the Iodent heterotic group. F7082, which was chosen for its high digestibility, derived from a cross between a line from the Stiff-Stalk group and a line from the Minnesota 13 group, known for its good digestibility. Each biparental family of one group was crossed with all the biparental families of the other group (Table S2). To evaluate as many lines as possible, most lines contributed to only one hybrid (699 in the Dent and 732 in the Flint), but about 20% of them contributed to two hybrids (163 in the Dent group and 146 in the Flint group) to allow estimation of GCA and SCA variance components. Note that only one Dent and one Flint line contributed to three or four hybrids, respectively. Founder lines of each group were crossed to produce 16 Dent–Flint hybrids, further referred to as “founder hybrids,” that were used as checks in field trials. All biological materials are described in detail in Giraud et al. (2017a).
Genotypic data
Founder and parental lines were genotyped with an 18480 SNPs Affymetrix® array provided by Limagrain. For QTL detection, we considered 9548 SNP markers that were mapped on the Dent–Flint consensus map established for the population (Giraud et al. 2017a). This map had a total length of 1578.6 cM and 5216 unique positions. Missing genotypes were imputed with Beagle v3.0 (Browning and Browning 2007). At each marker, probabilities of the four Flint (respectively, Dent) founder alleles transmitted to the parental lines were inferred using PlantImpute (Hickey et al. 2015). Detailed information on the map, quality control of the data, imputation and computation of founder allele probabilities is given in Giraud et al. (2017a). All the genotypic data and the map positions are available as supplemental files of Giraud et al. (2017a).
Field trial design and analysis
Hybrids were evaluated for silage in 8 different environments over two years (4 in 2013 and 4 others in 2014) in northern France and in Germany (Table S3). Field experiments were laid out as an augmented p-rep design (Williams et al. 2011) and were constituted of 1088 elementary plots, each containing two rows of five meters length. Plots were split into 68 incomplete blocks of 16 elementary plots each. Most of experimental hybrids were evaluated only once in each trial. Founder hybrids and around 17% of experimental hybrids were evaluated twice. In each block, five to six plots were used for repeated genotypes and hybrids were allocated to block to form an efficient incomplete block design. At harvest, a sample was taken from each plot, dried and ground. Silage quality traits were predicted using near-infrared reflectance spectrometry (NIRS) equations on silage powder. For six over the eight field trials, NIRS predictions were made at INRA from spectra supplied by experimenters using the 2013 equation of Gembloux University (version “135b”). For the two remaining field trials, predictions were directly supplied by experimenters and spectra were not transmitted. Seven silage quality traits were considered: cell wall content of the harvested dry matter measured by the neutral detergent fiber content (NDF in % of dry matter), cellulose, hemicellulose and lignin contents in the cell wall NDF (CELL, HCELL and LIGNIN in % of NDF) evaluated according to the method of Goering and Van Soest (1970), feeding value [MFU in Milk Fodder Unit per kilogram of dry matter (Andrieu 1995)], cell wall in vitro digestibility of the “non-starch and non-soluble carbohydrates” part of silage (DINAG in %) and cell wall in vitro digestibility of the “non-starch, non-soluble carbohydrates and non-crude protein” part of silage (DINAGZ in %). These two digestibility criteria were proposed by Argillier et al. (1995). Data from all eight environments were available for MFU and DINAG; for NDF, cellulose, hemicellulose and lignin, data from seven trials were available and six for DINAGZ. For trials for which we did not have access to spectra, we checked that the predictions of silage quality traits were well correlated to those obtained at INRA for the other trials (results not shown). In this study, we also considered silage yield (DMY in tons of dry matter per ha), dry matter content at harvest (DMC in %) and female flowering time (DtSILK in days after January the first) to complete the analyses previously done by Giraud et al. (2017a, b) by estimating (1) variance components including G × E interactions and (2) correlations with digestibility traits.
An exploratory analysis was carried out for each trait and each trial to eliminate outlying observations (7 observations in total). Then, all data of hybrids with dry matter content at silage harvest below 25% (33 observations) and above 45% (53 observations) were considered as missing data because NIRS predictions are unreliable for samples presenting extreme moisture (Baker et al. 1994; Stuth et al. 2003; Méchin and Reymond, personal communication). 951 experimental hybrids, corresponding to hybrids with validated phenotypic records and for which both parental lines had genotypic data consistent with their pedigree (821 Flint and 801 Dent parental lines), were considered for further analyses. The number of analyzed hybrids derived from each biparental family varied between 126 (for family D6) and 178 (for family F1). For each Flint–Dent family combination, between 15 (Dent family D6–Flint family F3) and 34 hybrids (Dent family D2–Flint family F1) were evaluated (Table S2).
Variance component analysis
Before performing QTL detection, we estimated heritabilities, GCA and SCA variance components and the contribution of the family structure to the hybrid variation for silage quality traits. Genetic variance decomposition was done on the single-plot performances using the ASReml-R package (Butler et al. 2007; R Core Team 2013). We used models that included a genetic effect of the hybrids, a fixed trial effect and some random effects controlling spatial heterogeneities within trials (using either a row–column effects or block effects).
where \(Y_{hlxyz}\) is the phenotypic value of the hybrid \(h\) evaluated in the trial \(l\) at the plot located at row \(x\), column \(y\) and in block \(z\). \(\mu\) is the intercept, and \(\lambda_{l}\) is the fixed effect of the trial \(l\). To distinguish between the checks and the experimental hybrids, two genetic effects were included: \(\tau_{h}\) was a fixed genetic effect for the checks which had a specific level for each check and the same level for all experimental hybrids; \(H_{h}\) was a random genetic effect with one level per experimental hybrid and the same level for all checks. \(\left( {\tau \lambda } \right)_{hl}\) and \((H\lambda )_{hl}\) were the corresponding genotype by trial interaction effects, considered as fixed for the checks and as random for the experimental hybrids. \(P_{xyz\left( l \right)}\) was the effect of the plot within the trial l. Depending on the trial, we used either the sum of a random row and a random column effect or a random block effect to model \(P_{xyz\left( l \right)}\). \(E_{hlxyz}\) was the residual of the model assumed to be independent and identically distributed (iid) within a given trial but with a different variance in each trial. In this generic model, the hybrid value was decomposed into GCA and SCA components with or without considering the family structure. In a first model (model 1) the family structure was not considered:
where \(H_{{h\left( {kk^{\prime}} \right)}}\) is the genetic effect of the experimental hybrid h between the Flint parental line \(k\) and the Dent parental line \(k'\). \(P_{k}\) (respectively, \(P^{\prime}_{{k^{\prime}}}\)) is the random effect of the parental line \(k\) (respectively, \(k'\)) issued from the Flint founder lines \(i\) and \(j\) (respectively, Dent founder lines \(i'\) and \(j'\)), corresponding to the GCAs of the parental lines with \(P_{k} \hookrightarrow N\left({0, \sigma_{{GCA_{d}}}^{2}} \right)\) and assumed to be iid (respectively \(P^{\prime}_{{k^{\prime}}} \hookrightarrow N\left({0, \sigma_{{GCA_{f}}}^{2}} \right)\), iid). \(\left( {PP^{\prime}} \right)_{{kk^{\prime}}}\) is the random effect of the interaction between the inbred line \(k\) and the inbred line \(k'\), corresponding to the SCA with \(\left({PP^{\prime}} \right)_{{kk^{\prime}}} \hookrightarrow N\left({0, \sigma_{SCA}^{2}} \right)\) iid. The same decomposition of \(H_{{h\left( {kk^{\prime}} \right)}}\) was done for the interaction with the environment \((H\lambda )_{{h\left( {kk^{\prime}} \right)l}}\):
With \(\left({P\lambda} \right)_{kl} \hookrightarrow N\left({0, \sigma_{{GCA_{d} \times E}}^{2}} \right)\) iid. \(\left({P^{\prime}\lambda} \right)_{kl} \hookrightarrow N\left({0, \sigma_{{GCA_{f} \times E}}^{2}} \right)\) iid and \(\left({PP^{\prime}\lambda} \right)_{kl} \hookrightarrow N\left({0, \sigma_{SCA \times E}^{2}} \right)\) iid.
In the second model, hybrid value was decomposed into family effects and within-family GCA and SCA effects. Thus, \(H_{{kk^{\prime}}}\) was decomposed into:
where \(\alpha_{ij}\) (respectively, \(\beta_{{i^{\prime}j^{\prime}}}\)) is the fixed effect of the Flint (respectively, Dent) family origin of the Flint (respectively, Dent) parental line \(k\) (respectively, \(k'\)) and \(\left( {\alpha \beta } \right)_{{iji^{\prime}j^{\prime}}}\) is the fixed interaction effect between the Flint and Dent family origins of the parental lines. \(P_{k}^{*}\), \(P^{*'}_{{k^{\prime}}}\) and \(\left( {PP^{\prime}} \right)_{{kk^{\prime}}}^{*}\) are the within-family equivalents of \(P_{k}\), \(P^{\prime}_{{k^{\prime}}}\) and \(\left( {PP^{\prime}} \right)_{{kk^{\prime}}}\) in model (1). Similarly, the genotype by environment interaction \((H\lambda )_{{h\left( {kk^{\prime}} \right)l}}\) is decomposed as the sum of the interactions between the family and the environment, considered as fixed, and the interactions between the within-family GCA and SCA components and the environment, considered as random.
The percentage of within-family hybrid variance was estimated from the comparison of variance components issued from models (1) and (2). Broad-sense heritability \(H^{2}\) at the whole design level was calculated for each trait from model (1) as: \(H^{2} = \frac{{\sigma_{H}^{2} }}{{\sigma_{H}^{2} + \frac{{\sigma_{H \times E}^{2} }}{{n{\text{site}}_{H} }} + \frac{{\sigma_{E}^{2} }}{{n{\text{rep}}_{H} }}}}\) where \(\sigma_{H}^{2}\) is the genetic variance of the hybrids computed as the sum of the GCA and SCA components, \(n{\text{site}}_{H}\) and \(n{\text{rep}}_{H}\) are the average number of trials and the average number of plots per experimental hybrid in the whole design, respectively, \(\sigma_{H \times E}^{2}\) is the hybrid by environment interaction (equaled to the sum of \(\sigma_{{{\text{GCA}}_{\text{d}} \times E}}^{2}\), \(\sigma_{{{\text{GCA}}_{\text{f}} \times E}}^{2}\) and \(\sigma_{{{\text{SCA}} \times E}}^{2}\)), and \(\sigma_{E}^{2}\) is the average residual variance of the model over the different environments. The within-family heritability was calculated similarly but using the within-family variance components from model (2). Standard errors of estimation of heritabilities were obtained with the delta method implemented in the “pin” function of the R package “nadiv” (Wolak 2012). For each model we also estimated the percentages of SCA in the hybrid variance (with and without including SCA × E and GCA × E variance components) and the ratio of the G × E variance components over the total genetic variance component [100 × \(\sigma_{H \times E}^{2} /(\sigma_{H}^{2}\) + \(\sigma_{H \times E}^{2} )\)]. Note that in model 2, genetic variance components were assumed to be identical in all families. We relaxed this assumption by considering family-specific variances, but this did not improve significantly the likelihood of the models at a 5% level risk (results not shown).
QTL detections were based on the least square means (lsmeans) of each hybrid over the different environments. To obtain lsmeans, we first corrected the individual single-plot performances by the best linear unbiased predictions (BLUPs) of the spatial effects (block or row–column effects depending on their significance for the trait and trial considered) obtained with model (2). Then for each trait, lsmeans of hybrids were derived from a fixed effect model including a hybrid and a trial effect and considering residual variances specific of each trial. Pearson’s correlations between the different traits and their significance level were calculated based on the lsmeans of experimental hybrids used for QTL detection. Plot performances of experimental hybrids, corrected for spatial heterogeneities, were analyzed with a bivariate model to estimate genetic variances and covariances between traits and derive genetic correlations. In the bivariate model, we considered a fixed trial effect and a random hybrid effect and took into account the heterogeneity of error variances among trials. Due to convergence issues, G × E interactions were not included in this model. The R “nadiv” (Wolak 2012) package was used to compute standard errors of estimation of the genetic correlations.
QTL detection
In addition to the possibility of estimating SCA/dominance effects at QTL, an advantage of the studied population is to enable different allele codings for QTL detection, making it possible to test whether QTL are specific or common to the two heterotic groups and/or if there is an allelic series at each QTL. Following Giraud et al. (2017a, b), three models that differed in the allele coding were tested for QTL detection of silage quality traits. The general QTL detection model can be written as:
where \(y\) is a (N × 1) vector of the lsmeans of the hybrids with \(N\) being the number of experimental hybrids phenotyped for the considered trait; \(\mu\) is the intercept; \(1\) is a (N × 1) vector of 1. α (respectively, β) is a (6 × 1) vector of the fixed effects of the Flint (respectively, Dent) family of origin of the Flint (Dent) parental line; (αβ) is the (36 × 1) vector of the fixed interaction effects between the Flint and Dent families of parental lines. A, B and are the corresponding design matrices. \(u_{f}\) (respectively, \(u_{d}\)) is the (\(N_{f}\) × 1) [respectively, (\(N_{d}\) × 1)] vector of the random effects of the \(N_{f}\) Flint (respectively, \(N_{d}\) Dent) parental lines, with \(u_{f} \hookrightarrow N \left({0, I\sigma_{{u_{f}}}^{2}} \right)\), (respectively, \(u_{d} \hookrightarrow N \left({0, I\sigma_{{u_{d}}}^{2}} \right)\), ). \(\gamma_{q}\) is the vector of QTL effects for locus q, and \(X_{q}\) is the incidence matrix that includes information based on marker genotypes (see below for the three different QTL models implemented). \(Z_{f}\) and \(Z_{d}\) are the design matrices that relate the \(N\) experimental hybrids to the \(N_{f}\) Flint and \(N_{d}\) Dent parents. These genetic effects corresponding to the parents of the hybrids are included to account for the fact that some hybrids were derived from the same parental inbred lines. is the ( × 1) column vector of the residuals of the model with \( e\hookrightarrow N \left({0, {\text{I}}\sigma_{e}^{2}} \right)\) .
The first QTL model, called “Founder alleles,” considers that each of the eight founder lines carries a different allele and decomposes QTL effect into its GCA and SCA components. In this model \(\gamma_{q}\) contains 24 effects, four corresponding to the Flint parental GCA allelic effects, 4 to the Dent GCA parental allelic effects and 16 pairwise SCA interactions between the Dent and Flint allelic effects. \(X_{q}\) is the matrix including the founder allele probabilities. The first four columns correspond to the probabilities (between 0 and 1) that a given hybrid received a given Flint founder line allele, and the next four contain the founder allele probabilities for the Dent founder lines. The last 16 columns correspond to the Hadamard product of the four columns of \(X_{q}\) corresponding to the four Dent alleles with the ones corresponding to the four Flint alleles. This model has 3 degrees of freedom (df) for the Dent and the Flint GCA allelic effects, respectively, and 9 df for the interactions corresponding to SCA effects.
The second model, called “SNP within group,” considers observed alleles at SNP received from each parental line, assuming different effects in the two heterotic groups. This model assumes that two inbred lines from the same group that share the same allele at SNP are IBD (identical by descend). \(\gamma_{q}\) contains three parameters, one GCA SNP effect per group and one SCA SNP effect. \(X_{q}\) has three columns. The first two correspond to the SNP genotypes of the Dent and Flint parental lines, respectively, each coded as 0, 1 or 2 for genotypes AA, AB and BB, respectively (if B is the reference allele). The last column is the product of the first two and corresponds to the SCA effect. This model has 1 df for the allelic effect of each group and 1 df for the interaction. These first two models decompose QTL effects into GCA and SCA components by considering the parental group origin of the alleles.
The third model called “Hybrid genotype” directly considers the genotype of the hybrid and makes the strong assumption that QTL effects are the same in both heterotic groups. For this model, \(X_{q}\) has two columns, one with 0, 1 and 2 if the hybrid genotype at marker q is AA, AB and BB, respectively, and the second one with 0 for hybrids that are homozygotes and 1 for heterozygotes. This model decomposes the hybrid QTL effect \(\gamma_{q}\) into an additive and a dominance term, with 1 df each. More details can be found in Giraud et al. (2017b).
QTL analysis was performed with ASReml-R (Butler et al. 2007). The “SNP within group” and the “Hybrid genotype” models were run on the 4758 mapped markers which were polymorphic in both heterotic groups and with a minor allele frequency (MAF) superior to 5% in both heterotic groups to avoid identifiability issues. The “Founder alleles” model was performed on the 9548 mapped markers. For each model, we considered a 5% genome-wide significance threshold based on the number of efficient markers (Gao et al. 2008) and determined by Giraud et al. (2017a, b) as being equal to 3.84 for the “Founder alleles” model, 4.40 for the “SNP within group” model and 4.53 for the “Hybrid genotype.” A multimarker procedure was implemented using a forward and backward marker selection process. At each step of the forward process, the most significant maker (based on the total locus effect or on one of the GCA/additive or SCA/dominance effect) was added to the model as cofactor until no marker had a significant effect at the 5% genome-wide risk level. Then all markers were jointly tested, and we removed step by step, in a backward process, each marker with nonsignificant effect until we only kept markers for which the total effect or one of its components (GCA or SCA/additivity or dominance) was significant. Graphics for visualizing QTL were based on the − log (p value) of marker Wald test in a model that considered as cofactor only markers located at more than 10 cM from the tested position.
For each model we estimated the proportion of the phenotypic variance explained by both QTL and family effects (R2_global) that was defined as the squared correlation between predicted (based on these two parameters) and observed hybrid performances (lsmeans). The percentage of phenotypic variance explained by the family effects (\(R_{\text{fam}}^{2}\)) and the percentage of variance jointly explained by the detected QTL (\(R_{\text{QTL}}^{2}\)) and by each individual QTL were estimated by adapting the \(R^{2}\) estimation process proposed by Nakagawa and Schielzeth (2013) for mixed models. From these parameters, we estimated the percentage of within-family phenotypic variance explained by the QTL as \(R_{\text{QTL}}^{2*} = \frac{{R_{\text{QTL}}^{2} }}{{1 - R_{\text{fam}}^{2} }}\).
The quality of prediction of these models was evaluated by cross-validation using 80% of the data as calibration set and the remaining 20% as validation set. Sampling was stratified by families and repeated 100 times. To reduce computation time, we did not perform a de novo QTL detection for each calibration set. Instead, we considered the QTL detected in the whole dataset, removed the ones that were not significant in the calibration set and re-estimated the effects of the remaining QTL. The percentages of variance explained by the models were estimated by the squared correlation between the hybrid value predictions and lsmeans of the validation set. To evaluate the interest of adding SCA/dominance to the model, the cross-validations were done with and without considering SCA/dominance QTL effect (using an individual significance risk level of 5%). A model including only the family effects was also implemented and used as a benchmark.
Using Biomercator (Sosnowski et al. 2012), we compared our QTL with those of silage productivity traits detected by Giraud et al. (2017a, b), considering an arbitrary 20 cM confidence interval for each detected QTL. To facilitate comparison with QTL found in the literature, we indicated for each QTL the chromosome bin where it was detected, using the bins defined in the MaizeGDB database (Lawrence et al. 2004). QTL were also projected on the physical map of maize reference genome v3 (Lawrence et al. 2004). We positioned on this physical map meta-QTL of traits related to cell wall biomass composition and cell wall biomass digestibility published by Truntzler et al. (2010), and QTL of in vitro digestibility of NDF (IVNDFD) summarized in a recent study of Barrière et al. (2016). We also compared QTL positions with those of genes involved in the biosynthesis of cell wall as well as those of the maize brown midrib mutant genes (bm), well known to affect lignin content and cell wall digestibility.
Results
Hybrid variance components
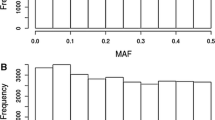
Broad-sense heritabilities at the design level (H2) were high for all traits and ranged between 0.69 (NDF) and 0.87 (LIGNIN) for silage quality traits and between 0.82 (DMY) and 0.89 (DMC and DtSILK) for agronomical traits (Table 1). When family effects were not considered (model 1), hybrid variance decomposition (Table 1, Fig. 1 and Table S4) showed a predominance of GCA components over SCA. SCA variance explained from less than 1% of the hybrid genetic variance \(\sigma_{H}^{2}\) for DINAG, DINAGZ, CELL and HCELL to 10% for NDF and was not significant. Proportions of SCA were higher for agronomical traits and varied between 14% for DMC and 19% for DMY. It was significant at 5% level for DMY and DtSILK. Dent and Flint GCA were significant for all traits (Table S4), but Dent GCA (GCAd) variance was globally larger than Flint GCA (GCAf) variance for all traits but NDF and DMC. The Dent GCA variance explained up to 70% of the hybrid variance for HCELL and LIGNIN (Table S4).
Hybrid genetic variance components for silage quality traits and agronomical traits. For each trait, the graphic on the left corresponds to the model that does not consider family effects (“without fam”; model 1) and the graphic on the right represents the model that takes family effects into account (“with fam”; model 2). The different colors correspond to the different variance components (GCAd, GCAf and SCA in green, orange and red, respectively) with lighter shade showing the corresponding interactions with the environmental conditions. The gray bars correspond to the proportion of genetic variance absorbed by family fixed effects
Family effects were highly significant for all traits (model 2) but were globally more important for silage quality traits than for agronomical traits. Except for NDF and DMC, the Dent family effects were more significant than the Flint family effects. The interaction between the Flint and Dent family effects were only significant for HCELL and DMC (results not shown). Family fixed effects absorbed more than 50% of the hybrid genetic variance for silage quality traits (except MFU and NDF), and they absorbed less than 25% of the hybrid genetic variance for agronomical traits (Table 1). Within-family heritabilities (H2*) were lower than H2 and varied between 0.54 (CELL) and 0.72 (LIGNIN) for silage quality traits and between 0.77 (DMC) and 0.88 (DtSILK) for agronomical traits. When family effects were considered (model 2), the proportion of hybrid variance due to GCAs decreased. This affected more the Dent GCA than the Flint GCA, so that Flint and Dent within-family GCA variance components were of the same magnitude. Including family effects slightly increased the SCA variance component for most traits (except MFU and NDF), but this component remained low for all silage quality traits [between less than 1% (DINAG) and 19% (HCELL)] and moderate for agronomical traits [between 23% (DtSILK) and 27% (DMY)]. Likelihood ratio test on random effects showed that none of the SCA variance components was significant for silage quality traits, whereas they were significant for the three agronomical traits.
Genotype by environment interactions explained between 12% (DtSILK, LIGNIN) and 35% (NDF) of the total genetic variance (Fig. 1, Table 1). They were globally small for silage quality traits except for MFU and NDF that both showed a large SCA × E variance component, significant at 5% risk level (Table S4). Considering SCA × E variance component increased the proportion of SCA in the total hybrid variance for these traits (Table 1), but the estimation error of these SCA × E variance components was large. Genotype by environment interactions were moderate for the agronomical traits DMY and DMC. For these traits, interactions were mostly associated with GCA × E interactions significant at 0.1% risk level (Table S4). None of the SCA × E interactions was significant for agronomical traits. In model 2, family × E interaction effects were significant for all traits (results not shown). As for main effects, including family effects in the model reduced the hybrid by environment variance components, but this reduction was small.
Adjusted means and correlations between traits
Experimental hybrid performances were on average close to founder hybrid performances but showed a variation that exceeded that of founder hybrids for all traits (Table 2). On average, experimental hybrids derived from one or both founder lines F7088 and F7082 that were chosen for their good digestibility had the best performance for DINAG (+ 0.43 points for F7088 and + 0.44 for F7082). For MFU, founder lines with the best performances were F373 for the Flint group (+ 0.004 MFU kg−1) and F7082 for the Dent group (+ 0.006 MFU kg−1). The hybrid between the founder lines F7088 and F7082 had the best digestibility profile among founder hybrids, with high values for MFU, DINAG, DINAGZ and HCELL and low values for LIGNIN and CELL. Founder hybrids derived from crosses involving the Dent founder line F98902 had the lowest digestibility profile, except the one derived from the Flint line chosen for its digestibility (F7088 × F98902) which showed intermediate performances (Fig S1). Hybrids derived from F98902 had the best performances for DMY, illustrating potential antagonism between silage quality and productivity.
Pearson’s correlations based on lsmeans and genetic correlations showed consistent results (Fig. 2 for correlations based on lsmeans, and Table S5 for the genetic correlations). The MFU of experimental hybrids was positively correlated with the two digestibility traits DINAG and DINAGZ, these two traits being highly correlated. These three traits were positively correlated with HCELL and negatively correlated with NDF, CELL and LIGNIN. The two digestibility traits (DINAG and DINAGZ) were strongly correlated with LIGNIN, HCELL and CELL and to a lesser extent with NDF. MFU, which is the main target in selection, was strongly correlated with HCELL (0.81 based on lsmeans; 0.87 for the genetic correlation) and NDF (− 0.80 on lsmeans; − 0.79 for the genetic correlation). Among cell wall traits, HCELL, CELL and LIGNIN were highly correlated and showed moderate correlations with NDF.
Pearson’s correlation matrix based on experimental hybrid lsmeans for silage biomass quality traits and for silage biomass productivity traits. The histograms of these traits are shown on the diagonal. Symbols “***”, “**”, “*” correspond to the level of significance of the Person’s correlation corresponding to p values of 0.001, 0.01 and 0.05, respectively
Silage quality traits showed moderate to weak correlations with agronomical traits based on lsmeans. More particularly, DMY showed negative correlations with MFU (− 0.53) and to a lesser extent with digestibility traits (− 0.39 with DINAG and − 0.28 with DINAGZ). Higher genetic correlations were found between DMY and MFU (− 0.72) and digestibility traits (− 0.53 with DINAG and − 0.48 with DINAGZ). Correlations between silage dry matter content (DMC) and silage quality traits were low (< 0.3 in absolute value for lsmeans; < 0.4 for the genetic correlations).
QTL detection
In total, the “SNP within group” model detected 80 QTL against 76 and 74, respectively, for the “Founder alleles” and “Hybrid genotype” models (Table 3, Tables S6-8). “SNP within group” model detected more QTL for all traits, but MFU and HCELL for which more QTL were detected with the “Hybrid genotype.” Among all the detected QTL, a majority (39) was detected by all three models (Fig S2). However, some QTL were model specific. The “SNP within group” model shared a lot of QTL with the “Founder alleles” model (54) and the “Hybrid genotype” model (50), which shared together fewer QTL (43).
Most of the QTL detected with the “Founder alleles” and/or the “SNP within group” model showed GCA effects that were group specific (Table 3 and example of chromosome 1 for DINAG in Fig S3). A small number of QTL (between 0 and 3) were significant for both their GCAf and GCAd components. Few QTL which were significant for one component (mostly one GCA or the additive component) were not significant for the global effect. We detected more QTL for GCAd than for GCAf with the “SNP within group” model and more QTL for GCAf than for GCAd with the “Founder alleles” model. Very few QTL were detected for SCA/dominance effects. At the whole-genome 5% risk level, only one QTL with significant SCA effect was detected by the “Founder alleles” model for LIGNIN and one QTL with a significant dominance effect was detected by the “Hybrid genotype” model for DINAG. Among all detected QTL, on average, between 0 and 2 QTL had significant SCA/dominance effects at 5% individual marker risk level, depending on the traits and models. Globally over the different traits, the “Founder alleles” model detected more significant SCA/dominant QTL at this risk level than the other models (12 compared to 8 and 7 for the “SNP within group” and “Hybrid genotype” models, respectively).
QTL models explained between 33% (NDF, “Hybrid genotype” model, without dominance) and 77% (LIGNIN, “Founder alleles” model, with SCA effects significant at 5% individual risk level) of the total phenotypic variance (Table 4). These high R2 are mainly explained by the strong family structure effects on biomass quality traits ranging between 14.71% for the less affected trait (NDF) and 56.77% for the most affected one (LIGNIN). The detected QTL explained between 21.02% (NDF, “Hybrid genotype” model, without dominance) and 46.23% (LIGNIN, “Founder alleles” model, with SCA effects when significant) of the within-family phenotypic variance. Compared to the other models, the “Founder alleles” model explained a larger part of the phenotypic variance for all traits, whereas the “Hybrid genotype” explained a lower one (except for HCELL). When dominance/SCA was considered, the increase in percentage of explained phenotypic variance compared to models including only GCA/additive effect was small. It was lower for the “SNP within group” model (from + 0.2 for MFU to + 1.3 for NDF) and for the “Hybrid genotype” model (+ 0.2 for CELL to + 2 for NDF) than for the “Founder alleles” model (+ 2.8 for HCELL to + 6.5 for NDF). When considered separately, individual QTL explained a small percentage of phenotypic variance ranging between < 1% and 4.5% (Table S6 to S8).
For each trait, all founder lines carried both favorable and unfavorable alleles (results not shown). The two founder lines chosen for their good digestibility (F7082 for the Dent and F7088 for the Flint) carried more favorable alleles than the other ones for the digestibility trait DINAG, but not for MFU (Fig. 3).
QTL allelic effects of founder inbred lines (from the “Founder alleles” model) for cell wall digestibility (DINAG, a) and silage feeding value (MFU, b). For each QTL, associated marker, chromosome location, genetic position and the proportion of total phenotypic variance explained are indicated. For each founder inbred line, the genotypic value based on QTL was calculated as the sum of its allelic effects over all the detected QTL
R 2: cross-validation results
A cross-validation procedure was carried out to assess the predictive abilities of the different QTL-based models and correct for potential bias due to an over-fitting of the QTL model when the same data are used for QTL detection and R2 estimation. The QTL detection model leading to the highest R2 varied depending on the trait (Table 5). As expected, the phenotypic variance explained by QTL based on the cross-validation was lower than that obtained on the whole dataset: for instance, 20.64% instead of 34.14% for NDF with the “Hybrid genotype” model and 67.33% instead of 76.21% for LIGNIN with the “Founder alleles” model. Based on cross-validation results, the best predicting model was generally not the model that detected the highest number of QTL. The “Founder alleles” model led to the highest cross-validation R2 only for HCELL and LIGNIN. The “SNP within group” gave the highest cross-validation R2 for DINAG, MFU and NDF, whereas the highest cross-validation R2 for DINAGZ and CELL was obtained with “Hybrid genotype” model. Adding SCA/dominance in prediction models generally decreased R2. (Only a slight increase of about + 0.1 was found for NDF with all models.) Considering the best model for each trait, cross-validation R2 ranged between 20.64% for NDF, “Hybrid genotype” with dominance effects model, and 67.33% for LIGNIN, “Founders alleles” with no SCA in the model.
Digestibility and productivity QTL co-localization
Using Biomercator (Sosnowski et al. 2012), we plotted the QTL detected for silage quality traits and added QTL for silage agronomical traits detected by Giraud et al. (2017a, b). For each trait we plotted the QTL found with the model that was considered as being the best one based on cross-validation. We considered an arbitrary 20 cM confidence interval (CI).
We observed many co-localizations between QTL detected for the different quality traits (Fig. 4). Highly correlated traits (DINAG vs. DINAGZ, CELL vs. HCELL, etc.) usually had QTL located in the same genomic regions. About 25% of the QTL were mapped on chromosome 1 into 4 non-overlapping regions. One of these regions from bin 1.02 to 1.04 gathered QTL for all silage quality traits (but NDF). Except for MFU, the QTL mapped in this region are the most significant ones for each trait. This region therefore seems to play an important role in the variation of silage quality.
Some regions appeared specific to silage quality or cell wall composition traits and did not carry QTL for agronomical traits. Among these regions, some affected only one trait (QTL NDF2 on bin 1.07, QTL DINAG4 on bin 2.10, QTL NDF8 on 5.07 and QTL LIGNIN9 on bin 7.00), while other regions displayed QTL for several quality traits (bin 6.06 for DINAG and MFU, bin 7.02 for DINAG and all cell wall components, and bin 10.07 for HCELL and LIGNIN). Reciprocally, some regions with QTL for silage agronomical traits (such as DMY3 on bin 3.06, DMC5 on bin 3.07, DMC9 on bin 6.05, DtSILK13 on bin 7.05 and DMC4-DtSILK5 on bin 2.09) showed no QTL for silage quality or composition traits. Except these above-mentioned specific regions, all remaining regions showed co-localizations of QTL for silage quality and silage agronomical traits. Interestingly, two genomic regions located on chromosome 8 (from bin 8.03 to 8.05) and on chromosome 10 (from bin 10.03 to 10.04) displayed co-localizations of all traits.
For regions which showed an overlap between QTL for DMY and those for MFU, DINAG or DINAGZ, we looked at the relationship between the GCA parental allelic effects estimated for these different traits with the “Founder alleles” model. In all the cases, we observed a negative correlation between the QTL allelic effects estimated for MFU and DMY (Fig. 5). The same tendency was observed for some of the QTL detected for DINAG and DINAGZ (Fig S4 and S5) but not all. So, in these regions where QTL for DMY overlapped with QTL for quality traits, the parental alleles increasing DMY usually had a negative effect on MFU (and to a lesser extent on DINAG and DINAGZ).
Relationship between QTL allelic effects of the 8 founder lines (in orange for Flint ones and in green for Dent ones) for silage feeding value (MFU) and silage productivity (DMY) for regions carrying QTL for these two traits. For each region, the chromosome bin location, and the phenotypic variation explained for each trait and each region (between brackets) are indicated. Within each genetic group, the founder line that was chosen for its silage quality is plotted with the “*” symbol
Discussion
Predominance of QTL showing group-specific GCA effects
To our knowledge, this is the first study that estimated GCA and SCA variance components in a Dent–Flint factorial for maize silage quality traits. The proportion of SCA and within-family SCA variance for silage quality traits was small and lower than that observed for agronomical traits. Our results for agronomical traits confirmed the first estimations done by Giraud et al. (2017a) using a model that did not consider G × E interactions. They showed that SCA explained about 20% of the hybrid variance for DMY, while the highest proportion of SCA obtained for silage quality traits was 10% (for NDF). These results agree with the limited contribution of SCA effects observed by Argillier et al. (2000) for digestibility-related traits in a factorial. Despite the small number of experimental hybrids derived per parental line, our population enables the simultaneous detection of QTL involved in the GCA of each group and in the SCA component of hybrid value. We did not find strong evidence of SCA QTL for silage quality traits. The low SCA variance and the small number of QTL showing SCA effects can be either (1) due to the fact that these traits involved QTL with biological additive effects or (2) due to the divergence between Flint and Dent heterotic groups for QTL showing dominance. In this second case, additive and dominance effects are confounded and both contribute to GCA (Reif et al. 2007). In the literature, strong correlations have been found between per se and test-cross values (Argillier et al. 2000; Torres et al. 2015) for silage quality traits, which supports the first hypothesis. However, recently Leng et al. (2018) found only moderate correlations between per se and test-cross silage quality performances for a Dent and a Flint segregating population. This result and the fact that we detected QTL specific of the Dent and the Flint heterotic groups in our study using the “SNP within group” and the “Founder alleles” models suggest that the lack of SCA for silage quality traits in our experiment may also be due, at least partly, to group divergence for QTL showing dominance.
Even if SCA seemed to be limited when considering average performances of hybrids in our experimental design, the SCA × E variance component was found to be relatively important for NDF, MFU, which is the main targeted trait for improving silage quality, and also, to a lesser extent, for DINAG, DMY and DMC. Even if this variance component was not estimated with a high accuracy in our experiment, this suggests that the best hybrid combinations for silage might depend on the environment, confirming breeder observations (Carolo, personal communication).
In this study, we performed QTL detection on the average hybrid performances. This strategy is clearly justified since for most traits G × E interactions explained a small part of the variance. Nevertheless, at least for some traits, it would clearly be interesting to extend QTL models to identify regions involved in GCA × E and SCA × E interactions. This is beyond the scope of this paper, but this is an interesting prospect for future studies. The factorial population we developed between unselected biparental families is well adapted for this objective.
Relationship between feeding value, digestibility and biomass yield
Animal performances (milk or meat yield) are related to both silage feeding value (MFU) and the dry matter intake (DMI) which is the amount of silage ingested by animals. This latter criterion is known to vary with starch/NDF ratio in whole maize plant (Khan et al. 2015) and reaches its optimum value at silage physiological maturity (30% < DMC < 35%). Barrière and Emile (2000) showed that silage feeding value was mainly explained by NDF digestibility (r2 = 0.59) and to a lesser extent by starch content (r2 = 0.36). In this study, we did not investigate starch content. This component is supposed to be completely digestible, and as it impacts not only the MFU but also the dry matter intake it is not the main target for improving MFU. We chose to focus more on cell wall digestibility that is known to be the limiting factor for improving maize silage feeding value (Barrière and Emile 2000).
Our experimental hybrids showed performances for silage quality traits consistent with those described in the literature for early and semi-early hybrids. The MFU value in our study varied between 0.83 and 0.96, while that of registered hybrids in the French Official Catalog of Species and Varieties ranged from 0.79 to 0.97 MFU kg−1 (Barrière and Emile 2000; Baldy et al. 2017). Also, Baldy et al. (2017) observed DINAG performances of registered hybrids ranging from 42 and 48%, while that of our study ranged from 45 to 56%.
We found a negative correlation of − 0.53 between MFU and DMY lsmeans and a genetic correlation of − 0.72 between them. This correlation is thus stronger than the negative correlation (− 0.50) reported by Barrière and Emile (2000) and Surault et al. (2005). Argillier et al. (2000) found no correlation between DMY and digestibility traits (closely linked to MFU) in a factorial between lines from different origins (issued from different Dent and Flint groups). Baldy et al. (2017) reported even a positive relationship between DMY and MFU when analyzing recently registered hybrids of the French catalogue (the most recent hybrids showing increased DMY and better digestibility compared to older ones). This confirms that these correlations depend on the material tested. In our case, three of the Dent founder lines (except F7088) belong to the Iodent group, known to have a good combining ability with Flint lines for DMY but also a poor digestibility (Barrière et al. 2003). This variability among the Dent founder lines is consistent with the importance of Dent GCA variance component relative to the Flint GCA for silage quality traits and DMY. It may also explain most of the observed correlations between these traits. The heterotic groups we used and the choice in each group of a founder line based on its good digestibility may have reinforced the negative correlation between DMY and silage quality traits. Better understanding of the genetic bases of this correlation (pleiotropy versus linkage) is therefore of key importance for a breeding prospect as the objective is to improve simultaneously DMY and MFU.
Co-localization between QTL of silage quality and production traits
Graphical representation of all QTL along the genome is consistent with the correlations between silage quality traits. Indeed, QTL of correlated traits were often detected in same genomic regions. These multitrait QTL co-localizations suggest a complex genetic determinism implying pleiotropic QTL. For instance, 9 of the 14 regions with QTL of cell wall digestibility (DINAG and DINAGZ) traits also carried QTL of lignin content. This is consistent with the strong negative correlation between lignin content and digestibility observed in our study and also often reported in the literature (see, for instance, Jung et al. 1997; Giordano et al. 2014). Lignin being not digestible and impeding cellulose and hemicellulose digestion (Jung and Vogel 1986; Cornu et al. 1994; Moore and Jung 2001), it is reasonable to assume that QTL for lignin content may directly affect digestibility. However, all cell wall digestibility QTL did not co-localize with those of lignin content and reciprocally. Other parameters than lignin content affect cell wall digestibility, and selecting for varieties with low lignin content is not the only way to select for digestibility, as stated, for instance, in Méchin et al. (2005), Zhang et al. (2012) or El hage et al. (2018). In this study, we also measured two other cell wall components: the cellulose and the hemicellulose contents that were found to be highly correlated with the lignin content. All QTL detected for CELL and HCELL co-localized with LIGNIN QTL, except one on chromosome 5. It would be interesting to evaluate other traits such as the structure and composition of the lignin in its different subunits as well as the content of p-hydroxycinnamic acids (ferulic and p-coumaric acids) that are involved in cross-linkages between cell wall components and found to explain part of the cell digestibility by Zhang et al. (2012). Recently, El Hage et al. (2018) found that beyond lignin content and cell wall composition, the distribution of lignified cell types in maize internodes was highly variable and may play an important role in cell wall digestibility. This is consistent with the meta-analysis results of Truntzler et al. (2010) who found only a partial overlap between digestibility MQTL and cell wall component MQTL. Thus, identifying QTL for other cell wall components, for lignin structure as well as for the repartition of different tissues in stem would be of interest for forage quality breeding. This calls for further development of easy ways to measure all these parameters on a large number of hybrids evaluated in field conditions (using NIRS calibration, for instance).
Adding QTL detected by Giraud et al. (2017a) for agronomical traits, we observed 15 regions with co-localizations between silage quality and silage agronomical traits among all the 28 QTL regions that were detected in this study. Among them, co-localizations between QTL for all silage quality and all agronomical traits were found on chromosome 8 (bins 8.03 to 8.05) and chromosome 10 (bins 10.3 to 10.04). These two regions are known to carry cloned QTL involved in maize flowering time (Salvi et al. 2002; Ducrocq et al. 2009). Consistent with trait correlations, founder allelic effects on DtSILK at these QTL were negatively correlated with allelic effects on MFU and to a lesser extent with digestibility effects (results not shown). Cell wall digestibility traits (DINAG and DINAGZ) and feeding value (MFU) were often found in regions involving silage dry matter yield (DMY). Five among the 14 regions involved in DINAG or DINAGZ and five among the 11 regions involved in MFU also contained DMY QTL. The negative correlation between overlapping QTL effects for DMY and MFU suggests that at least some QTL may have pleiotropic effects on both traits. Such pleiotropy may limit the possibility of selecting early maize varieties with both high DMY and good digestibility. The biological relationship between digestibility-related traits and biomass production could be explained by the known negative impact of cell wall degradability on traits such as plant resistance to stalk lodging, pest, diseases and abiotic stress (Buendgen et al. 1990; Flint-Garcia et al. 2003; Cardinal and Lee 2005; Barros-Rios et al. 2011; Santiago et al. 2013; Le Gall et al. 2015; Li et al. 2016; Badji et al. 2018) that contribute to higher productivity.
Nevertheless, some silage quality QTL (for instance, on bins 1.07, 2.10, 5.07, 6.06 and 7.02) seem physically distant from silage productivity QTL and thus may be used to improve silage quality without impacting silage productivity. Results from Baldy et al. (2017) showed that in recent varieties, breeders succeeded in improving simultaneously yield and digestibility. It would be interesting to know which regions have been selected for during this process.
Comparison with QTL detected in the literature
We compared the detected QTL with those already published for related traits, and more specifically those issued from a meta-analysis of QTL for traits related to cell wall biomass composition and cell wall biomass digestibility (Truntzler et al. 2010), and from Barrière et al. (2016) that assembled QTL of in vitro digestibility of NDF (IVDNDF) and identified their co-localizations with different families of candidate genes. Most of the QTL we identified are localized in the same regions than those cited in at least one of these publications. Only one QTL for the NDF was detected in a region (bin 7.05 by the “Hybrid genotype” model) where no meta-QTL and no IVDNDF QTL were identified. This QTL only co-localized with female flowering time (DtSILK) and might therefore correspond to an indirect effect of flowering time on NDF. Reciprocally, most of the QTL regions reported in these syntheses are identified in our analysis. This illustrates the power of this multiparental hybrid population for detecting QTL and suggests that most QTL with sizeable effects in these genetic pools have now been detected.
Interestingly, we found QTL co-localization close to regions where some maize bm mutations were mapped. Maize bm mutations are known to affect the lignin composition and its degradability (Vermerris et al. 2010). For instance, the bm3, that codes for the COMT gene, involved in the monolignol pathway is located in bin 4.05 where we detected QTL for all silage quality traits and where MQTL for both cell wall biomass composition and cell wall biomass digestibility were identified (Truntzler et al. 2010) as well as a QTL for IVDNDF (Barrière et al. 2016). Also, the bm1, bm5 and bmr2 were positioned in bin 5.04 and co-localized with QTL of quality and productivity traits of this study as well as MQTL for both cell wall biomass composition and digestibility. As none of our parents carried any of the bm mutations, these QTL might thus correspond to allelic variation at the same genes, or in regions involved in their regulation. All co-localizations between our QTL and those published by Barrière et al. (2016) correspond to regions where these authors identified candidate genes that encode transcription factors such as MYB and NAC, involved in the lignin biosynthesis pathway. The resolution of our QTL mapping does not allow to go further in the identification of the causal genes. Our results can be used to initiate fine mapping, but this process might be difficult since our QTL generally present small effects. One notable exception is the region on chromosome 1 (bin 1.02) which displays large effects for several quality traits and no effect on biomass production. Interestingly, the same region was also identified by Barrière et al. (2010) who pointed out its interest for improving maize cell wall digestibility.
In recent studies, Wang et al. (2016) and Li et al. (2016) performed genome-wide association analysis of forage quality in maize mature stalk of inbred lines evaluated for per se value. They investigated cell wall composition (NDF, ADF, LIGNIN, CELL and HCELL) and its digestibility via the in vitro dry matter (IVDMD) of stalk using a diverse maize population of 368 inbred lines genotyped for 560K SNP. Significant associations between SNP and these traits were found with small effect on the phenotypic variation (4 to 7%). Interestingly, they found a strong association involving NDF, ADF, IVDMD and LIGNIN at 155.65 Mbp in the chromosome bin 6.06, a region where we found QTL co-localization between DINAG, DINAGZ and MFU and where major QTL of digestibility and lignin content were found by Roussel et al. (2002) in a RIL progeny. Furthermore, the associated SNP is contained in the first intron of the gene GRMZM2G140817 (ZmC3H2) that is involved in cell wall components biosynthesis pathway.
Implications on breeding strategies
Compared to tester-based evaluation in each heterotic group the use of an incomplete factorial can reduce by a factor 2 the phenotyping effort for a same number of tested lines. Also, compared to hybrid panels that have been used to perform GWAS, the family structure of our population makes it possible to trace founder alleles in the experimental hybrids and to easily control for spurious associations due to long-range linkage disequilibrium at the level of the founder lines.
QTL detection allows the estimation of allelic effects of the different founder lines. The founder lines chosen for their digestibly within each group (F7088 for Flint and F7082 for Dent) have a genotypic score of + 1.38 and + 0.75 points of digestibility (DINAG), respectively (Fig. 3). This is consistent with the good performance of the founder hybrid F7088 × F7082 and the fact that each of these lines led to the best experimental hybrids based on lsmeans for digestibility criteria. Interestingly, the founder line that carries the most interesting allele within each group is not the same for all QTL. For DINAG, the best Flint founder line (F7088) has a predicted value at detected QTL which represents only 32% of the best Flint ideotype (obtained by summing the best allelic values over detected QTL). The corresponding score for the best Dent founder line (F7082) corresponds to less than 20%. For MFU, these values are 24% for the best Flint founder line (F02803) and 26% for the best Dent founder line (F04401). This suggests that there is room for possible improvement of future inbred lines within each group for these traits.
Using marker-assisted selection (MAS), QTL offer the possibility to carry out early screening of the available genetic material based only on molecular marker information. Furthermore, QTL permit to follow alleles of interest in the progeny at different stages of the selection scheme. Inbred lines complementary at the QTL level can be easily identified to produce the best intergroup hybrid combinations for both silage productivity and silage digestibility performances. QTL information can also be used within each group to define genotypic ideotypes, select the best inbred lines and then guide their intercrossing with the aim of producing future parental inbred lines combining the best favorable alleles. This can complement genome-wide predictions by allowing a specific monitoring of genomic regions with main effects.
Author contribution statement
C.B., A.C. and L.M. initiated this project. L.M. and C.B coordinated it. C.B., A.C. and L.M. supervised this work. H.G. and A.I.S. designed the statistical models and their implementation. A.I.S. analyzed the data and prepared the manuscript. M.R. and V.M. supervised analyses and interpretations related to silage quality traits. All authors discussed the results and contributed to the final manuscript. All authors revised and approved the manuscript.
References
Andrieu J (1995) Prévision de la digestibilité et de la valeur énergétique du maïs fourrage à l’état frais. INRA Prod Anim 8(4):273–274
Argillier O, Barrière Y, Hebert Y (1995) Genetic-variation and selection criterion for digestibility traits of forage maize. Euphytica 82(2):175–184
Argillier O, Mechin V, Barrière Y (2000) Inbred line evaluation and breeding for digestibility-related traits in forage maize. Crop Sci 40(6):1596–1600
Badji A, Otim M, Machida L, Odong T, Bomet Kwemoi D, Okii D, Agbahoungba S et al (2018) Maize combined insect resistance genomic regions and their co-localization with cell wall constituents revealed by tissue-specific QTL meta-analyses. Front Plant Sci 9:895
Baker CW, Givens DI, Deaville ER (1994) Prediction of organic matter digestibility in vivo of grass silage by near infrared reflectance spectroscopy: effect of calibration method, residual moisture and particle size. Anim Feed Sci Technol 50(1):17–26
Baldy A, Jacquemot MP, Griveau Y, Bauland C, Reymond M, Mechin V (2017) Energy values of registered corn forage hybrids in France over the last 20 years rose in a context of maintained yield increase. Am J Plant Sci 08(06):1449–1461
Barrière Y, Emile JC (2000) Le maïs fourrage: III. évaluation et perspectives de progrès génétiques sur les caractères de valeurs alimentaire. Fourrages 163:221–238
Barrière Y, Guillet C, Goffner D, Pichon M (2003) Genetic variation and breeding strategies for improved cell wall digestibility in annual forage crops—a review. Anim Res 52(3):193–228
Barrière Y, Riboulet C, Méchin V, Maltese S, Pichon M, Cardinal A, Lapierre C, Lubberstedt T, Martinant JP (2007) Genetics and genomics of lignification in grass cell walls based on maize as model species. Genes Genomes Genomics 1:133–156
Barrière Y, Thomas J, Denoue D (2008) QTL mapping for lignin content, lignin monomeric composition, p-hydroxycinnamate content, and cell wall digestibility in the maize recombinant inbred line progeny F838 × F286. Plant Sci 175(4):585–595
Barrière Y, Méchin V, Denoue D, Bauland C, Laborde J (2010) QTL for yield, earliness, and cell wall quality traits in topcross experiments of the F838 × F286 early maize RIL progeny. Crop Sci 50(5):1761–1772
Barrière Y, Courtial A, Chateigner-Boutin A-L, Denoue D, Grima-Pettenati J (2016) Breeding maize for silage and biofuel production, an illustration of a step forward with the genome sequence. Plant Sci 242(January):310–329
Barrière Y, Guillaumie S, Denoue D, Pichon M, Goffner D, Martinant JP (2017) Investigating the unusually high cell wall digestibility of the old INRA early flint F4 maize inbred line. Maydica 62:M31
Barros-Rios J, Malvar RA, Jung H-JG, Santiago R (2011) Cell wall composition as a maize defence mechanism against corn borers. Phytochemistry 72(4):365–371
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81(5):1084–1097
Buendgen MR, Coors JG, Grombacher AW, Russell WA (1990) European corn borer resistance and cell wall composition of three maize populations. Crop Sci 30(3):505–510
Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2007) ASReml-R reference manual. The State of Queensland, Department of Primary Industries and Fisheries, Brisbane
Cardinal AJ, Lee M (2005) Genetic relationships between resistance to stalk-tunneling by the european corn borer and cell wall components in maize population B73 × B52. Theor Appl Genet 111(1):1–7
Cardinal A, Lee M, Moore K (2003) Genetic mapping and analysis of quantitative trait loci affecting fiber and lignin content in maize. Theor Appl Genet 106(5):866–874
Cornu A, Besle JM, Mosoni P, Grenet E (1994) Lignin–carbohydrate complexes in forages: structure and consequences in the ruminal degradation of cell wall carbohydrates. Reprod Nutr Dev 34(5):385–398
Courtial A, Thomas J, Reymond M, Méchin V, Grima-Pettenati J, Barrière Y (2013) Targeted linkage map densification to improve cell wall related QTL detection and interpretation in maize. Theor Appl Genet 126(5):1151–1165
Courtial A, Méchin V, Reymond M, Grima-Pettenati J, Barrière Y (2014) Colocalizations between several QTLs for cell wall degradability and composition in the F288 × F271 early maize RIL progeny raise the question of the nature of the possible underlying determinants and breeding targets for biofuel capacity. Bioenergy Res 7(1):142–156
Ducrocq S, Giauffret C, Madur D, Combes V, Dumas F, Jouanne S, Coubriche D, Jamin P, Moreau L, Charcosset A (2009) Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics 183(4):1555–1563
El Hage F, Legland D, Borrega N, Jacquemot M-P, Griveau Y, Coursol S, Méchin V, Reymond M (2018) Tissue lignification, cell wall p-coumaroylation and degradability of maize stems depend on water status. J Agric Food Chem 66(19):4800–4808
Flint-Garcia SA, Jampatong C, Darrah LL, McMullen MD (2003) Quantitative Trait Locus analysis of stalk strength in four maize populations. Crop Sci 43(1):13
Fontaine AS, Bout S, Barrière Y, Vermerris W (2003) Variation in cell wall composition among forage maize (Zea Mays L.) inbred lines and its impact on digestibility: analysis of neutral detergent fiber composition by pyrolysis–gas chromatography–mass spectrometry. J Agric Food Chem 51(27):8080–8087
Gao X, Starmer J, Martin ER (2008) A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol 32(4):361–369
Giordano A, Liu Z, Panter SN, Dimech AM, Shang Y, Wijesinghe H, Fulgueras K et al (2014) Reduced lignin content and altered lignin composition in the warm season forage grass paspalum dilatatum by down-regulation of a cinnamoyl CoA reductase gene. Transgenic Res 23(3):503–517
Giraud H, Bauland C, Falque M, Madur D, Combes V, Jamin P, Monteil C et al (2017a) Linkage analysis and association mapping QTL detection models for hybrids between multiparental populations from two heterotic groups: application to biomass production in maize (Zea Mays L.). G3 7(11):3649–3657
Giraud H, Bauland C, Falque M, Madur D, Combes V, Jamin P, Monteil C et al (2017b) Reciprocal genetics: identifying QTL for general and specific combining abilities in hybrids between multiparental populations from two maize (Zea Mays L.) heterotic groups. Genetics 207(3):1167–1180
Goering HK, Van Soest PJ (1970) Forage fiber analyses (apparatus, reagents, procedures, and some applications). U.S. Agricultural Research Service Handbook, pp 1–379
Grabber JH, Quideau S, Ralph J (1996) P-coumaroylated syringyl units in maize lignin: implications for β-ether cleavage by thioacidolysis. Phytochemistry 43(6):1189–1194
Hartley RD (1972) p-Coumaric and ferulic acid components of cell walls of ryegrass and their relationships with lignin and digestibility. J Sci Food Agric 23(11):1347–1354
Hickey JM, Gorjanc G, Varshney RK, Nettelblad C (2015) Imputation of single nucleotide polymorphism genotypes in biparental, backcross, and topcross populations with a hidden markov model. Crop Sci 55(5):1934
Jung HG, Vogel KP (1986) Influence of lignin on digestibility of forage cell wall material. J Anim Sci 62(6):1703–1712
Jung HG, Mertens DR, Payne AJ (1997) Correlation of acid detergent lignin and Klason lignin with digestibility of forage dry matter and neutral detergent fiber. J Dairy Sci 80(8):1622–1628
Khan NA, Yu P, Ali M, Cone JW, Hendriks WH (2015) Nutritive value of maize silage in relation to dairy cow performance and milk quality. J Sci Food Agric 95(2):238–252
Krakowsky MD, Lee M, Beeghly HH, Coors JG (2003) Characterization of quantitative trait loci affecting fiber and lignin in maize (Zea Mays L.). Maydica 48(4):283–292
Krakowsky MD, Lee M, Coors JG (2005) Quantitative trait loci for cell wall components in recombinant inbred lines of maize (Zea Mays L.) I: stalk tissue. Theor Appl Genet 111(2):337–346
Krakowsky MD, Lee M, Coors JG (2006) Quantitative trait loci for cell wall components in recombinant inbred lines of maize (Zea Mays L.) II: leaf sheath tissue. Theor Appl Genet 112(4):717–726
Lawrence CJ, Dong Q, Polacco ML, Seigfried TE, Brendel V (2004) MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res 32(Database issue):D393–D397
Le Gall H, Philippe F, Domon J-M, Gillet F, Pelloux J, Rayon C (2015) Cell wall metabolism in response to abiotic stress. Plants 4(1):112–166
Leng P, Ouzunova M, Landbeck M, Wenzel G, Eder J, Darnhofer B, Lübberstedt T (2018) Quantitative trait loci mapping of forage stover quality traits in six mapping populations derived from European elite maize germplasm. Plant Breed 137(2):139–147
Li K, Wang H, Hu X, Liu Z, Wu Y, Huang C (2016) Genome-wide association study reveals the genetic basis of stalk cell wall components in maize. PLoS ONE 11(8):e0158906
Lübberstedt T, Melchinger AE, Schön CC, Utz HF, Klein D (1997a) QTL mapping in testcrosses of European flint lines of maize: I. Comparison of different testers for forage yield traits. Crop Sci 37(3):921–931
Lübberstedt T, Melchinger AE, Klein D, Degenhardt H, Paul C (1997b) QTL mapping in testcrosses of European flint lines of maize: II. Comparison of different testers for forage quality traits. Crop Sci 37(6):1913–1922
Lübberstedt T, Melchinger AE, Fähr S, Klein D, Dally A, Westhoff P (1998) QTL mapping in testcrosses of flint lines of maize: III. Comparison across populations for forage traits. Crop Sci 38(5):1278–1289
Méchin V, Argillier O, Menanteau V, Barrière Y, Mila I, Pollet B, Lapierre C (2000) Relationship of cell wall composition to in vitro cell wall digestibility of maize inbred line stems. J Sci Food Agric 80(5):574–580
Méchin V, Argillier O, Hébert Y, Guingo E, Moreau L, Charcosset A, Barrière Y (2001) Genetic analysis and QTL mapping of cell wall digestibility and lignification in silage maize. Crop Sci 41(3):690–697
Méchin V, Argillier O, Rocher F, Hébert Y, Mila I, Pollet B, Barriére Y, Lapierre C (2005) In search of a maize ideotype for cell wall enzymatic degradability using histological and biochemical lignin characterization. J Agric Food Chem 53(15):5872–5881
Moore KJ, Jung H-JG (2001) Lignin and fiber digestion. J Range Manag 54(4):420–430
Nakagawa S, Schielzeth H (2013) A general and simple method for obtaining R² from generalized linear mixed-effects models. Methods Ecol Evol 4:133–142
Penning BW, Sykes RW, Babcock NC, Dugard CK, Held MA, Klimek JF, Shreve JT et al (2014) Genetic determinants for enzymatic digestion of lignocellulosic biomass are independent of those for lignin abundance in a maize recombinant inbred population1. Plant Physiol 165(4):1475–1487
R Core Team (2013) R: a language and environment for statistical computing. R foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org
Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung H-JG (1994) Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J Am Chem Soc 116(21):9448–9456
Reif JC, Gumpert F-M, Fischer S, Melchinger AE (2007) Impact of interpopulation divergence on additive and dominance variance in hybrid populations. Genetics 176(3):1931–1934
Riboulet C, Fabre F, Dénoue D, Martinant JP, Lefevre B, Barrière Y (2008) QTL mapping and candidate gene research from lignin content and cell wall digestibility in a top-cross of a flint maize recombinant inbred line progeny harvested at silage stage. Maydica 53(1):1
Roussel V, Gibelin C, Fontaine A-S, Barrière Y (2002) Genetic analysis in recombinant inbred lines of early dent forage maize. II—QTL mapping for cell wall constituents and cell wall digestibility from per se value and top cross experiments. Maydica 47(January):9–20
Salvi S, Tuberosa R, Chiapparino E, Maccaferri M, Veillet S, van Beuningen L, Isaac P, Edwards K, Phillips RL (2002) Toward positional cloning of vgt1, a QTL controlling the transition from the vegetative to the reproductive phase in maize. Plant Mol Biol 48(5–6):601–613
Santiago R, Barros-Rios J, Malvar RA (2013) Impact of cell wall composition on maize resistance to pests and diseases. Int J Mol Sci 14(4):6960–6980
Sosnowski O, Charcosset A, Joets J (2012) BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28(15):2082–2083
Stuth J, Jama A, Tolleson D (2003) Direct and indirect means of predicting forage quality through near infrared reflectance spectroscopy. Field Crops Res 84(1–2):45–56
Surault F, Emile JC, Briand M, Barrière Y, Traineau R (2005) Variabilité génétique de la digestibilité in vivo d’hybrides de maïs. Bilan de 34 années de mesures. Fourrages 183:459–474
Torres AF, Noordam-Boot CMM, Dolstra O, van der Weijde T, Combes E, Dufour P, Vlaswinkel L, Visser RGF, Trindade LM (2014) Cell wall diversity in forage maize: genetic complexity and bioenergy potential. Bioenergy Res 8(1):187–202
Torres AF, Noordam-Boot CMM, Dolstra O, Vlaswinkel L, Visser RGF, Trindade LM (2015) Extent of genotypic variation for maize cell wall bioconversion traits across environments and among hybrid combinations. Euphytica 206(2):501–511
Truntzler M, Barrière Y, Sawkins MC, Lespinasse D, Betran J, Charcosset A, Moreau L (2010) Meta-analysis of QTL involved in silage quality of maize and comparison with the position of candidate genes. Theor Appl Genet 121(8):1465–1482
Vermerris W, Sherman DM, McIntyre LM (2010) Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J Exp Bot 61(9):2479–2490
Wang H, Li K, Hu X, Liu Z, Wu Y, Huang C (2016) Genome-wide association analysis of forage quality in maize mature stalk. BMC Plant Biol 16:227
Williams E, Piepho H-P, Whitaker D (2011) Augmented p-rep designs. Biom J 53(1):19–27
Wolak ME (2012) Nadiv : an R package to create relatedness matrices for estimating non-additive genetic variances in animal models. Methods Ecol Evol 3(5):792–796
Zhang Y, Culhaoglu T, Pollet B, Melin C, Denoue D, Barrière Y, Baumberger S, Méchin V (2011) Impact of lignin structure and cell wall reticulation on maize cell wall degradability. J Agric Food Chem 59(18):10129–10135
Zhang Y, Yang D-Q, Wang X-M, Feng M, Brunette G (2012) Process for fungal modification of lignin and preparing wood adhesives with the modified lignin and wood composites made from such adhesives. Patent: WO 2012/113058 A1
Acknowledgements
A. I. Seye’s Ph.D. was funded by the Senegalese Institute of Agricultural Research (ISRA) through a Scholarship from the West Africa Agricultural Productivity Program (WAAPP) given by the National Institute of Higher Education in Agricultural Sciences—Montpellier SupAgro. We are grateful to Caussade Semences, Euralis Semences, Limagrain Europe, Maïsadour Semences, Pioneer Genetics, R2n, Syngenta Seeds and KWS grouped in the frame of the ProMais “SAM-MCR” program for the funding, inbred lines development, hybrid production and phenotyping. We also thank scientists from these companies for helpful discussions on the results. We thank Magali Joannin of the INRA station of GQE Le Moulon for seed management and D. Madur and V. Combes for DNA extraction and genotyping analyses. We thank C. Palaffre and the INRA experimental unit of Saint-Martin-de-Hinx for seed production and seed management. We thank Dominique Kermarrec and the INRA station of Ploudaniel for their support to ProMaïs in phenotyping. We thank Pierre Dardenne from CRA-Wallonie for the NIRS prediction equations of silage quality traits.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments comply with the current laws of the countries in which the experiments were performed.
Additional information
Communicated by Thomas Lubberstedt.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seye, A.I., Bauland, C., Giraud, H. et al. Quantitative trait loci mapping in hybrids between Dent and Flint maize multiparental populations reveals group-specific QTL for silage quality traits with variable pleiotropic effects on yield. Theor Appl Genet 132, 1523–1542 (2019). https://doi.org/10.1007/s00122-019-03296-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03296-2