Abstract
Key message
Population structure affects genomic selection efficiency as well as the ability to forecast accuracy using standard GBLUP.
Abstract
Genomic prediction models usually assume that the individuals used for calibration belong to the same population as those to be predicted. Most of the a priori indicators of precision, such as the coefficient of determination (CD), were derived from those same models. But genetic structure is a common feature in plant species, and it may impact genomic selection efficiency and the ability to forecast prediction accuracy. We investigated the impact of genetic structure in a dent maize panel (“Amaizing Dent”) using different scenarios including within- or across-group predictions. For a given training set size, the best accuracies were achieved when predicting individuals using a model calibrated on the same genetic group. Nevertheless, a diverse training set representing all the groups had a certain predictive efficiency for all the validation sets, and adding extra-group individuals was almost always beneficial. It underlines the potential of such a generic training set for dent maize genomic selection applications. Alternative prediction models, taking genetic structure explicitly into account, did not improve the prediction accuracy compared to GBLUP. We also investigated the ability of different indicators of precision to forecast accuracy in the within- or across-group scenarios. There was a global encouraging trend of the CD to differentiate scenarios, although there were specific combinations of target populations and traits where the efficiency of this indicator proved to be null. One hypothesis to explain such erratic performances is the impact of genetic structure through group-specific allele diversity at QTLs rather than group-specific allele effects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, new breeding methods emerged grouped under the term genomic selection (GS). Their aim is to predict breeding values using all the genomic markers jointly rather than testing the significance of each of them (Meuwissen et al. 2001). Several models have been proposed in the literature, making different hypotheses on the distribution of QTL effects such as GBLUP, BayesA, BayesB or BayesC\(\pi\) (Heslot et al. 2012). Most of the time, genomic predictions are calibrated on a training set (TS) and then applied to a population to be selected, for which the genotypic data is the only source of information available. This scenario is interesting for traits that are expensive or difficult to evaluate, but genomic prediction accuracy may be limited when the genetic distance is large between the TS and the validation set (VS) (Pszczola et al. 2012). Other applications have recently emerged such as a better monitoring of field trials by limiting repetitions (Endelman et al. 2014).
Along with the development of genomic prediction models, there has been a significant effort to develop a priori indicators of precision. The forecast of genomic prediction accuracy could allow breeders to evaluate the interest of a generic TS to predict breeding values in a given breeding population. It would also be possible to optimize the TS constitution in order to maximize genomic prediction accuracy and more generally to optimize breeding programs.
A first set of approaches using deterministic equations were developed involving different parameters such as the trait heritability, the population size and the effective number of chromosome segments linked to the effective population size (Daetwyler et al. 2008; Goddard et al. 2011; Erbe et al. 2013; Elsen 2016). Their efficiency depends largely on the ability to estimate this latter parameter accurately, which proved complicated in practice (Brard and Ricard 2015). An other set of approaches, further referred to as CD, used mixed model equations requiring only the relationships between individuals using genomic or pedigree data and estimates of heritability (VanRaden 2008; Rincent et al. 2012; Rabier et al. 2016).
The genomic prediction models and their corresponding indicators of precision were first developed while considering one homogeneous population. However, this assumption is often violated as genetic structure is a common feature in human, animal and plants. Genetic structure arises when the allele frequencies of subgroups of individuals differ when compared to the ancestral population from which they originate. It might be due to a reproductive isolation followed by an independent drift in each group. In maize, genetic structure is found at the level of heterotic groups, that have been selected for their complementarity in order to maximize heterosis of inter-group hybrids, but may also be observed within each of these heterotic groups (Rincent et al. 2014).
When doing GS, genetic structure can impact the accuracy of predictions (Guo et al. 2014; Albrecht et al. 2014). Most of the models assume a single population, presupposing the conservation of QTL effects between individuals. However, when a genetic structure is observed, differences between QTLs effects can be observed as well as differences in terms of LD extent (Wientjes et al. 2015c) and linkage phase between SNPs and QTLs across populations (Wientjes et al. 2015b). If the same structure is found within the TS and the VS, it is well taken into account by the kinship, when using a GBLUP model, and contributes to genomic prediction accuracy (Guo et al. 2014). But if the structure is different, the accuracy can be strongly impacted. In dairy cattle, when trying to predict breeding values of a breed with a small population size using information coming from a distantly related breed with a larger size, the gain in prediction accuracy is generally very low and may even be negative (de Roos et al. 2009). Likewise, in maize, a TS combining dent and flint lines allowed marginal gains in terms of accuracy compared to pure dent or pure flint TS (Technow et al. 2013). A substantial gain was nevertheless observed when combining related dairy cattle breeds (Brøndum et al. 2011).
To improve genomic prediction accuracy in structured populations, it is possible to adjust both the experimental design and data modeling. Concerning the experimental design, it is possible to improve accuracy by creating hybrids or admixed individuals allowing to connect the different groups (Toosi et al. 2013; Esfandyari et al. 2015). Concerning the models, several alternatives have been proposed such as specifying the structure as a fixed effect (Guo et al. 2014) or modeling genetic covariances between individuals from different groups by adapting multi-trait models. The latter led to improvement of genomic prediction accuracy in dairy cattle (Olson et al. 2012; Karoui et al. 2012) and dairy goat (Carillier et al. 2014) when the genetic correlations between groups were sufficiently high. In maize, such types of models were also applied and allowed very limited gains (Lehermeier et al. 2015). In the case of populations resulting from the admixture between groups, there were attempts to take into account the quantitative assignment of individuals to groups by applying random regression but this allowed limited gains (Strandén and Mäntysaari 2013; Makgahlela et al. 2013).
Structure does not only affect genomic prediction accuracy, but also the ability to forecast this accuracy using indicators. When the population features a strong genetic structure, a priori indicators proved to be inefficient to forecast genomic prediction accuracy in multi-breed dairy cattle populations (Hayes et al. 2009) and to optimize TS in rice populations (Isidro et al. 2015). To tackle this issue, new indicators were recently developed in order to take into account such structures, proving their efficiency on simulated data (Wientjes et al. 2015a, 2016).
The objectives of this article are first to study the impact of genetic structure on genomic prediction accuracy within a diversity panel of maize dent lines. An important question for breeders is whether or not one should combine groups in TS in order to increase accuracy. A second objective is to evaluate the gain in accuracy one may expect when applying models that explicitly account for genetic structure rather than using a standard GBLUP analysis. The last objective is to compare different indicators of precision to study their ability to forecast GBLUP accuracy in the presence of genetic structure.
Materials and methods
Genetic material and genotypic data
Genetic material is a panel of 389 dent maize lines assembled within the “Amaizing” project and aiming at representing the diversity of the dent heterotic group that can be used in European breeding. This panel includes most lines from panels assembled for previous projects: “CornFed” (Rincent et al. 2014) and “Drops” (Millet et al. 2016). This panel was constructed for Genome Wide Association Studies and to apply genomic prediction for traits that are expensive to evaluate. One originality of the panel is to include 49 elite lines coming from seven breeding companies (Fig. 1a), all members of the Amaizing project.
The genotyping data, initially assembled for GWAS studies, included SNPs from different technologies: the 50K Illumina MaizeSNP50 BeadChip (Ganal et al. 2011), the 600K Affymetrix Axiom Maize Genotyping Array (Unterseer et al. 2014) and Genotyping-By-Sequencing (Elshire et al. 2011; Glaubitz et al. 2014). The lines from public origin were all genotyped with the three SNP technologies. The lines from private origin were all genotyped with the 50K chip, 28 were also genotyped with the 600K but none with GBS. At each SNP, allele 0 was attributed to the allele carried by B73, the maize inbred line used as reference for sequencing, or to the allele carried by the first line in alphabetic order if B73 genotype was missing or heterozygous. A quality control on SNP data was applied, removing markers featuring heterozygosity above 15% and missing value rate above 20% for 50K and 600K SNPs. For GBS data, heterozygous were transformed into missing values and markers with more than 70% of missing data were discarded. After merging the three datasets, duplicated SNPs were removed from the dataset based on physical position information leaving 986,045 SNPs. The imputation of missing values was done on the whole dataset using Beagle v.3.3.2 and default parameters (Browning and Browning 2009).
Structure analysis
We performed a structure analysis using the ADMIXTURE software (Alexander et al. 2009) for different numbers of groups, ranging from 2 to 8 (Supplementary Figs. S1, S2 and S3). The cross-validation (CV) error criterion proposed by ADMIXTURE showed an improvement while increasing the number of groups. For the following analyses, we considered three groups which could be linked to well-defined groups in maize breeding which are A: Lancaster and other dent lines (207 lines), B: Stiff-Stalk (98 lines) and C: Iodent (84 lines). Subdividing in more groups would have led to define families or specific pedigree structures rather than well-known genetic groups and to insufficient number of lines per group to perform analyses. A PCoA was performed on genetic distances computed as \(D_{i,j}=1-K_{i,j}^0\) with \(K^0_{i,j}\) being the kinship coefficient between lines i and j in Eq. (1, see below). This analysis clearly separated individuals based on their maximal admixture coefficient (Fig. 1b).
Phenotypic data
All the lines were crossed to the same tester UH007 to produce hybrid progenies for phenotypic evaluation. The 2014 field trials (Supplementary Table S1) were conducted in seven locations in standard agronomic conditions including Blois, Mons, Niederhergheim, Souprosse, Villampuy (France), Bernburg (Germany) and Graneros (Chile). Each trial was a latinized alpha design where every genotype was repeated 2 times on average. Grain moisture (in % of humidity), grain yield at 15% of humidity (quintals per hectare) and male flowering time were recorded for each plot. Male flowering time was converted into growing degree-days, considering a base temperature of 6 Celsius degrees, using the mean daily air temperature measured at each location. An economic index, called yield index, was also computed as: \(\mathrm{Yield\,Index} = \mathrm{Grain\,Yield} - 2.5 \times \mathrm{Grain\,Moisture}\). This index corresponds to grain yield penalized for an excess of humidity at harvest, which would require an expensive drying process, and is used for variety registration in France.
We started the analysis from data collected after correction for within trial spatial effects using different models (Supplementary Table S1). Then, we computed least-square means (LS-means) over the whole design using model: \(Y_{ij}=\mu _i+T_j+E_{ij}\) where \(Y_{ij}\) is the performance of individual i in the location j, \(\mu _i\) is the intercept for individual i, \(T_j\) is the \(j^{th}\) random trial effect where \(T_j\sim \mathcal {N}(0,\sigma ^2_T)\) are all independent and identically distributed (i.i.d.), \(E_{ij}\) is the error were \(E_{ij}\sim \mathcal {N}(0,\sigma ^2_E)\) i.i.d. and \(E_{ij}\) and \(T_j\) are assumed to be independent.
Genomic prediction models
All the genomic prediction models used in this study can be written as:
where \(\varvec{y}\) is the vector of LS-means which will be further referred to as phenotypes, \(\varvec{X}\) is the incidence matrix for fixed effects, \(\varvec{\beta }\) is the vector of fixed effects, \(\varvec{Z}\) is an incidence matrix linking observations to breeding values, \(\varvec{g}\) is the vector of breeding values and \(\varvec{e}\) is the vector of errors. All models assume independence between \(\varvec{g}\) and \(\varvec{e}\).
GBLUP
We used a standard additive GBLUP model as a base model \(\mathbf {M_0}\) with the following assumptions: \(\varvec{X}=\varvec{1}_N\) was a vector of 1 of length N (with N the number of individuals), \(\varvec{\beta }=\mu\) the general mean of performances, \(\varvec{g}\sim \mathcal {N}(0,\varvec{K}\sigma _G^2)\) and \(\varvec{e}\sim \mathcal {N}(0,\varvec{I}\sigma _E^2)\) with \(\sigma _G^2\) and \(\sigma _E^2\) being the genetic and residual variances, respectively. The kinship between individuals i and j, \(K^0_{i,j}\), was estimated using VanRaden (2008):
where \(W_{im}\) is the genotype of individual i at locus m (coded 0 ; 0.5 ; 1) and \(f_m\) is the allele frequency of allele “1” at locus m, estimated on the whole dataset.
Structured GBLUP
The standard kinship estimation combines relatedness and genetic structure information. In order to test whether modeling the structure as a fixed effect could improve the predictions, we used two adapted GBLUP models.
Model \(\mathbf {M_1}\) followed the same assumptions as \(\mathbf {M_0}\) except that population structure was added as a fixed effect where \(\varvec{X}\) was the \((N\times Q)\) incidence matrix for fixed effects with \(X_{iq}=1\) if i was assigned to the \(q^{th}\) genetic group (otherwise \(X_{iq}=0\)), \(Q=3\) is the number of genetic groups and \(\varvec{\beta }=(\mu _A,\mu _B,\mu _C)^T\) is a vector of fixed group effects. For \(\mathbf {M_1}\), the kinship was estimated following Plieschke et al. (2015), by centering the genotypes using group-specific allele frequencies to remove the structure from the kinship and to avoid a redundancy of information in the model:
where \(p_{im}=\sum _{q=1}^QX_{iq}f_{mq}\) and \(f_{mq}\) is the allele frequency of group q as provided by ADMIXTURE.
Model \(\mathbf {M_2}\) followed the same assumptions as \(\mathbf {M_1}\) except that it considered quantitative assignments of individuals to groups in the prediction model and in the kinship. Thus \(X_{iq}\) became the admixture coefficient of individual i for group q. The kinship \(K_{i,j}^2\) used in \(\mathbf {M_2}\) was estimated using the same expression as for \(K_{i,j}^1\) in Eq. (2) but with \(p_{im}\) being a weighted mean of ancestral group-specific allele frequencies with weights corresponding to admixture coefficients (Thornton et al. 2012).
Multi-group GBLUP
We also used a multivariate model \(\mathbf {M_3}\) considering categorical assignments to genetic groups, which is an adaptation of a multi-trait model to the analysis of one trait in different groups proposed by Lehermeier et al. (2015).
In this model, \(\varvec{X}\) and \(\varvec{\beta }\) are the same as in \(\mathbf {M_1}\), \(\varvec{Z}\) is an incidence matrix linking phenotypes to the corresponding group-specific breeding value (categorical assignments), \(\varvec{g}=\begin{bmatrix}\varvec{g^*}_A\\\varvec{g^*}_B\\\varvec{g^*}_C\end{bmatrix}\) is the expanded vector of breeding values of each individual in each group with a size of 3N and \(\varvec{e}=\begin{bmatrix}\varvec{e}_A\\\varvec{e}_B\\\varvec{e}_C\end{bmatrix}\) is the vector of errors of size N where:
with \(\sigma _{G_{X,Y}}\) being the genetic covariance between groups X and Y (the letters X, Y and Z were further used as group names when not specifically designating group A, B or C). In this model, the kinship between individuals i and j, \(K_{i,j}^{0'}\) (Eq. 3), was computed following Astle and Balding (2009) as recommended by Lehermeier et al. (2015), although results were very consistent using the kinship defined by VanRaden (2008) (Eq. 1).
Note that the genetic covariance between groups results from the genetic covariance between allele effects in each group as described in Karoui et al. (2012) and Lehermeier et al. (2015).
We also defined \(r_{X,Y}=\dfrac{\sigma _{G_{X,Y}}}{\sigma _{G_{X}}\sigma _{G_{Y}}}\) where \(r_{X,Y}\) is the genetic correlation between groups X and Y.
For each model, the Genomic Estimated Breeding Values (GEBV) of the VS were computed as: \(\mathbf {\widehat{y}}_{VS}=\mathbf {X}_{VS}\varvec{\widehat{\beta }}+\mathbf {\widehat{g}}_{VS}\)
Model parameters were estimated using ASReml-R (Butler et al. 2009) for models \(\mathbf {M_0}\), \(\mathbf {M_1}\) and \(\mathbf {M_2}\), using restricted maximum likelihood method. For the last model \(\mathbf {M_3}\), a Gibbs sampler implemented in R was used to estimate the parameters.Footnote 1 The choice of hyper-parameters was the same as described in Lehermeier et al. (2015). A total of 300,000 MCMC samples were collected with 100,000 discarded as burn-in and thinning was done by keeping one every two samples. The parameter estimates were obtained by computing posterior means.
Evaluation of the precision of genomic predictions
The precision of the models was evaluated using four different CV procedures either neglecting genetic structure or aiming at evaluating its impact on the precision of genomic predictions.
The first CV procedure was an averaged Holdout (HO) method and allowed us to study the level of precision that can be obtained when neglecting the role of genetic structure. The initial dataset was split with proportions \(\frac{4}{5}\) and \(\frac{1}{5}\) for the TS and the VS, respectively. The splitting was repeated 100 times and the precision criteria were averaged over repetitions.
The second CV procedure was a Leave-One-Out method (LOO) where every individual was predicted with a model calibrated using all the remaining individuals. It allowed a simple graphic representation of the quality of prediction of each individual using all the other individuals from the panel, whatever their group of origin. We also used this approach to evaluate the link between the CD (see below) and the prediction of each individual.
The third CV procedure, named Structured Holdout (SHO), allowed us to study the impact of genetic structure in genomic prediction accuracy using different scenarios. It considered samples of restricted sizes where 18 individuals are predicted using the model calibrated with 66 other individuals, repeating sampling 100 times. Those numbers were chosen in order to fit with all the scenarios (Table 1), knowing that group C is limited to 84 individuals. The individuals were assigned to the three groups according to their maximal admixture coefficient. All the scenarios are designated as TS_VS, TS and VS referring to the groups represented in the TS and VS, respectively. When there were more than one group in the TS or the VS, the composition was always perfectly balanced between groups. As an example, ABC_A referred to a TS equally composed of individuals from the three groups and a VS composed of lines from group A only. Note that the across-group SHO scenarios (i.e., when no individual from the group forming the VS is present in the TS) cannot be evaluated using model \(\mathbf {M_1}\) and \(\mathbf {M_3}\). They require a TS where all the genetic groups are represented in order to estimate all the group-specific intercept and variance parameters.
The fourth CV procedure, referred to as Structured Holdout + (SHO\(^+\)), aimed at evaluating the benefits of extra-group individuals (individuals from a group absent of the VS) to improve genomic prediction accuracy. It considered the same samples as in SHO for intra-group predictions (scenarios X_X) complemented with 66 individuals of each of the two other groups, reaching a size of 198 individuals. For instance, ABC\(^+\)_A referred to the SHO\(^+\) scenario to predict group A.
Three criteria of precision were used to compare models and scenarios. The first was the predictive ability, defined as the correlation between GEBV and the phenotypes. The second was the accuracy which was computed by dividing the predictive ability by the square root of the heritability. Here, the heritability was computed using the estimated variances obtained by applying \(\mathbf {M_0}\) to the whole panel. The third criterion was the Root Mean Square error of Prediction (RMSP), defined as the root mean square of the differences between LS-means and the GEBV.
A priori estimation of accuracy
In mixed models, the accuracy related to the prediction of individual i can be quantified by its associated Coefficient of Determination (CD), using the general formula:
where \(\varvec{g}_i\) and \(\widehat{\varvec{g}}_i\) are the breeding value of individual i and its corresponding BLUP, respectively, \(\varvec{G}_{i,TS}\) is the covariance matrix between breeding values of i and the TS, \(\varvec{G}_{i,i}\) is the genetic variance of i and \(\varvec{\varSigma }_{TS,TS}\) is the covariance matrix between phenotypic values within the TS.
The standard CD in a GBLUP model assumes an unstructured population, as described in model \(\mathbf {M_0}\) and is computed using Eq. (4), where:
-
\(\varvec{G}_{i,TS}=\varvec{K^0}_{i,TS}\widehat{\sigma }^2_G\)
-
\(\varvec{G}_{i,i}=\varvec{K^0}_{i,i}\widehat{\sigma }^2_G\)
-
\(\varvec{\varSigma }_{TS,TS}=\varvec{K^0}_{TS,TS}\widehat{\sigma }^2_G+\varvec{I}\widehat{\sigma }^2_E\)
-
\(\widehat{\sigma }^2_G\) and \(\widehat{\sigma }^2_E\) are the genetic and residual variances, respectively, estimated using \(\mathbf {M_0}\) calibrated with all the individuals.
We also considered the multi-group CD as proposed by Wientjes et al. (2015a) and derived from \(\mathbf {M_3}\). When considering scenario XY_Z, the elements of Eq. (4) become:
-
\(\varvec{G}_{i,TS}=\begin{bmatrix}\varvec{K}_{Z,X}\widehat{\sigma }_{G_{Z,X}}&\varvec{K}_{Z,Y}\widehat{\sigma }_{G_{Z,Y}}\end{bmatrix}_{i,TS}\)
-
\(\varvec{G}_{i,i}=\begin{bmatrix}\varvec{K}_{Z,Z}\widehat{\sigma }^2_{G_{Z}}\end{bmatrix}_{i,i}\)
-
\(\varvec{\varSigma }_{TS,TS}=\begin{bmatrix}\varvec{K}_{X,X}\widehat{\sigma }_{G_{X}}^2 +\varvec{I}\widehat{\sigma }_{E_{X}}^2&\varvec{K}_{X,Y}\widehat{\sigma }_{G_{X,Y}}\\ \varvec{K}_{Y,X}\widehat{\sigma }_{G_{X,Y}}&\varvec{K}_{Y,Y}\widehat{\sigma }_{G_{Y}}^2 +\varvec{I}\widehat{\sigma }_{E_{Y}}^2\end{bmatrix}_{TS,TS}\)
-
\(\widehat{\sigma }_{G_{Z,X}}\), \(\widehat{\sigma }_{G_{X}}^2\) and \(\widehat{\sigma }_{E_{X}}^2\) are the genetic covariance between group Z and X, the genetic variance in group X and the residual variances in group X, respectively, estimated on the whole dataset.
Two versions of this multi-group CD were computed using different kinships \(\varvec{K}\). The first version called \(CDgp^1\) used \(\varvec{K}^0\) defined in Eq. (1) while the second called \(CDgp^2\) used a new estimator recommended by Wientjes et al. (2017):
with \(p_{im}=\sum _{q=1}^QX_{iq}f_{mq}\) where \(X_{iq}\) considered categorical assignment of individuals to groups as defined in \(\mathbf {M_1}\).
\(CDgp^1\) is computed using variances estimated with \(\mathbf {M_3}\) and \(\varvec{K^{0'}}\) (computed using Eq. (3) as recommended by Lehermeier et al. 2015) on the whole dataset, although parameters estimates were very consistent using \(\varvec{K^{0}}\) (Eq. 1). \(CDgp^2\) is computed using variances estimated with \(\mathbf {M_3}\) and \(\varvec{K^3}\) on the whole dataset as summarized in Table 2.
After computing the CD values of every individual of the VS, we averaged them and computed the square root to obtain an a priori indicator of accuracy.
The a priori estimates of accuracy were compared to empirical accuracies, obtained from model \(\mathbf {M_0}\) with the SHO method, for the different scenarios described above using two criteria of precision. The first criterion was the correlation between the a priori estimates of accuracy and the empirical accuracies. The second criterion was the Root Mean Square error of Estimation (RMSE) which is defined as the root mean square of the differences between a priori and empirical accuracies.
We also computed standard CD for each predicted individual in the context of LOO CV.
Results
Global, within- and across-group precision of genomic predictions
We first estimated variances for the four traits by applying model \(\mathbf {M_0}\). The estimated heritabilities were very high (Table 3), between 0.86 and 0.95 and consistent with the high heritabilities computed in each trial without considering kinship (Supplementary Table S1).
The estimates of accuracy obtained with the HO method and \(\mathbf {M_0}\) model ranged from 0.77 for yield index to 0.84 for grain yield (Table 4). The accuracy estimates were close to the predictive abilities as a consequence of high heritabilities. We also studied the ability of all the lines from public origin to predict all the private lines. The accuracies obtained were 0.64, 0.49, 0.33 and 0.76 for grain moisture, grain yield, yield index and male flowering respectively.
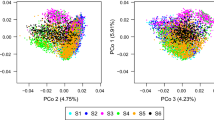
In Fig. 2, where predictions were obtained with LOO, we observed a differentiation between groups for the four traits. For instance, groups B and C were almost perfectly separated along the two axes for male flowering. These mean differences may be partly responsible for the high level of correlation obtained in Table 4.
In order to study more carefully the impact of genetic structure on genomic predictions, we performed a third type of CV named SHO using \(\mathbf {M_0}\) with different scenarios defined in Table 1. Accuracies (Table 5) and predictive abilities (Supplementary Table S2) showed similar trends. Scenario ABC_ABC displayed in general a higher accuracy than scenarios ABC_X but its RMSP was generally not the lowest (Table 6).
Considering group-specific VS, the best predictions, both in terms of accuracy and RMSP, were achieved when predicting one group with individuals from the same group. The only exception was grain moisture and male flowering in group C for which the lowest RMSP and the highest accuracies were obtained by using a TS consisting of individuals from the three groups (scenario ABC_C). The worst predictions were always achieved when trying to predict one group using only one other group (scenarios X_Y) while using the two other groups allowed intermediate accuracies and RMSP (scenarios XY_Z). Except for yield index, trying to predict group C using scenario AB_C was among the best options, conversely to what was observed for the symmetric scenarios in the other group for which BC_A and AC_B were outperformed by A_A and B_B, respectively.
In general, group-specific accuracies tended to be higher in group A than in group B and C, regardless of the trait considered. The opposite was generally observed when considering RMSP, for which group A presented a higher prediction error, meaning a lower precision.
In order to study the impact of adding extra-group individuals on the accuracy of genomic predictions, we performed SHO\(^+\) CV (Table 7). Adding individuals always increased accuracy and decreased RMSP except in group C for yield index. Generally, the gain in precision was greater in group C than in group B, itself greater than in group A.
Accounting for structure in genomic prediction models
We tested three other models, taking into account genetic structure, to compare them to model \(\mathbf {M_0}\) on scenario ABC_ABC and on scenarios ABC_X (SHO CV). In general, the four models tended to reach similar performances when considering accuracy as a criterion (Fig. 3). Model \(\mathbf {M_3}\) reached performances below the other models in all the scenarios for male flowering and for some scenarios in the other traits such as scenario ABC_C for grain moisture. However it allowed better accuracies in scenario ABC_ABC and ABC_C for yield index. The same conclusions could be made considering predictive ability or RMSP as criteria (Supplementary Figs. S4 and S5). The across-group scenarios were also tested to compare \(\mathbf {M_0}\) and \(\mathbf {M_2}\) (model considering quantitative assignment to groups) showing no improvement when using the latter (Supplementary Figs. S6, S7 and S8).
A priori estimation of precision
To compute the CD, variances were estimated within the whole population using \(\mathbf {M_0}\) (Table 3). To compute \(CDgp^1\) and \(CDgp^2\), variances and genetic correlations were estimated between groups using \(\mathbf {M_3}\) with \(\varvec{K^{0'}}\) (Table 8) and \(\varvec{K^3}\) respectively (Supplementary Table S3). For all traits, the genetic variance estimates were lower in group C than in the other groups. The genetic correlations between groups were very high, around 0.90, except for grain moisture where they ranged from 0.72 to 0.76 using \(\varvec{K^{0'}}\) and from 0.62 to 0.72 using \(\varvec{K^3 }\). The group-specific heritabilities obtained from these estimates were also high (results not shown).
Before studying the ability of CD values to reflect the accuracy in the VS, we observed how CD values were connected to the prediction of individual performances obtained with LOO CV (Fig. 4). For grain moisture and male flowering (Fig. 4a, d), the individuals featuring low CD values were predicted close to the mean and were more likely to have important observed errors of prediction. Conversely, individuals featuring high CD values had a broader range of predicted values and the predictions were more accurate. A different situation was observed for grain yield and yield index which are submitted to directional selection (Fig. 4b, c). For these traits, individuals with low CD values were predicted to have low performances. Conversely, those with high CD values were predicted to have high performances.
The a priori accuracy of the different SHO scenarios were estimated by computing the square root of the average of the CDs over the individuals of the VS. The a priori estimates of accuracy were compared to the empirical accuracies using the correlation between a priori estimates and empirical accuracies and the RMSE between these two accuracies.
When first looking at the different plots between a priori and empirical accuracies (Supplementary Figs. S9, S10, S11 and S12), one could notice that there was a high variability of the empirical accuracies for a defined scenario (see also Table 5). All the indicators (CD, \(CDgp^1\) and \(CDgp^2\)) led to either positive or null values of correlation between the a priori estimates of accuracy and the empirical accuracies (Table 9). There were different abilities to forecast accuracy depending on the trait and the group considered. It was harder to predict the level of accuracy in group C than in groups A and B for all the traits except yield index. For instance, the correlation was almost null regardless of the indicator used for grain moisture in this group. In contrast, the ability to forecast the level of precision was up to 0.56 in group C for yield index. When comparing the three indicators using the correlation between empirical and a priori accuracies, it was difficult to assess which one performed best. The differences were very low between CD, \(CDgp^1\) and \(CDgp^2\) which might be explained by the high genetic correlation between groups (except for grain moisture), as well as a limited impact of the kinship used to compute the CDs. Along with the correlation, using the RMSE between a priori and empirical accuracies did not allow us to identify a better indicator of accuracy (Supplementary Table S4).
Discussion
The impact of genetic structure on genomic prediction accuracy
We investigated genomic prediction accuracy using LS-means corrected for trial effects as observed phenotypes to minimize environmental effects. As a consequence, the estimates of heritabilities obtained when fitting additive model \(\mathbf {M_0}\) were very high and consistent with the high heritabilities obtained for each trial without considering kinship. Along with the high heritabilities, the predictive abilities and the accuracies were high when neglecting population structure, revealing both the relevance of model \(\mathbf {M_0}\) to make predictions and the quality of the data.
In this dataset, structure participated to genomic prediction accuracy, as the accuracy was generally higher in scenario ABC_ABC than in scenarios ABC_X for a given TS size. The standard kinship matrix contains information about the structure of the population in genetic groups. When there is a difference of mean between groups and the same structure is found in the TS and the VS, this difference is well taken into account by the GBLUP model and participates to the accuracy (Guo et al. 2014). At the extreme, one could imagine a trait for which there would be a global positive correlation between predicted and true breeding values but with a null accuracy within each group composing the VS. The RMSP criterion is not impacted by genetic structure like the accuracy, as RMSP is not lower in scenario ABC_ABC than in scenarios ABC_X. RSMP is thus complementary to the accuracy to evaluate the precision of the predictions.
The main interest of a plant breeder is to know the level of precision that can be reached within each genetic group, as selection will be often applied on individuals derived from crosses between related elite lines from a same group. In this dataset, to predict a group-specific VS for a given size of TS, it was generally better to use a TS from the same group (scenarios X_X), as previously shown in soybean (Duhnen et al. 2017). Depending on the trait, accuracy could be severely impacted by not representing relatives of the VS in the TS (scenarios X_Y and XY_Z). This suggests an inconsistency of allele effects between groups, or different LD extent between SNPs and QTLs. However, these hypotheses are not supported by the high genetic correlations estimates between groups for all the traits.
Group C showed interesting results as it was best predicted with a diverse TS, except for yield index. Simultaneously, the accuracy of scenario AB_C was as high or higher than the accuracy achieved in scenario C_C. We can hypothesize that the allele effects are conserved between groups and that there are none or few QTLs specifically polymorphic in group C for these three traits. This hypothesis is supported by SNP data as group C features less specifically polymorphic SNP and a lower genome-wide genetic diversity than the other two groups (results not shown). As this group is the less diverse, with a high degree of relatedness between individuals, it might be beneficial to calibrate a model on a diverse set of individuals. Indeed, from a statistical point of view, the precision of the estimation of the effects of each locus or each breeding value gets worse as the diversity decreases within the TS. The situation is different for groups A and B which possibly presented specifically polymorphic QTLs. Thus, when trying to predict group A or B by the two other groups (using scenarios B_A, C_A, BC_A, A_B, C_B and AC_B), the effects associated to these group-specific polymorphisms cannot be taken into account by the model. The particular situation of group C (Iodents) is supported by its recent history as it was initially derived from individuals from group A for its complementarity with group B (Stiff Stalks) 50 to 70 years ago. Yield index presented a different behavior and possibly involves yield QTLs independent from precocity that are specifically polymorphic in each group. We also checked that this different behavior was not due to predicting yield index directly rather than computing it using the individual predictions of grain yield and grain moisture (results not shown).
One could notice that QTLs specifically polymorphic in one group is the extreme case of QTLs having group-specific allele diversities.
In a configuration where allele effects seem conserved between groups (based on genetic correlations estimates in Table 8), which is consistent with a moderate and recent genetic structure, one should recommend the constitution of a TS as diverse as possible where all the genetic groups are represented. This is supported by the high level of accuracy reached for scenarios ABC_X and by the accuracy gains obtained by adding extra-group individuals to the TS in SHO\(^+\) CV. Such diverse TS should be efficient to calibrate prediction models for a wide range of genetic material. This underlines its value as a generic TS for expensive traits evaluated on high-throughput phenotyping platforms or through extensive field trials (Millet et al. 2016). We showed that the accuracies, obtained when predicting the elite private lines by lines from public origin, were moderate or high depending on the trait. The accuracy was lower for traits submitted to directional selection such as grain yield or yield index for which the variability is limited among the elite lines. As elites lines were distributed in the three groups, we checked that accuracies were not entirely explained by the genetic structure (results not shown). As more data were imputed for elite lines, we also checked the impact of imputation by performing a subset of analyses (within-group or across-group SHO scenarios) on 50K data that were available for all the lines and found very limited changes in terms of accuracy.
Modeling genetic structure to improve predictions
When performing genomic predictions within a structured population, one may wish to improve accuracy by using specific models taking into account this structure. There are different possibilities such as specifying structure as a fixed effect considering categorical or quantitative assignments of individuals to groups. It is also possible to model group-specific random effects, with group-specific variances and covariances between groups.
For this dataset, applying different models did not allow to improve the accuracy on the scenarios tested (ABC_ABC and ABC_X). For model \(\mathbf {M_1}\) and \(\mathbf {M_2}\), structure was removed from the kinship in Eq. (2) after being included as a fixed effect thanks to a genotypic centering using group-specific allele frequencies. One could expect that modeling groups as fixed effect would be advantageous if the differences between groups are larger than what can be attributed to differences in allele frequencies (Plieschke et al. 2015). Such benefits were not observed on our data. In \(\mathbf {M_3}\) the assumption is not only that groups differ in terms of mean but that allele effects may be potentially correlated between groups, leading to group-specific genetic variances and specific covariances between groups. This model did not improve genomic prediction accuracy and sometimes reached substantially lower accuracies than \(\mathbf {M_0}\). When applying \(\mathbf {M_3}\), there was a variability of genetic correlations estimates probably due to small TS sizes (results not shown) which might explain its poorer performances. One should also notice that the inference procedure differed between model \(\mathbf {M_3}\) (Bayesian inference) and the three other models (REML), which could possibly explain part of the differences in terms of performance.
It is important to note that for \(\mathbf {M_1}\) and \(\mathbf {M_3}\) which consider categorical assignments, all the parameters are not estimable for across-group predictions. For instance, scenario AB_C requires to estimate \(\mu _C\) for both models, and \(\sigma ^2_{G_{C}}\), \(\sigma _{G_{AC}}\) and \(\sigma _{G_{BC}}\) for model \(\mathbf {M_3}\). These quantities cannot be estimated if group C is absent of the TS. In such a context, one should either neglect genetic structure or take advantage of the admixed individuals to connect groups. We tested model \(\mathbf {M_2}\) which take into account such admixture as fixed effects showing no clear improvement in terms of accuracy compared to \(\mathbf {M_0}\).
Is it possible to forecast accuracy using CD?
Being able to forecast the accuracy of predictions would allow many applications such as the optimization of TS or the anticipation of genetic gain in breeding programs. Many indicators were developed in order to get an a priori estimation of accuracy. Among them, the CD is well known (VanRaden 2008) and can be easily derived as the square correlation between the breeding value of an individual and its corresponding BLUP in a standard GBLUP model. The estimate obtained is supposed to quantify the amount of information available from the TS to correctly predict the breeding values, with a value scaled between 0 and 1. In theory, when the CD value is low, the prediction is more likely to be inaccurate (high expected errors of prediction) and will be strongly shrunk toward the mean and conversely for a high CD value. This situation was observed for grain moisture and male flowering on the LOO plot. A different situation was observed for grain yield and yield index and could be interpreted as an effect of modern breeding. In this panel, the lines featuring a high CD were recent and related to several older ones featuring lower CDs. As those two traits are those of major interest in breeding and this panel included lines that have been under directional selection, it yielded this gradient of CD values along the prediction axis. Individuals featuring high CD values were predicted to have high performances and conversely for low CD values. Male flowering and grain moisture on opposite were not submitted to directional selection. Thus individual CD value is an interesting criterion in breeding but one could question whether it could be informative about the accuracy of a set of individuals.
The CD value is linked to the accuracy as it represents its square value. For a defined TS and VS, one can easily compute each individual CD of the VS and compute the square root of the average CD to get an a priori estimation of accuracy. Such CD-based accuracy usually succeeded in differentiating the structure-based scenarios in terms of accuracy. However, there were differences depending on the trait and the genetic group considered. For instance the null correlation between empirical and a priori accuracies encountered in group C for grain moisture was probably due to the overestimation of within-group a priori accuracy (Supplementary Fig. S9 and scenario C_C).
The use of standard CD in the presence of a genetic structure was already criticized in the literature for TS optimization (Isidro et al. 2015) or to forecast accuracy (Hayes et al. 2009). One hypothesis to explain the poor performance of the CD in such context is the existence of different but correlated allele effects between groups. To tackle this problem, a new CD indicator was derived from a multi-group model (Wientjes et al. 2015a). The authors also recommended the use of an alternative Kinship estimator, using group-specific allele frequencies to standardize the genotypic data. In this study, we tested two versions of this indicator, one using a standard Kinship: \(CDgp^1\), and the other using the Kinship \(\varvec{K^3}\) in Eq. (5) recommended by Wientjes et al. (2017): \(CDgp^2\).
Both indicators required estimates of genetic correlations between groups. These correlations were originally defined as resulting from the correlation between allele effects in the different groups (Karoui et al. 2012; Lehermeier et al. 2015). Each correlation may also be considered at the level of one individual where its breeding value in one group could be correlated to its potential breeding value in another group, assuming that these two groups showed correlated allele effects. The estimated correlations obtained in this study by applying \(\mathbf {M_3}\) using \(\varvec{K^{0'}}\) for \(CDgp^1\) or \(\varvec{K^3}\) for \(CDgp^2\), were very high except for grain moisture. One could wonder how accurate they were as these values did not allow us to explain the differences in prediction accuracies observed between traits. In a recent study, Wientjes et al. (2017) investigated the impact of different genomic relationship matrices on the estimation on genetic correlations between groups. While they showed that using the genomic relationship matrix \(\varvec{K^0}\) of VanRaden (2008) or \(\varvec{K^3}\) of Wientjes et al. (2017) estimated genetic correlations between groups unbiasedly, they warned about the use of \(\varvec{K^{0'}}\) defined in Astle and Balding (2009) that we used in \(\mathbf {M_3}\). However, we could check that the kinship of VanRaden (2008) and the one of Astle and Balding (2009) gave very close estimates (results not shown).
As a consequence of high genetic correlations, CD and \(CDgp^1\) gave very similar results. One could notice that using genetic correlations of 1 and equal genetic and residual variances for each group would result in \(CDgp^1\) and CD being perfectly equal. Using \(\varvec{K^3}\) and a set of parameters estimated using \(\mathbf {M_3}\) with \(\varvec{K^3}\) to compute \(CDgp^2\) also led to very similar results which indicated us that the kinship estimator did not have much of an impact to forecast accuracy in this dataset. Grain moisture is the only trait featuring lower genetic correlations between groups but both multi-group CD did not outperform standard CD for this trait.
As \(CDgp^1\) and \(CDgp^2\) assume an \(\mathbf {M_3}\) genetic model, one could wonder whether they would not be better correlated to empirical accuracies obtained with the \(\mathbf {M_3}\) model instead of \(\mathbf {M_0}\). As mentioned in the previous part of the discussion, \(\mathbf {M_3}\) cannot be used as a predictive model for across-group scenarios. However, we predicted breeding values and computed empirical accuracies obtained with \(\mathbf {M_3}\), using parameters estimated on the whole dataset. Multi-group CDs did not better forecast these empirical accuracies (Supplementary Figs. S13 and S14).
Once again, these results supported the hypothesis that the allele effects were indeed highly correlated between groups and the impact of genetic structure would mostly be due to different group-specific allele diversity at QTLs. The CD indicators are based on macro-parameters such as the global genetic variance, but they do not take into account more detailed information like the number of QTLs and their localization along the genome. Simple simulations, using real genotypic data, of traits similar in terms of genetic variance and heritability with the ones measured in our study, showed an important variability of the impact of genetic structure on accuracy considering SHO results (Supplementary Fig. S15). The differences between traits were only due to allele effects sampling and they could not be captured by CD indicators, thus supporting this hypothesis. The impact of QTLs specifically polymorphic in DH bi-parental families on genomic prediction accuracy was recently shown by Schopp et al. (2017) when performing across family predictions. The authors also discussed the impact of such QTLs on CD-based estimations of accuracy and recommended to use \(\varvec{K^3}\), the kinship estimator we used in \(CDgp^2\). However, we showed that \(CDgp^2\) did not improve estimations of accuracy in our study. We also tested other CDs such as the CD of contrast (Rincent et al. 2012, 2017) between breeding values and the mean breeding value of the TS, or the new proxy developed by Rabier et al. (2016). Both approaches did not give better results than the individual CD (results not shown).
Conclusion
In conclusion, genetic structure impacted genomic prediction accuracy in this dent maize panel. For a given size of TS, the highest accuracies were often achieved when the TS and the VS were consistent in terms of group composition. However, a diverse TS remained efficient for every VS and adding extra-group individuals almost always improved accuracy. These results are encouraging concerning the use of this panel as a generic TS to be characterized on high-throughput phenotyping platforms or through extensive field trials. Using alternative prediction models, taking genetic structure into account, did not allow any precision gain compared to GBLUP. Finally, the use of CD, a priori indicator derived from mixed model equations, proved to be sometimes but not always effective to forecast the level of precision in a set of predicted individuals. New indicators taking structure into account did not achieve better performances. This study has highlighted that, in groups that diverged recently, the impact of group structure is likely due to differences in group-specific allele diversity instead of differences in allele effects that cannot be captured by global parameters such as genetic covariances between groups used in indicators proposed so far. As the distribution of allele effects along the genome is probably of great importance, new a priori indicators of precision taking such information into account need to be developed.
Author contribution statement
AC perceived the idea of investigating the topic. SR, TMH, LM and AC wrote the manuscript. SR conducted the phenotypic data analyses. SR performed the genomic analyses and wrote the early version of the manuscript. All authors read and reviewed the manuscript.
References
Albrecht T, Auinger H-J, Wimmer V, Ogutu JO, Knaak C, Ouzunova M, Piepho H-P, Schön C-C (2014) Genome-based prediction of maize hybrid performance across genetic groups, testers, locations, and years. Theor Appl Genet 127(6):1375–1386
Alexander D, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Astle W, Balding DJ (2009) Population structure and cryptic relatedness in genetic association studies. Stat Sci 24(4):451–471
Brard S, Ricard A (2015) Is the use of formulae a reliable way to predict the accuracy of genomic selection? J Anim Breed Genet 132(3):207–217
Brøndum R, Rius-Vilarrasa E, Strandén I, Su G, Guldbrandtsen B, Fikse W, Lund M (2011) Reliabilities of genomic prediction using combined reference data of the Nordic Red dairy cattle populations. J Dairy Sci 94:4700–4707
Browning B, Browning S (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223
Butler DG, Cullis BR, Gilmour AR, Gogel BJ (2009) ASReml-R reference manual
Carillier C, Larroque H, Robert-Granié C (2014) Comparison of joint versus purebred genomic evaluation in the french multi-breed dairy goat population. Genet Select Evol 46(1):67
Daetwyler HD, Villanueva B, Woolliams JA (2008) Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS ONE 3(10):1–8
de Roos APW, Hayes BJ, Goddard ME (2009) Reliability of genomic predictions across multiple populations. Genetics 183(4):1545–1553
Duhnen A, Gras A, Teyssèdre S, Romestant M, Claustres B, Daydé J, Mangin B (2017) Genomic selection for yield and seed protein content in soybean: a study of breeding program data and assessment of prediction accuracy. Crop Sci 57:1–13
Elsen J-M (2016) Approximated prediction of genomic selection accuracy when reference and candidate populations are related. Genet Select Evol 48(1):18
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6(5):1–10
Endelman JB, Atlin GN, Beyene Y, Semagn K, Zhang X, Sorrells ME, Jannink J (2014) Optimal design of preliminary yield trials with genome-wide markers. Crop Sci 54:48–59
Erbe M, Gredler B, Seefried FR, Bapst B, Simianer H (2013) A function accounting for training set size and marker density to model the average accuracy of genomic prediction. PLoS ONE 8(12):1–11
Esfandyari H, Sørensen AC, Bijma P (2015) A crossbred reference population can improve the response to genomic selection for crossbred performance. Genet Select Evol 47(1):76
Ganal MW, Durstewitz G, Polley A, Bérard A, Buckler ES, Charcosset A, Clarke JD, Graner E-M, Hansen M, Joets J, Le Paslier M-C, McMullen MD, Montalent P, Rose M, Schön C-C, Sun Q, Walter H, Martin OC, Falque M (2011) A large maize (zea mays l.) snp genotyping array: development and germplasm genotyping, and genetic mapping to compare with the b73 reference genome. PLoS ONE 6(12):1–15
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) Tassel-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9(2):1–11
Goddard M, Hayes B, Meuwissen T (2011) Using the genomic relationship matrix to predict the accuracy of genomic selection. J Anim Breed Genet 128(6):409–421
Guo Z, Tucker DM, Basten CJ, Gandhi H, Ersoz E, Guo B, Xu Z, Wang D, Gay G (2014) The impact of population structure on genomic prediction in stratified populations. Theor Appl Genet 127(3):749–762
Hayes BJ, Bowman PJ, Chamberlain AC, Verbyla K, Goddard ME (2009) Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet Select Evol 41(1):51
Heslot N, Yang H, Sorrells ME, Jannink J (2012) Genomic selection in plant breeding: a comparison of models. Crop Sci 52:146–160
Isidro J, Jannink J-L, Akdemir D, Poland J, Heslot N, Sorrells ME (2015) Training set optimization under population structure in genomic selection. Theor Appl Genet 128(1):145–158
Karoui S, Carabaño MJ, Díaz C, Legarra A (2012) Joint genomic evaluation of French dairy cattle breeds using multiple-trait models. Genet Select Evol 44(1):39
Lehermeier C, Schön C-C, de los Campos G (2015) Assessment of genetic heterogeneity in structured plant populations using multivariate whole-genome regression models. Genetics 201(1):323–337
Makgahlela M, Mäntysaari E, Strandén I, Koivula M, Nielsen U, Sillanpää M, Juga J (2013) Across breed multi-trait random regression genomic predictions in the nordic red dairy cattle. J Anim Breed Genet 130(1):10–19
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157(4):1819–1829
Millet EJ, Welcker C, Kruijer W, Negro S, Coupel-Ledru A, Nicolas SD, Laborde J, Bauland C, Praud S, Ranc N, Presterl T, Tuberosa R, Bedo Z, Draye X, Usadel B, Charcosset A, Van Eeuwijk F, Tardieu F (2016) Genome-wide analysis of yield in Europe: allelic effects vary with drought and heat scenarios. Plant Physiol 172(2):749–764
Olson KM, Van Raden PM, Tooker ME (2012) Multibreed genomic evaluations using purebred Holsteins, Jerseys, and Brown Swiss. J Dairy Sci 95(9):5378–5383
Plieschke L, Edel C, Pimentel EC, Emmerling R, Bennewitz J, Götz K-U (2015) A simple method to separate base population and segregation effects in genomic relationship matrices. Genet Select Evol 47(1):53
Pszczola M, Strabel T, Mulder H, Calus M (2012) Reliability of direct genomic values for animals with different relationships within and to the reference population. J Dairy Sci 95(1):389–400
Rabier C-E, Barre P, Asp T, Charmet G, Mangin B (2016) On the accuracy of genomic selection. PLoS ONE 11(6):1–23
Rincent R, Charcosset A, Moreau L (2017) Predicting genomic selection efficiency to optimize calibration set and to assess prediction accuracy in highly structured populations. Theor Appl Genet 130(11):2231–2247
Rincent R, Laloë D, Nicolas S, Altmann T, Brunel D, Revilla P, Rodríguez V, Moreno-Gonzalez J, Melchinger A, Bauer E, Schoen C-C, Meyer N, Giauffret C, Bauland C, Jamin P, Laborde J, Monod H, Flament P, Charcosset A, Moreau L (2012) Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (zea mays l.). Genetics 192(2):715–728
Rincent R, Nicolas S, Bouchet S, Altmann T, Brunel D, Revilla P, Malvar RA, Moreno-Gonzalez J, Campo L, Melchinger AE, Schipprack W, Bauer E, Schoen C-C, Meyer N, Ouzunova M, Dubreuil P, Giauffret C, Madur D, Combes V, Dumas F, Bauland C, Jamin P, Laborde J, Flament P, Moreau L, Charcosset A (2014) Dent and flint maize diversity panels reveal important genetic potential for increasing biomass production. Theor Appl Genet 127(11):2313–2331
Schopp P, Müller D, Wientjes YCJ, Melchinger AE (2017) Genomic prediction within and across biparental families: means and variances of prediction accuracy and usefulness of deterministic equations. G3 Genes Genom Genet 7(11):3571–3586
Strandén I, Mäntysaari EA (2013) Use of random regression model as an alternative for multibreed relationship matrix. J Anim Breed Genet 130(1):4–9
Technow F, Burger A, Melchinger A E (2013) Genomic prediction of northern corn leaf blight resistance in maize with combined or separated training sets for heterotic groups. Genes–Genomes–Genetics 3(2):197–203
Thornton T, Tang H, Thomas J, Heather M, Bette J, Risch N (2012) Estimating kinship in admixed populations. Am J Hum Genet 91:122–138
Toosi A, Fernando R, Dekkers J (2013) Genomic selection in admixed and crossbred populations. J Anim Sci 130(1):10–19
Unterseer S, Bauer E, Haberer G, Seidel M, Knaak C, Ouzunova M, Meitinger T, Strom TM, Fries R, Pausch H, Bertani C, Davassi A, Mayer KF, Schön C-C (2014) A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genomics 15(1):823
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91(11):4414–4423
Wientjes YC, Veerkamp RF, Bijma P, Bovenhuis H, Schrooten C, Calus MP (2015a) Empirical and deterministic accuracies of across-population genomic prediction. Genet Select Evol 47(1):5
Wientjes YC, Veerkamp RF, Calus MP (2015b) Using selection index theory to estimate consistency of multi-locus linkage disequilibrium across populations. BMC Genet 16(1):87
Wientjes YCJ, Bijma P, Vandenplas J, Calus MPL (2017) Multi-population genomic relationships for estimating current genetic variances within and genetic correlations between populations. Genetics 207(2):503–515
Wientjes YCJ, Bijma P, Veerkamp RF, Calus MPL (2016) An equation to predict the accuracy of genomic values by combining data from multiple traits, populations, or environments. Genetics 202(2):799–823
Wientjes YCJ, Calus MPL, Hayes BJ, Goddard ME, Hayes BJ (2015c) Impact of QTL properties on the accuracy of multi-breed genomic prediction. Genet Select Evol 47(1):42
Acknowledgements
This research was supported by the “Investissement d’Avenir” project “Amaizing”. S. Rio is jointly funded by the program AdmixSel of the INRA metaprogram SelGen and by the breeding companies partners of the Amaizing project: Caussade-Semences, Euralis, KWS, Limagrain, Maisadour, RAGT and Syngenta. We thank Valerie Combes, Delphine Madur and Stephane Nicolas for DNA extraction, analysis and assembly of genotypic data. We thank Cyril Bauland and Carine Palaffre (INRA Saint-Martin de Hinx) for the panel assembly and the coordination of seed production, all breeding companies partners of the Amaizing project and Biogemma for field trials and Pierre Dubreuil (Biogemma) for the assembly and the analysis of phenotypic data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments comply with the current laws of the countries in which the experiments were performed.
Additional information
Communicated by Benjamin Stich.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rio, S., Mary-Huard, T., Moreau, L. et al. Genomic selection efficiency and a priori estimation of accuracy in a structured dent maize panel. Theor Appl Genet 132, 81–96 (2019). https://doi.org/10.1007/s00122-018-3196-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-018-3196-1