Abstract
Key message
A diverse collection of barley lines was phenotyped with three North American Pyrenophora teres f. teres isolates and association analyses detected 78 significant marker-trait associations at 16 genomic loci.
Abstract
Pyrenophora teres f. teres is a necrotrophic fungal pathogen and the causal agent of the economically important foliar disease net form net blotch (NFNB) of barley. The deployment of effective and durable resistance against P. teres f. teres has been hindered by the complexity of quantitative resistance and susceptibility. Several bi-parental mapping populations have been used to identify QTL associated with NFNB disease on all seven barley chromosomes. Here, we report the first genome-wide association study (GWAS) to detect marker-trait associations for resistance or susceptibility to P. teres f. teres. Geographically diverse barley genotypes from a world barley core collection (957) were genotyped with the Illumina barley iSelect chip and phenotyped with three P. teres f. teres isolates collected in two geographical regions of the USA (15A, 6A and LDNH04Ptt19). The best of nine regression models tested were identified for each isolate and used for association analysis resulting in the identification of 78 significant marker-trait associations (MTA; −log10p value >3.0). The MTA identified corresponded to 16 unique genomic loci as determined by analysis of local linkage disequilibrium between markers that did not meet a correlation threshold of R 2 ≥ 0.1, indicating that the markers represented distinct loci. Five loci identified represent novel QTL and were designated QRptts-3HL, QRptts-4HS, QRptts-5HL.1, QRptts-5HL.2, and QRptts-7HL.1. In addition, 55 of the barley lines examined exhibited a high level of resistance to all three isolates and the SNP markers identified will provide useful genetic resources for barley breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Net blotch is a damaging foliar disease of barley (Hordeum vulgare) and infects the crop in most major production regions across the world. Caused by the necrotrophic fungal pathogen Pyrenophora teres, net blotch impacts the supply of quality barley for the malting industry due to its impact on yield and malting quality (Murray and Brennan 2010). Two forms of the pathogen exist, including Pyrenophora teres f. teres and Pyrenophora teres f. maculata, causal agents of net form net blotch (NFNB) and spot form net blotch (SFNB), respectively. The two forms are morphologically identical but can be differentiated by the unique symptoms they elicit on the host (Smedegård-Petersen 1971; McLean et al. 2009). In addition, phylogenetic analysis using mating-type (MAT) gene sequence from net form and spot form isolates revealed a distinct genetic isolation (Rau et al. 2007). Several management strategies are recommended to combat this disease, including the destruction of crop residue and crop rotation to reduce the presence of primary inoculum, as well as fungicide applications. However, the deployment of genetic resistance is considered the most desirable means of disease management (Mathre 1997).
Sexual populations of P. teres f. teres occur worldwide and contribute to a highly variable and diverse pathogen population (Peever and Milgroom 1994; Jonsson et al. 2000; Rau et al. 2005). A recent diversity survey utilized 75 isolates collected from North Dakota, assayed on 22 differential lines, identifying 49 different pathotypes. In addition, molecular analysis using simple-sequence repeat (SSR) markers showed 40 unique genotypes and the MAT genes were present in statistically equal proportions, indicative of a diverse sexual population in part due to the occurrence of random mating (Liu et al. 2012). Tekauz (1990) had previously sampled 182 P. teres f. teres isolates in another North American population study to investigate the diversity in western Canada. Inoculation of the 182 isolates onto 12 barley differentials revealed 45 distinct pathotypes. More recently, Akhavan et al. (2016) analyzed the virulence of 39 P. teres f. teres isolates collected from western Canada using nine differential lines and identified 16 unique pathotypes. In addition, an apparent shift in virulence in P. teres f. teres populations in this region has occurred as evidenced by the appearance of seven novel pathotypes and the absence of three pathotype groups identified by Tekauz (1990). However, because of the low number of isolates utilized in this study, it is probable that these pathotypes could still be present in the population (Akhavan et al. 2016). These population studies of North American P. teres f. teres populations show the diversity available and how dynamic the populations can be, given the ability to recombine to form new combinations of virulence genes coupled with the selection pressure exerted by popular local barley varieties. This pathogen has the high propensity to rapidly adapt its virulence repertoire to overcome resistances present in popular barley cultivars being grown locally (Liu et al. 2012). The variable nature of the pathogen indicates that a thorough understanding of the virulence present in the population and multiple sources of resistance or lack of susceptibility need to be deployed to efficiently control this disease.
Traditional linkage mapping methods utilizing recombinant inbred lines (RILs) or doubled haploid (DH) populations have been widely used to identify quantitative trait loci (QTL) providing resistance or susceptibility to P. teres f. teres at seedling and adult plant stages. Seedling resistance QTL have been identified on all seven barley chromosomes (reviewed by Liu et al. 2011), including the detection of a common major QTL on or near the centromere of barley chromosome 6H. (Steffenson et al. 1996; Graner et al. 1996; Richter et al. 1998; Raman et al. 2003; Cakir et al. 2003; Ma et al. 2004; Emebiri et al. 2005; Yun et al. 2005; Manninen et al. 2000, 2006; Friesen et al. 2006; Grewal et al. 2008; Abu Qamar et al. 2008, St. Pierre et al. 2010). In addition, this pathosystem appears remarkably complex, as resistance has been observed to be dominant, incomplete, and recessive suggesting different mechanisms of host-parasite interaction leading to compatibility (susceptibility) or incompatibility (resistance).
The use of bi-parental mapping populations for the identification of QTL requires the time-consuming task of developing mapping populations and only captures the diversity stemming from the two founding parents, but does have the benefit of detecting rare alleles with functional polymorphisms between the two parents (Zhu et al. 2008; Rafalski 2010). Association mapping (AM), or linkage disequilibrium (LD) mapping, provides an alternative to traditional linkage mapping through the use of diverse panels of host genotypes to identify marker-trait associations (MTAs). The use of diverse germplasm collections instead of bi-parental populations allows for the exploitation of ancestral recombination events occurring over the plants evolutionary history (Zhu et al. 2008). Association mapping has several advantages, including the ability to detect QTL at a higher resolution, less time required to conduct the genetic analyses, and the potential to identify a greater number of loci associated with the phenotype (Yu et al. 2006; Zhu et al. 2008). Successful association mapping requires the selection of a correct statistical model that suitably controls the detection of false-positive associations. Several methods have been described to control this type I error rate, such as the inclusion of population structure (Zhao et al. 2007) and familial relatedness (Yu et al. 2006). In addition, methods, such as the compressed mixed linear model (CMLM) (Zhang et al. 2010) and the enriched compressed mixed linear model (ECMLM), utilize a clustering of individuals to create a compressed kinship matrix (Li et al. 2014). These methods promise an increase in statistical power as well as a decrease in computation time.
Association mapping has emerged as a powerful tool to identify loci controlling quantitative traits in various economically important crop species. Using a nested association mapping (NAM) population, loci associated with complex traits including leaf architecture and southern leaf blight resistance have been identified throughout the maize genome (Tian et al. 2011; Kump et al. 2011). In addition, Huang et al. (2010) used 517 rice land races with highly saturated genotypic data of ~3.6 million single nucleotide polymorphisms (SNPs) to successfully identify genomic regions associated with 14 different genetically complex agronomic traits. The use of genome-wide association studies (GWAS) has also been applied to barley, with significant loci being identified for association with resistance to Fusarium head blight, stem rust, spot blotch, and leaf rust (Massman et al. 2011; Zhou et al. 2014; Zhou and Steffenson 2013; Ziems et al. 2014). In addition, a diverse panel of barley accessions was evaluated for disease reaction to P. teres f. maculata (Neupane et al. 2015) and the GWAS identified 21 novel loci associated with resistance or susceptibility to P. teres f. maculata, as well as six previously identified QTL (Tamang et al. 2015).
This study utilized a GWAS approach to identify loci associated with disease resistance or susceptibility to North American isolates of the barley pathogen P. teres f. teres. The utilization of the world barley core collection allowed for the identification of MTA and the underlying SNP markers that can be utilized by barley breeders to incorporate resistances or eliminate susceptibility from elite barley germplasm providing more tools and knowledge to begin effectively combating this economically important yet shifty and dynamic pathogen.
Materials and methods
Biological materials
A total of 1050 accessions from the globally diverse barley core collection, including cultivars, breeding lines, landraces, and genetic stocks, were acquired from the National Small Grain Collection, Aberdeen, Idaho (Muñoz-Amatriaín et al. 2014). Three North American P. teres f. teres isolates (15A, 6A and LDNH04Ptt19) were used to phenotype the barley core collection accessions. P. teres f. teres isolate 15A was collected in Fresno County, California (Wu et al. 2003), isolate 6A was collected from Fresno County, California (Steffenson and Webster 1992), and isolate LDNH04Ptt19 (hereafter referred to as LDN) was collected in North Dakota. These isolates were chosen for use in a GWAS due to geographical diversity within the United States (California and North Dakota), differential virulence on a NFNB differential set (data not shown), and previous use in genetic mapping studies using bi-parental populations (Abu Qamar et al. 2008; Liu et al. 2015).
Phenotyping
Barley accessions were arranged into blocks consisting of 60 lines per block. Three seeds per accession were planted in cones, placed in a rack bordered with the susceptible barley cultivar Tradition, and grown under greenhouse conditions until the 2–3 leaf stage, approximately 14 days. Inoculations were performed as described in Friesen et al. (2006). Disease reactions were evaluated 7 day post-inoculation using a 1–10 rating scale (Tekauz 1985). A total of three replicates were completed for each isolate per block. The average disease score of the three replicates was used in the association analyses.
Genotyping
A total of 2417 barley lines, consisting of landraces, breeding lines, and cultivars from the National Small Grains Collection, were genotyped utilizing the 9k Illumina Infinium iSELECT assay (Muñoz-Amatriaín et al. 2014). Genotypic data for 998 accessions, which were phenotyped with the aforementioned Ptt isolates, with less than 30% missing data were downloaded from The Triticeae Toolbox (T3) barley database (https://triticeaetoolbox.org/barley/). In addition, genetic positions of the SNP markers on the iSelect consensus map were used for this analysis (Muñoz-Amatriaín et al. 2014). Due to some markers not being anchored to the iSelect consensus map, sequences of significant SNP markers were used in BLAST searches of the barley genome to identify genetic positions based on population sequencing (POPSEQ) from which the relative marker position could be estimated (IBSC 2012; Mascher et al. 2013).
Imputation, allele similarity, and linkage disequilibrium
As heterozygous genotypes were present at extremely low levels (~0.06%), and due to the subjectivity of calling heterozygotes from the Infinium iSelect assay, these genotypes were recoded as missing data and missing genotypic data were imputed in fastPhase 1.3 (Scheet and Stephens 2006) with default settings. An allele similarity matrix was calculated in JMP Genomics v6.1 (SAS Institute Inc.) using the imputed genotypic data to eliminate any redundant barley genotypes. Linkage disequilibrium was calculated as squared allele frequency correlations (R 2) between all intrachromosomal marker pairs in JMP Genomics v6.1.
Population structure
The software STRUCTURE v.2.3.4 (Pritchard et al. 2000) was used to determine the population structure of the selected core collection lines and assign membership to subpopulations. Using genetic positions derived from the barley iSelect consensus map (Muñoz-Amatriaín et al. 2014), a single marker was selected from each locus, resulting in 1744 markers to be used in STRUCTURE analysis. Initially, an admixture model was used with a burn-in of 10,000, followed by 25,000 Monte Carlo Markov Chain (MCMC) replications for k = 1 to k = 10 with five iterations. Δk method (Evanno et al. 2005) was used in STRUCTURE HARVESTER (Earl and von Holdt 2012) to identify the ideal level of subpopulations. Following the identification of the optimum k value, individuals were placed into subpopulations by running a new STRUCTURE analysis with a burn-in of 100,000 and 100,000 MCMC iterations at the optimum k value. An individual was considered to be a member of a subpopulation if its membership probability was greater than 0.80. STRUCTURE output was then used as a covariate (Q) in several association models. Principal components analysis (PCA) was conducted in the R package Genome Association and Prediction Integrated Tool (GAPIT) (Lipka et al. 2012; Tang et al. 2016) and TASSEL 5.0 (Bradbury et al. 2007) with default settings. Principal components explaining at least 25% were used as a covariate in association analyses (PC25). A kinship matrix (K) was constructed using the Loiselle algorithm (Loiselle et al. 1995) in GAPIT by setting the ‘group.from’ and ‘group.to’ parameters equal to the population size and the ‘group.by’ parameter equal to one. In addition, using the enriched compressed mixed linear model (ECMLM) proposed by Li et al. (2014), an optimal compressed kinship matrix (K comp) was calculated in GAPIT using the Loiselle algorithm and the parameters of ‘group.from = 300’, ‘group.to = 957’, and ‘group.by = 50’. Kinship clustering methods tested were average, ward, and complete. Kinship group summary methods used were mean and maximum.
Marker-trait association models
Nine models were tested for the identification of MTAs and conducted in TASSEL 5.0 and GAPIT (Bradbury et al. 2007; Lipka et al. 2012; Tang et al. 2016). A naïve model utilizing only genotypic and phenotypic data and not accounting for population structure or relatedness was conducted using the general linear model (GLM) procedure in TASSEL 5.0. Correcting for population structure, an additional model using population structure (Q) as a fixed effect was also analyzed using the GLM procedure in TASSEL 5.0. In addition, in a similar manner, another model accounting for population structure using the first three principal components (PC25) was analyzed. A fourth model, accounting for relatedness, utilized the kinship matrix (K) as a random effect in a mixed linear model (MLM) using GAPIT. Another model accounting for relatedness used the compressed kinship matrix (K comp) as a random effect in an MLM. Finally, four additional models accounting for population structure and kinship (Q + K, PCA25 + K, Q + K comp, and PC25 + K comp) were analyzed as an MLM with Q and PC25 as fixed effects and kinship as a random effect in GAPIT. Heritability for each trait was also calculated in GAPIT. The mean-squared deviation (MSD) was calculated for each model (Mamidi et al. 2011), and the model with the lowest MSD value was selected for further analysis. Marker-trait associations with a p value <0.001 were declared significant. In addition, p values from the optimal model for each trait were adjusted using a false discovery rate (FDR) multiple testing correction procedure in GAPIT and markers with a p value <0.1 were considered highly significant. Manhattan plots were generated in the R package ‘qqman’ (Turner 2014).
QTL identification
The genetic positions of the significant markers detected were determined via BLAST searches of the barley genome (http://webblast.ipk-gatersleben.de/barley/viroblast.php) to anchor them to a POPSEQ position (Mascher et al. 2013). Loci were initially declared distinct if separated by more than 5 cM. As LD can be variable throughout the genome, to ensure that loci were indeed distinct, local LD was examined between the most significant markers at each intrachromosomal locus. If R 2 values between significant marker pairs were less than 0.10, the loci were considered distinct.
Results
Phenotypic analysis
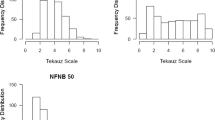
A total of 1050 barley accessions from the barley core collection (Muñoz-Amatriaín et al. 2014) were evaluated for disease reaction to three North American P. teres f. teres isolates (15A, 6A, and LDN) (Online Resource 1). Average disease reaction scores for isolate 15A ranged from highly resistant 1 to highly susceptible 9.67 with an overall mean of 4.40 (Online Resource 2). Similarly, average disease reaction scores for isolate 6A ranged from 1 to 9.33 with an overall mean of 4.10 (Online Resource 3). Average disease reaction scores for isolate LDN ranged from 1 to 9.67 with an overall mean of 5.40 (Online Resource 4). A total of 55 barley accessions were highly resistant to all three isolates with an average disease reaction score of less than or equal to 2.5 for each isolate. Examination of the alleles of the highly resistant lines for 19 significant markers revealed 16 markers with a predominant allele being present in over 75% of the highly resistant lines (Online Resource 5). Characteristics, such as geographic origin, row type, and growth habit of 52 highly resistant lines, which have available genotypic data, are available in Online Resource 6.
Marker properties
Among the 1050 barley lines that were phenotyped with P. teres f. teres, 998 accessions had genotypic data that met our quality thresholds. This resulted in a total of 6525 high-quality SNPs with a minimum minor allele frequency (MAF) ≥5% and missing data per individual marker ≤50% to be used in association analyses. In addition, allele similarity analysis eliminated 41 lines from the association panel due to genotypic redundancy, resulting in a total of 957 barley accessions used in subsequent analyses.
Linkage disequilibrium, PCA, and structure
PCA identified three principal components that accounted for at least 25% of the cumulative variation, with the first three principal components accounting for approximately 14, 7, and 5%, respectively (Online Resource 7). Initial STRUCTURE analysis indicated an optimal k value of 2, with 234 and 552 individuals belonging to subpopulation one and two, respectively (Online Resource 8). The remaining 171 lines had membership probabilities of less than 0.80 to either of the populations and are considered admixture. Visualization of the results of PCA (Online Resource 7) and STRUCTURE analysis (Online Resource 9) reveals a similar pattern of barley lines being clustered into groups corresponding to 2- or 6-row classes. Population one from the STRUCTURE analysis corresponded to two-row barley lines and population two represented six-row barley lines.
Association mapping
A total of nine models were used for association analyses, and the optimal model was chosen by the calculation of the MSD. Very small differences were observed in the MSD values for models incorporating a kinship matrix, indicating that the inclusion of relatedness in association analyses appropriately corrected for the possibility of spurious associations. Results from models, including kinship, were very similar, but the model with the lowest MSD was chosen for further analysis (Table 1).
Isolate 15A
The best model for association tests using average disease scores for isolate 15A only included an uncompressed kinship matrix (K) and had an MSD of 0.00006 (Table 1). Heritability was estimated to be 0.717. A total of 41 markers with MAF ranging from 0.054 to 0.489 were significantly associated with average disease score at the threshold of p < 0.001 (Table 2). The –log10(p) of the significant markers ranged from 3.00 to 8.80 and explained between 0.73–2.51% of the phenotypic variation (R 2) per marker (Table 2). Significant markers were located on five barley chromosomes corresponding to eight unique loci. Two markers were located on 2H, two on 3H, one on 4H, 35 on 6H, and one on 7H (Table 2). The unique genomic regions identified include two on 2H (120.04 and 125.35 cM), two on 3H (47.1 and 150 cM), one on 4H (1.13 cM), two on 6H (48.94–49.79 cM, 53.6–55.52 cM), and one on 7H (131.02 cM) (Table 2). BLAST searches of the barley genome with marker SCRI_RS_188420 did not result in the anchoring to a POPSEQ genetic position; however, its iSelect consensus map position places it near the telomere of the long arm of chromosome 3H at ~150 cM. In addition, three markers on chromosome 6H (11_20936, SCRI_RS_111556, and SCRI_RS_111820) were unable to be anchored to a POPSEQ position via BLAST searches of the barley genome; however, comparison of their respective consensus map positions and the POPSEQ positions of other markers at a similar consensus map locus allowed for the inclusion of them into the corresponding unique loci. Marker 11_20936 was placed in the 6H 48.94–49.79 cM bin and markers, SCRI_RS_111556 and SCRI_RS_111820 were placed into the 6H 53.6–55.52 cM bin.
Isolate 6A
An association model incorporating both population structure (Q) and a compressed kinship matrix (K comp) was found to be the optimal model with an MSD of 0.00008 (Table 1). Heritability of this trait was estimated to be 0.709. A total of 24 significant markers were detected with –log10(p) values ranging from 3.00 to 8.70 and MAF ranging from 0.08 to 0.47. Phenotypic variation attributed to each marker (R 2) ranged from 0.55 to 1.85%. Significant markers were located on four barley chromosomes corresponding to seven unique loci. Two markers were located on chromosome 4H, two on 5H, 17 on 6H, and three on 7H (Table 3). The distinct genomic regions detected include one on 4H (52.69 cM), one on 5H (93.4 cM), three on 6H (49.79 cM, 54.82–55.38 cM, and 59.92 cM), and two on 7H (70.54 and 131.02 cM). In addition, a marker (SCRI_RS_158011) was unmapped in the iSelect consensus map and was assigned to chromosome 6H through BLAST searches of the barley genome; however, no specific genetic locus was obtained through this method. BLAST searches of an additional marker (11_21310) from chromosome 6H did not yield POPSEQ positions; however, due to its consensus map position in comparison with nearby markers, it was placed into the chromosome 6H 59.92–60.48 cM bin. Marker SCRI_RS_4717 was unmapped on the iSelect consensus map; however, BLAST searches of the barley genome anchored it to the short arm of chromosome 5H.
Isolate LDN
A model, including the first three principal components explaining approximately 25% of the cumulative variation (PC25) and an uncompressed kinship matrix (K), was selected as the best model for the isolate LDN with an MSD of 0.00001 (Table 1). Heritability was estimated to be 0.732 for this trait. A total of 23 significant markers were identified with seven markers located on chromosome 3H, one on 4H, one on 5H, 13 on 6H, and one on 7H. The –log10(p) of the significant markers ranged from 3.03 to 8.22 and had an MAF ranging from 0.15 to 0.49. Phenotypic variation (R 2) of individual markers varied from 0.79 to 2.48% (Table 4). A total of nine distinct genomic loci were detected, including three on 3H (2.41–2.69 cM, 46.03 cM, and 83.99 cM), one on 4H (52.69 cM), one on 5H (69.31 cM), three on 6H (49.08–49.22 cM, 53.60–55.38 cM, 65.86 cM), and one on 7H (131.02 cM). In addition, one marker (SCRI_RS_182648) was not anchored on the iSelect consensus map, but was assigned to chromosome 6HL via BLAST searches of the barley genome.
Discussion
Net form net blotch of barley has the potential to cause severe economic impacts in all barley production regions of the world. Thus, the identification of barley resistance/susceptibility QTL that interact with the effectors present within the highly diverse pathogen populations is important to effectively deploy durable genetic resistance against this adaptable enemy. Regions of the barley genome associated with resistance or susceptibility to P. teres f. teres have previously been identified through traditional QTL analysis methods utilizing bi-parental populations (reviewed by Liu et al. 2011). Although this method provides a powerful genetic approach to characterize these complex interactions, it does have limitations that can be overcome using the GWAS approach. These limitations include the time required to develop bi-parental populations and the ability to select parents representing the greatest amount of functional polymorphism. The amount of functional polymorphism captured by each host population only represents a very small proportion available in the primary barley germplasm pool because of the high variability of effectors in pathogen populations and the diverse interacting host resistance or necrotrophic effector susceptibility targets. Here, we utilize GWAS as an alternative to identify NFNB resistance/susceptibility QTL. Using a diverse panel of barley germplasm, GWAS identified 16 distinct loci associated with disease reaction to three North American P. teres f. teres isolates.
Although GWAS is a very powerful approach, it also has its own limitations. A major concern when conducting GWAS is the detection of false associations, but the error can be reduced by accounting for population structure and kinship in the linear models. A total of nine models were tested in this study, utilizing population structure derived from Bayesian clustering in STRUCTURE (Q) (Pritchard et al. 2000) and the use of PCs accounting for at least 25% of cumulative variation (PC25), as well as accounting for familial relatedness using a kinship matrix produced using the Loiselle algorithm (K and K comp) (Loiselle et al. 1995). In addition, both uncompressed and compressed kinship matrices were used to evaluate any differences in the reduction of spurious associations or an increase in statistical power (Zhang et al. 2010; Li et al. 2014). Overall, the performance of models that included kinship (K or K comp) was very similar, regardless of the inclusion of population structure. This indicated that using relatedness was sufficient for the control of false-positive MTA and that using population structure alone was not stringent enough in the diversity panel used.
The lowest MSD value was used as the model selection criteria and selected a different model for each isolate (Table 1). GWAS using the best model for each isolate revealed 16 unique genomic regions represented by a total of 78 significant markers. Following FDR p value adjustment, seven loci did not meet the adjusted p value threshold of <0.10. However, further examination reveals that the FDR adjustment may be too stringent for these analyses. One of the loci on chromosome 4H fell below the significance threshold after FDR adjustment for isolate LDN but remained significant for isolate 6A and this QTL region had been previously identified via bi-parental mapping (Steffenson et al. 1996; Grewal et al. 2008). In addition, a locus on chromosome 7H, which was common to all three isolates and represented the previously identified locus QTLUHs-7H in bi-parental population analyses (König et al. 2014), also did not meet the adjusted FDR p value criteria. In addition, a distinct locus near the commonly mapped chromosome 6H centromere (Steffenson et al. 1996) did not meet the adjusted p value threshold for isolates 6A and LDN, but remained significant for isolate 15A. The data suggest that these are not false associations due to the fact that loci appeared significant at unadjusted p values of <0.001 between more than a single isolate and corresponded to QTL previously mapped in bi-parental populations, thus, indicating that FDR adjustment may be too stringent at some loci resulting in the loss of true positive associations. Therefore, the unadjusted p values were used as the determinant of significance for the association analyses.
Two markers on the long arm of chromosome 2H, 12_30690, and 12_10579, were found to be significantly associated with disease reaction to P. teres f. teres isolate 15A and are approximately 5.31 cM apart based on genetic anchoring via POPSEQ (Mascher et al. 2013). Analysis of local LD between these two markers revealed a very low level of correlation with an R 2 value of 0.004, indicating that these two markers may represent two distinct loci and that significance did not arise due to tight linkage. Previously, Raman et al. (2003) identified a QTL on the long arm of chromosome 2H designated QRpts2L which accounted for approximately 7% of the phenotypic variation in a bi-parental population. Due to the low resolution of the mapping in the Raman and colleagues study and the use of AFLP and RFLP markers, it was not possible to determine a POPSEQ position for the QTL, and they reported with any certainty. Thus, one or both of the MTA detected here may be linked to the resistance gene/genes underling the previously identified QRpts2L QTL, and thus, we have designated these loci, QRpts2L.1 and QRpts2L.2.
A total of four distinct loci were detected on chromosome 3H with two corresponding to disease reaction to either P. teres f. teres isolate LDN or isolate 15A. A strongly associated locus comprised of five significant markers was identified at approximately 2.41–2.69 cM. A QTL was previously identified at this locus in a Hector × NDB112 RIL population explaining approximately 20% of the phenotypic variation (Liu et al. 2015). In addition, the isolate used to detect this QTL in the Hector × NDB112 population was collected from the same region as LDN, indicating that they likely possess the same necrotrophic effector which interacts with a host gene at this locus. An additional locus was identified that was unique to isolate LDN at approximately 83.99 cM on chromosome 3H. QTL had been previously detected on chromosome 3H at approximately 75 cM (QRpts3La) and 89 cM with R 2 values of 0.16 and 0.07, respectively (Raman et al. 2003; Liu et al. 2015). Since the locus detected in our association analysis falls within this region, it may be the same as those previously identified (Fig. 1). The analysis conducted using isolate 15A revealed a unique locus near the telomere of the long arm of chromosome 3H designated QRptts-3HL (Fig. 1). As the strongest associated marker (SCRI_RS_188420) at this locus was not anchored to a POPSEQ position, markers flanking this position (11_10935 and SCRI_RS_126369) from the iSelect consensus map were used in BLAST searches to estimate its position placing it in the approximate interval of 142.2–143.13 cM. The nearest QTL previously identified on the long arm of chromosome 3H are located at approximately 109 and 114 cM (Liu et al. 2015; Raman et al. 2003). This indicates that the significant association detected in this study represents a novel locus linked to disease reaction to isolate 15A and was given the designation QRptts-3HL (Fig. 1). A fourth locus was identified near the centromere on chromosome 3H in the interval from 46.03 to 47.01 cM and was common to both isolates LDN and 15A. A study had previously mapped a QTL designated Rpt-3H-4, to a similar locus (~46 cM) near the centromere of chromosome 3H with R 2 value of 0.12, indicating that this associated locus is likely the same QTL as Rpt-3H-4 (Fig. 1: Yun et al. 2005).
Association mapping analyses of disease reaction to P. teres f. teres isolates 15A (California), LDN (North Dakota), and 6A (California). Barley chromosomes are listed on the x-axis. A –log10(p) scale of significance is represented on the y-axis with the red horizontal line representing the significance threshold of –log10(p) = 3. The colored pixels represent individual SNP markers used in the association analyses. Both previously identified and newly designated QTL are listed at the top of the figure. Boxes are drawn around the distinct significant loci detected
A common locus was detected near the centromere of chromosome 4H at POPSEQ position 52.69 cM associated with disease reaction to isolates 6A and LDN. This centromeric locus had been previously identified in the bi-parental population of Steptoe × Morex (Steffenson et al. 1996) as evidenced by BLAST searches of the barley genome using the sequence of the nearby marker, ABG484, of the previously mapped QTL (LOD 11.1 and R 2 = 0.31). In addition, Grewal et al. (2008) identified the same QTL, designated QRpts4 (Fig. 1), in a CDC Dolly × TR251 DH population with an LOD of 4.2 and R 2 of 5%. Interestingly, it was also determined that this QTL was associated with resistance to spot form net blotch and represents a potential source of resistance to two closely related yet distinct pathogens (Tamang et al. 2015). An additional locus was identified near the telomere of the short arm of chromosome 4H associated with isolate 15A at POPSEQ position 1.13 cM designated QRptts-4HS (Fig. 1). No previously identified QTL have been reported near this locus, indicating that it is a novel QTL. Two distinct loci were detected on the long arm of chromosome 5H at POPSEQ positions 69.31 and 93.4 cM associated with disease reaction to isolates LDN and 6A designated QRptts-5HL.1 and QRptts-5HL.2 (Fig. 1), respectively. Previously, no QTL had been detected on the long arm of chromosome 5H, indicating that both these loci are novel. In addition, marker SCRI_RS_4717 was significantly associated with disease reaction to isolate 6A, and was localized to the short arm of chromosome 5H. Although no POPSEQ position could be derived, it is possible that this marker belongs to the locus described by Liu et al. (2015) at the ~45 cM position using the same isolate.
The centromeric region of barley chromosome 6H has long been associated with resistance or susceptibility to NFNB. Various studies have mapped both dominant resistances and dominant susceptibilities to the same region using a diverse array of barley genotypes and pathogen isolates (Steffenson et al. 1996; Graner et al. 1996; Richter et al. 1998; Raman et al. 2003; Cakir et al. 2003; Ma et al. 2004; Emebiri et al. 2005; Yun et al. 2005; Manninen et al. 2000, 2006; Friesen et al. 2006; Grewal et al. 2008; Abu Qamar et al. 2008, St. Pierre et al. 2010). Unsurprisingly, 58 of the total 79 significant markers detected in this study were localized to the centromeric region of chromosome 6H within the interval of ~49–66 cM, with four distinct loci. These loci were detected at the approximate positions of 49, 55, 60, and 66 cM. The ~60 cM locus was specific to isolate 6A, while the ~65 cM locus was only associated with isolate LDN. In addition, the ~66 cM locus was detected by Liu et al. (2015) using a Hector × NDB112 bi-parental population and Japanese isolate JPT9901. This indicates that the North Dakota isolate LDN used in this GWAS and JPT9901, although geographically diverse may possess a common necrotrophic effector targeting a host gene at this locus. Both the ~49 and ~55 cM loci were significantly associated with disease reaction to all three isolates; however, 33 of the 58 significant MTAs at the centromeric 6H region belong to the ~55 cM locus. This indicates that underlying this region is likely a cluster of resistance or susceptibility genes. A recent high-resolution mapping study of the region at POPSEQ position corresponding to ~54 cM localized the dominant susceptibility gene Spt1 to a small ~0.24 cM region (Richards et al. 2016). Yet, surprisingly, it appeared that at least four different virulence genes (Shjerve et al. 2014) targeted a single susceptibility locus at this region suggesting that alleles of the same susceptibility gene are targeted by multiple distinct necrotrophic effectors or a tight cluster of susceptibility genes is present at this locus (Richards et al. 2016).
A common locus near the telomere of chromosome 7HL was detected in all three isolates at ~131 cM (Fig. 1). König et al. (2014) described a minor QTL, QTLUHs-7H, on the short arm of chromosome 7H in the DH population of Uschi × HHOR3073; however, BLAST searches of the sequences of the markers reported in their linkage group on the short arm of 7H are not co-linear with the POPSEQ consensus positions. In addition, marker GBM1464, which is tightly linked to the identified QTL, is positioned at ~130 cM, indicating that the previously detected QTL is likely the same as the MTA detected in this study, however, positioned incorrectly on the previously published map. In addition, a novel locus was detected at ~70 cM on the long arm of chromosome 6H designated QRptts-7HL.1 (Fig. 1) and is associated with disease reaction to isolate 6A.
Several loci, including Qrpts2L.1, Qrpts2L.2, Rpt-3H-4, Qrpts3La, Qrptts-5HL.1, Qrptts-5HL.2, and Qrptts-7HL.1, were supported by a single, significant SNP. This could be explained by a more rapid decay of linkage within these regions. In addition, three of these loci, Qrpts2L.1/Qrpts2L.2, Rpt-3H-4, and Qrpts3La, were previously identified in bi-parental populations and were observed to account for approximately 7, 12, and 16% of the phenotypic variability, respectively. Compared to the commonly identified centromeric 6H locus, where R 2 values have been reported as high as ~65% (Manninen et al. 2000; Cakir et al. 2003), these loci appear to contribute less to host resistance/susceptibility, which may explain the comparatively lower number of SNPs identified in these regions. Although the overall contribution to resistance/susceptibility of these loci harboring fewer associated SNPs may be lower compared to the highly significant centromeric 6H locus, the pyramiding of the resistant alleles of these minor effect QTL may provide effective and durable resistance.
This study reports on the first use of GWAS for the detection of markers significantly associated with disease reactions of barley to the necrotrophic fungal pathogen P. teres f. teres. A total of 78 MTAs were detected, corresponding to 16 unique genomic loci associated with resistance or susceptibility to three North American P. teres f. teres isolates, exemplifying the diversity of interactions occurring and complexity of this pathosystem. The loci identified could harbor either resistances or susceptibility targets, because both dominant gene-for-gene and recessive inverse gene-for-gene types of resistances have been previously described in barley-P. teres f. teres genetic interactions. Haplotype analysis of the significant SNP markers at each QTL of the highly resistant barley lines revealed a predominance of shared genotypes providing a useful resource for marker-assisted selection. The results provide foundational genetic information for the effective deployment of resistance or the elimination of host susceptibility factors from elite barley lines, providing a durable means of management for this economically important disease.
Author contribution statement
JR, TF, and RS designed the experiments. JR and TF performed the experiments. JR, TF, and RS analyzed the data. JR and RS wrote the manuscript.
References
Abu Qamar M, Liu ZH, Faris JD, Chao S, Edwards MC, Lai Z, Frankowiak JD, Friesen TL (2008) A region of barley chromosome 6H harbors multiple major genes associated with the net type net blotch resistance. Theor Appl Genet 117:1261–1270
Akhavan A, Turkington TK, Askarian H, Tekauz A, Xi K, Tucker JR, Kutcher HR, Strelkov SE (2016) Virulence of Pyrenophora teres populations in western Canada. Can J Plant Pathol. doi:10.1080/07060661.2016.1159617
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Cakir M, Gupta S, Platz GJ, Ablett GA, Loughman R, Embiri LC, Poulsen D, Li C, Lance RCM, Galway NW, Jones MGK, Appels R (2003) Mapping and validation of the genes for resistance to Pyrenophora teres f teres in barley (Hordeum vulgare L.). Aust J Agric Res 54:1369–1377
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Emebiri LC, Platz G, Moody DB (2005) Disease resistance genes in a doubled haploid population of two-rowed barley segregating for malting quality attributes. Aust J Agric Res 56(1):49–56
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Friesen TL, Faris JD, Lai Z, Steffenson BJ (2006) Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a barley doubled haploid population. Genome 409:855–859
Graner A, Foroughi-Wehr B, Tekauz A (1996) RFLP mapping of a gene in barley conferring resistance to net blotch (Pyrenophora teres). Euphytica 91:229–234
Grewal TS, Rossnagel BG, Pozniak CJ, Scoles GJ (2008) Mapping quantitative trait loci associated with barley net blotch resistance. Theor Appl Genet 116:529–539
Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang QF, Li J, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nature Genet 42(11):961–967
International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491(7426):711–716
Jonsson R, Sail T, Bryngelsson T (2000) Genetic diversity for random amplified polymorphic DNA (RAPD) markers in two Swedish populations of Pyrenophora teres. Can J Plant Pathol 22(3):258–264
König J, Perovic D, Kopahnke D, Ordon F, Léon J (2014) Mapping seedling resistance to net form of net blotch (f.) in barley using detached leaf assay. Plant Breeding 133(3):356–365
Kump KL, Bradbury PJ, Wisser R, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S, McMullen MD, Ware D, Balint-Kurti PJ, Holland JB (2011) Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet 43:163–168
Li M, Liu X, Bradbury P, Yu J, Zhang Y, Todhunter RJ Buckler ES, Zhang Z (2014) Enrichment of statistical power for genome-wide association studies. BMC Biol 12:73
Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28(18):2397–2399
Liu Z, Ellwood SR, Oliver RP, Friesen TL (2011) Pyrenophora teres: profile of an increasingly damaging barley pathogen. Mol Plant Pathol 12(1):1–19
Liu ZH, Zhong S, Stasko AK, Edwards MC, Friesen TL (2012) Virulence profile and genetic structure of a North Dakota population of Pyrenophora teres f teres, the causal agent of net form net blotch of barley. Phytopathology 102(5):539–546
Liu Z, Holmes DJ, Faris JD, Chao S, Brueggeman RS, Edwards MC, Friesen TL (2015) Necrotrophic effector-triggered susceptibility (NETS) underlies the barley-f. interaction specific to chromosome 6H. Mol Plant Pathol 16(2):188–200
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Ma ZQ, Lalpitan NLV, Steffenson B (2004) QTL mapping of net blotch resistance genes in a doubled-haploid population of six-rowed barley. Euphytica 137:291–296
Mamidi S, Chikara S, Goos JR, Hyten DL, Annam D, Mafi Moghaddam S, Lee RK, Cregan PC, McClean PE (2011) Genome-wide association analysis identifies candidate genes associated with iron deficiency chlorosis in soybean. Plant Genome 4:154–164
Manninen O, Kalendar R, Robinson J, Schulman AH (2000) Application of BARE-1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley. Mol Genet Genom 264:325–334
Manninen OM, Jalli M, Kalendar R, Schulman A, Afanasenko O, Robinson J (2006) Mapping of major spot-type and net-type net-blotch resistance genes in the Ethiopian barley line CI 9819. Genome 49:1564–1571
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Muñoz - Amatriaín M, Close TJ, Wise RP, Schulman AH, Himmelbach A, Mayer KF, Scholz U, Poland JA, Stein N, Waugh R (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76(4):718–727
Massman J, Cooper B, Horsley R, Neate S, Dill-Macky R, Chao S, Dong Y, Schwarz P, Muehlbauer GJ, Smith KP (2011) Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol Breed 27:439–454
Mathre DE (1997) Compendium of barley diseases, 2nd edn. The American Phytopathological Society, St. Paul
McLean MS, Howlett BJ, Hollaway GJ (2009) Epidemiology and control of spot form of net blotch (Pyrenophora teres f maculata) of barley: a review. Crop Pasture Sci 60:303–315
Muñoz-Amatriaín M, Cuesta-Marcos A, Endelman JB, Comadran J, Bonman JM, Bockelman HE, Chao S, Russell J, Waugh R, Hayes PM, Muehlbauer GJ (2014) The USDA barley core collection: genetic diversity, population structure, and potential for genome-wide association studies. PLoS One 94:e94688
Murray GM, Brennan JP (2010) Estimating disease losses to the Australian barley industry. Australas Plant Pathol 39(1):85
Neupane A, Tamang P, Brueggeman RS, Friesen TL (2015) Evaluation of a barley core collection for spot form net blotch reaction reveals distinct genotype-specific pathogen virulence and host susceptibility. Phytopathology 105(4):509–517
Peever TL, Milgroom MG (1994) Genetic structure of Pyrenophora teres populations determined with random amplified polymorphic DNA markers. Can J Bot 72:915–923
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Rafalski JA (2010) Association genetics in crop improvement. Curr Opin Plant Biol 13:174–180
Raman H, Platz GJ, Chalmers KJ, Raman R, Read BJ, Barr AR, Moody DB (2003) Mapping of genetic regions associated with net form of net blotch resistance in barley. Aust J Agric Res 54:1359–1367
Rau D, Maier FJ, Papa R, Brown AHD, Balmas V, Saba E, Schaefer W, Attene G (2005) Isolation and characterization of the mating-type locus of the barley pathogen Pyrenophora teres and frequencies of mating-type idiomorphs within and among fungal populations collected from barley landraces. Genome 48(5):855–869
Rau D, Attene G, Brown AHD, Nanni L, Maier FJ, Balmas V, Saba E, Schafer W, Papa R (2007) Phylogeny and evolution of mating-type genes from Pyrenophora teres, the causal agent of barley “net blotch” disease. Curr Genet 51:377–392
Richards J, Chao S, Friesen T, Brueggeman R (2016) Fine mapping of the barley chromosome 6H net form net blotch susceptibility locus. G3 6(7):1809–1818
Richter K, Schondelmaier J, Jung C (1998) Mapping of quantitative trait loci affecting Drechslera teres resistance in barley with molecular markers. Theor Appl Genet 97(8):1225–1234
Scheet P, Stephens M (2006) A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78(4):629–644
Shjerve RA, Faris JD, Brueggeman RS, Yan C, Zhu Y, Koladia V, Friesen TL (2014) Evaluation of a Pyrenophora teres f. teres mapping population reveals multiple independent interactions with a region of barley chromosome 6H. Fungal Genet Biol 70:104–112
Smedegård-Petersen V (1971) Pyrenophora teres f maculata f nov and Pyrenophora teres f. teres on barley in Denmark. Royal Veterinary and Agricultural University, Copenhagen, pp 124–144
St Pierre S, Gustus C, Steffenson B, Dill-Macky R, Smith KP (2010) Mapping net form net blotch and Septoria speckled leaf blotch resistance loci in barley. Phytopathol 100(1):80–84
Steffenson BJ. Webster RK (1992) Quantitative resistance to Pyrenophora teres f. teres in barley. Phytopathol 82 (4):407–411
Steffenson BJ, Hayes HM, Kleinhofs A (1996) Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor Appl Genet 92:552–558
Tamang P, Neupane A, Mamidi S, Friesen T, Brueggeman R (2015) Association mapping of seedling resistance to spot form net blotch in a worldwide collection of barley. Phytopathology 105(4):500–508
Tang Y, Liu X, Wang J, Li M, Wang Q, Tian F, Su Z, Pan Y, Lipka AE, Buckler ES, Zhang, Z (2016) GAPIT version 2: an enhanced integrated tool for genomic association and prediction plant genome. Plant Genome. doi:10.3835/plantgenome2015.11.0120
Tekauz A (1985) A numerical scale to classify reactions of barley to Pyrenophora teres. Can J Plant Pathol 7(2):181–183
Tekauz A (1990) Characterization and distribution of pathogenic variation in Pyrenophora teres f. teres and P teres f. maculata from western Canada. Can J Plant Pathol 12(2):141–148
Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Fling-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43:159–162
Turner SD (2014) qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. biorXiv. doi:10.1101/005165
Wu HL, Steffenson BJ, Zhong S, Li Y, Oleson AE (2003) Genetic variation for virulence and RFLP markers in Pyrenophora teres. Can J Plant Path 25(1):82–90
Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Yun SJ, Gyenis L, Hayes PM, Matus I, Smith KP, Steffenson BJ, Muehlbauer GJ (2005) Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci 45(6):2563–2572
Zhang Z, Ersoz E, Lai C, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355–360
Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M (2007) An Arabidopsis example of association mapping in structured samples. PLOS Genet. doi:10.1371/journal.pgen.0030004
Zhou H, Steffenson B (2013) Genome-wide association mapping reveals genetic architecture of durable spot blotch resistance in US barley breeding germplasm. Mol Breed 32:139–154
Zhou H, Steffenson BJ, Muehlbauer G, Wanyera R, Njau P, Ndeda S (2014) Association mapping of stem rust race TTKSK resistance in US barley breeding germplasm. Theor Appl Genet 127(6):1293–1304
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1(1):5–20
Ziems LA, Hickey LT, Hunt CH, Mace ES, Platz GJ, Franckowiak JD, Jordan DR (2014) Association mapping of resistance to Puccinia hordei in Australian barley breeding germplasm. Theor Appl Genet 127:1199–1212
Acknowledgements
The authors would like to thank Danielle Holmes for technical assistance at the USDA-ARS, Cereal Crops Research Unit, Fargo ND. This research was funded by NSF ND EPSCoR Track 1 Grant 11A-1355466 and USDA-NIFA-AFRI Grant #2011-68002-30029 (T-CAP).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest for this article.
Ethical standards
All experiments performed complied with the ethical standards of the USDA-ARS and North Dakota State University.
Additional information
Communicated by Kevin Smith.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Richards, J.K., Friesen, T.L. & Brueggeman, R.S. Association mapping utilizing diverse barley lines reveals net form net blotch seedling resistance/susceptibility loci. Theor Appl Genet 130, 915–927 (2017). https://doi.org/10.1007/s00122-017-2860-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2860-1