Abstract
Key Message
The Rsv1 - h gene in cultivar Suweon 97, which confers resistance to SMVs, was mapped to a 97.5-kb location (29,815,195–29,912,667 bp on chromosome 13) in the Rsv1 locus, thereby providing additional insights into the molecular nature underlying variations in resistance alleles in this particular locus.
Abstract
Soybean mosaic virus (SMV) is a well-known devastating pathogen of soybean (Glycine max (L.) Merrill.) causing significant yield losses and seed quality deterioration. A single dominant allele, Rsv1-h, which confers resistance to multiple SMV strains, was previously reported in the cultivar Suweon 97, but its exact location is unknown. In the present study, Suweon 97 was crossed with a SMV-sensitive cultivar, Williams 82. Inoculating 267 F 2 individuals with two Chinese SMV strains (SC6-N and SC7-N) demonstrated that one single dominant gene confers SMV resistance. Another 1,150 F 2 individuals were then screened for two simple sequence repeat (SSR) markers (BARCSOYSSR_13_1103 and BARCSOYSSR_13_1187) that flank the Rsv1 locus. Seventy-four recombinants were identified and 20 additional polymorphic SSR markers within the Rsv1 region were then employed in genotyping these recombinants. F 2:3 and F 3:4 recombinant lines were also inoculated with SC6-N and SC7-N to determine their phenotypes. The final data revealed that in Suweon 97, the Rsv1-h gene that confers resistance to SC6-N and SC7-N was flanked by BARCSOYSSR_13_1114 and BARCSOYSSR_13_1115, two markers that delimit a 97.5-kb region in the reference Williams 82 genome. In such region, eight genes were present, of which two, Glyma13g184800 and Glyma13g184900, encode the characteristic CC-NBS-LRR type of resistance gene and were considered potential candidates for Rsv1-h.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean mosaic virus (SMV) is widely known as a devastating pathogen of cultivated soybean (Glycine max (L) Merr.) that causes significant yield losses and seed quality deterioration in almost all soybean growing regions around the world (Hill et al. 1987; Ren et al. 1997; Yang et al. 2014). Based on the disease reactions on different soybean cultivars, SMV isolates have been classified into various strains. In the United States, using two sensitive and six resistant soybean cultivars, Cho and Goodman successfully classified 98 SMV isolates into seven strains, namely, G1 to G7 (Cho and Goodman 1979, 1982). In China, a higher level of SMV diversity was reported and the isolates collected between 2003 and 2010 were classified into 21 strains, which were designated as SC1 to SC21 (Guo et al. 2005; Li et al. 2010; Wang et al. 2003; Zhan et al. 2006). To explore the relationships among various SMVs, a recent study sequenced 18 Chinese SMV strains/isolates and further reconstructed a phylogeny encompassing 83 SMV taxa (Zhou et al. 2015). Four different clades were revealed, and the seven US strains were grouped into clade II (G1 and G3), clade III (G6 and G7), and clade IV (G2, G4, and G5), respectively, whereas the Chinese strains/isolates were generally distributed across basal clade I, which included SC3, SC6, SC15, and SC21, and clade II, e.g., SC7 (Zhou et al. 2015).
To prevent such diversified SMVs from causing outbreaks, a widely adopted strategy is to cultivate and plant SMV-resistant cultivars; however, this approach requires extensive understanding of the genetic basis of soybean resistance to SMV (Whitham et al. 2016). Genetic studies on resistant cultivars PI 96983, OX686, and V94-5152 have revealed three distinct SMV-resistant loci, which were named Rsv1, Rsv3, and Rsv4, respectively (Buss et al. 1997; Buzzell and Tu 1989; Kiihl and Hartwig 1979). Allelism tests then identified additional resistance alleles within these loci (especially for the Rsv1 locus) in other resistant cultivars, which exhibited various phenotypes upon inoculation of different SMV strains. Up to 10 different Rsv1 alleles have been proposed, including Rsv1 in PI 96983, Rsv1-t in Ogden, Rsv1-k in Kwanggyo, Rsv1-m in Marshall, Rsv1-y in York, Rsv1-s in LR1 (derived from PI 486355), Rsv1-h in Suweon 97, Rsv1-r in Raiden, Rsv1-d in FT-10 (likely derived from Davis), and Rsv1-n in PI 507389 (Buss et al. 1989; Chen et al. 1991, 2001, Chen et al. 2002; Kiihl and Hartwig 1979; Ma et al. 1995, 2003; Silva et al. 2004; Tucker et al. 2009). It has been a daunting task for soybean researchers to understand the molecular nature underlying different Rsv1 alleles, which can facilitate in the proper usage of these gene resources. Some clues have arisen from fine-mapping studies of cultivar PI 96983. By crossing with the sensitive cultivar Lee68, Yu et al. (1994) first mapped the Rsv1 allele in PI 96983 to a region close to the simple sequence repeat (SSR) marker SM176 on chromosome 13. Several subsequent studies then elegantly showed that the Rsv1 allele is closely linked to two cloned nucleotide-binding site-leucine-rich repeat (NBS-LRR) type resistance genes (R genes), 3gG2 and 5gG3, in PI 96983 (Gore et al. 2002; Hayes et al. 2004; Jeong et al. 2001). Because the recombinants between the two NBS-LRR genes (3gG2 and 5gG3) showed phenotypes that differed from PI 96983, it was speculated that different alleles observed in various cultivars may not be real alleles of a single gene, but represent different combinations (or haplotypes) of R genes (Hayes et al. 2004). After the release of the soybean reference genome of cultivar Williams 82 (Schmutz et al. 2010), it became apparent that the Rsv1 locus is located within a cluster of NBS-LRR genes (~1.1 Mb in range). A thorough evolutionary study involving four legume genomes showed that the NBS-LRR genes in the Rsv1 region belong to the legume non-Toll/Interleukin-1 receptor-NBS-LRR (nTNL) family 45 (Shao et al. 2014). This particular nTNL R gene family had further diverged into 15 subfamilies, and the soybean Rsv1 locus on chromosome 13 inherited six of these (subfamilies 1, 2, 3, 9, 10, and 11; Shao et al. 2014). Notably, the 5gG3 and 3gG2 genes cloned in PI 96983, are respectively, allelic to gene 13g190000 (alias: 13g25920) and 13g190800 (alias: 13g26000) in Williams 82, and phylogenetically belong to subfamily 1 (Shao et al. 2014). To date, whether other Rsv1 alleles are also located within the same region as Rsv1 in PI 96983 or other regions within the locus and involve R genes of different subfamilies have not been established. Interestingly, a recent study conducted by Yang et al. (2013) showed that in PI 96983, two different genes differentially confer SMV resistance, with one gene located between SSR markers 13_1128 and 13_1136 and is responsible for resistance to SC3, SC6 and SC17, and the other located between markers 13_1140 and 13_1156 providing resistance to SC7 (Yang et al. 2013).
Contrary to the fact that several different alleles have been reported for the Rsv1 locus, relatively few cultivars, except for PI 96983, have been utilized in extensive mapping studies. Among all the proposed alleles, the Rsv1-h in Suweon 97 is the only one that confers resistance to all seven US strains (Chen et al. 2002; Cho and Goodman 1982). Our recent study also showed that inoculating the cultivar Suweon 97 with four Chinese SMV strains/isolates results in complete resistance (Zhou et al. 2015). Thereby Suweon 97 could be utilized as a valuable source for SMV defense.
Different from the conclusion made by Chen et al. (2002) that only one single Rsv1 gene (Rsv1-h) is present in the cultivar Suweon 97, Jeong and Jeong (2014) reported that in cultivar Hwangkeum (a later-released cultivar for Suweon 97 in Korea), at least two genes at the Rsv1 locus and also a possible Rsv3 gene together confer resistances to SMV strains G1 and/or G7. To investigate the molecular nature of Rsv1-h allele, in the present study, we crossed Suweon 97 with the SMV-sensitive cultivar Williams 82. A total of 1,150 F 2 individuals were screened for recombinants, and several recombinant lines were developed to map the Rsv1-h gene into a 97.5-kb region that differed from those in PI 96983.
Materials and methods
Plant materials
A cross between the SMV-resistant cultivar Suweon 97 (PI 483084, Rsv1-h) and the sensitive cultivar Williams 82 (rsv1) was performed in a greenhouse of the Huaiyin Institute of Agricultural Science (Huai’an, Jiangsu Province, China) in the summer of 2013. The F 1 individuals were then self-pollinated to harvest F 2 seeds at the southern experimental base (Hainan Province) in the spring of 2014. A total of 267 F 2 individuals were planted in the laboratory, and their phenotypes and segregation ratios upon SMV inoculations were examined. A total of 1150 F 2 individuals were planted in field in the summer of 2014, and 20 individuals for each selected recombinant F 2:3 lines were then planted in 2015 to derive F 3:4 lines for further homozygote screening.
SMV strains and soybean inoculation
The two SMV strains used in the present study were kindly provided by Dr. De-Yue Yu of Nanjing Agricultural University in 2009. The strains were recovered and maintained in a sensitive soybean cultivar NanNong 1138-2 (NN1138-2). In our previous study (Zhou et al. 2015), we reported the genomic sequence of these two strains (GenBank Accession numbers KP710867 and KP710868), which closely resemble that of the previously published SC6 (HM590054) and SC7 (HM590055) sequences, respectively (Yang et al. 2011). The two strains were thus named SC6-N and SC7-N. Phylogenetically, the SC6-N was grouped with SC6 and exhibited only two amino acid differences in the entire encoded protein. Similarly, the SC7-N was grouped with SC7 and showed a 16-amino acid difference (Zhou et al. 2015).
The Rsv1-h allele was initially proposed to confer resistance to US SMV strains (G1–G7, Chen et al. 2002); however, in the present study, two Chinese strains were employed. Tightly linked but different genes may, respectively, confer resistance to various SMV strains. However, to elucidate the molecular mechanism underlying SMV resistance in Suweon 97, it is optimal to avoid a complicated naming system and simply assume that ‘Rsv1-h’ is one gene or a combination of genes (haplotype) that confers resistance to multiple SMV strains, including those of the US and China.
For inoculation, the infected NN1138-2 trifoliolate leaves infected by either SC6-N or SC7-N were prepared in a sterilized mortar and homogenized in ice-cold .01 M phosphate buffer (PH 7.4) at a rate of 1 g infected tissue per 10 mL of buffer. Ten- to twelve-day-old soybean plants were subjected to mechanical inoculation. A small amount of 600-mesh carborundum was dusted on the leaves that were to be inoculated, and the inoculum was applied by rubbing both primary leaves of soybean plants. After rinsing the inoculated leaves, the plants were kept in aphid-free growth chambers at 22 °C, with a photoperiod of 16 h. From day 7 after inoculation, disease symptoms were examined and recorded every other day for 2 weeks.
DNA extractions and polymorphic SSR marker screening
Genomic DNA was extracted from the leaves of the two parental cultivars, all 1150 F 2 individuals, and a number of recombinant F 2:3 lines using the EasyPure Plant Genomic DNA Kit (Transgen, Beijing, China). On chromosome 13 of soybean genome, a total of 128 SSR markers (from BARCSOYSSR_13_1099 to BARCSOYSSR_13_1226, 13_1099 and 13_1226 for short, same for below) were found across the Rsv1 region (Song et al. 2010). The primers for these markers were synthesized (GenScript, Nanjing, China) and further used to detect marker polymorphisms between Suweon 97 and Williams 82.
Polymerase chain reactions (PCRs) were set in 20 μL volumes and the reaction mixtures included 1.5 μL of genomic DNA (20 ng/μL), 10 μL of 2 × EasyTaq PCR SuperMix (Transgen, Beijing, China), 1 μL each of the forward and reverse primers (10 mM), and 6.5 μL of double distilled water. The PCR conditions were as follows: denaturation at 94 °C for 5 min; 40 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C for 30 s, extension at 72 °C for 30 s; a final extension at 72 °C for 10 min, and cooling to 15 °C. All obtained PCR products were separated on 3 % agarose gels with DNA Green Dye staining (Tiandz, Beijing, China).
The general strategy for mapping the Rsv1-h in Suweon 97
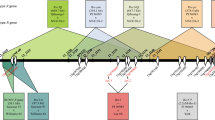
The general strategy employed in mapping the Rsv1-h gene in Suweon 97 is illustrated by Fig. 1. First, a total of 267 F 2 individuals were planted in laboratory and separately inoculated by SC6-N and SC7-N. Resistance controlled by one or more dominant genes was determined based on phenotype segregation ratios and examined by Chi-square tests.
Flow chart for mapping the Rsv1-h gene in Suweon 97 using two Chinese strains of soybean mosaic virus (SMV), SC6-N and SC7-N. ① Cross-breeding, ② self-pollinated, ③ screening of cross-over points with two SSR markers, ④ determining the cross-over points with 22 SSR markers, self-pollinated and harvested individually, ⑤ self-pollinated, harvested individually and screening of two sides of cross-over points
Then, the two outmost polymorphic SSR markers (13_1103 and 13_1187) flanking the Rsv1 locus were used to screen a population of 1,150 F 2 individuals. The F 2 individuals showing the same genotypes as Suweon 97 or Williams 82 were labeled as 1103SS1187SS and 1103WW1187WW, respectively. All F 3 seeds of 1103SS1187SS were harvested together. A hundred individuals were then randomly selected, planted, and inoculated separately by SC6-N or SC7-N (50 each). If the Rsv1-h allele conferring resistance to SC6-N and SC7-N were indeed flanked by markers 13_1103 and 13_1187 or closely linked with the two markers, then resistant phenotypes would be expected. Similar procedures were also performed on the F 3 seeds with 1103WW1187WWgenotypes and sensitive phenotypes would instead be expected.
The F 2 individuals showing recombinant genotypes on the two outmost SSR markers were further separated into three groups according to their genotypes on two intermediate polymorphic SSR markers (13_1124 and 13_1140). Three representatives from each group were then randomly chosen and 15 plants for each F 2:3 line were subjected to inoculation by either strain of SMVs. The genotype of F 2 individuals on Rsv1-h were inferred from the scored phenotypes on their corresponding F 2:3 lines. By doing so, the Rsv1-h gene was further narrowed down to one of the three blocks (block I1103–1124, block II1124–1140, and block III1140–1187). Next, for all the relevant F 2 recombinants within the narrowed block, 15 plants for each of their F 2:3 lines were inoculated to determine their phenotypes upon SC6-N or SC7-N inoculation and their genotypes were inferred. By doing so, the Rsv1-h gene was further limited to a smaller zone. Finally, a number of F 3 seeds were planted and screened to obtain homozygous F 3:4 lines, of which 15 plants for each line were inoculated by SC6-N or SC7-N again and the location of the Rsv1-h gene was double-examined.
Results
Inheritance of resistance of Suweon 97 to SMV strain SC6-N and SC7-N
To determine whether there is only one single dominant locus in Suweon 97 conferring resistance to the two Chinese strains, we inoculated 267 F 2 individuals with either SC6-N (146) or SC7-N (121) (Table 1). A satisfactory fit to 3:1 ratio (P > .05) was observed for both strains, thus supporting that one single dominant locus confers resistance to SC6-N and to SC7-N in Suweon 97, which is consistent with the results reported by Chen et al. (2002).
The Rsv1-h gene in Suweon 97 is flanked by or closely linked to SSR markers 13_1103 and 13_1187
A total of 128 SSR markers across the Rsv1 locus on chromosome 13 were investigated and 22 of these showed polymorphisms between the two parents, Suweon 97 and Williams 82 (Supplemental Table 1). The two outmost markers, 13_1103 and 13_1187, flanked a region of ~1.7 Mb in the reference Williams 82 genome, and all 19 members of the clustered NBS-LRR genes around Rsv1 locus were included in this region (Shao et al. 2014).
To test whether the Rsv1-h gene in Suweon 97 was indeed flanked by or closely linked to markers 13_1103 and 13_1187, we screened a population of 1,150 F 2 individuals for the two markers and identified 262 individuals that showed the same genotypes as Suweon 97 (labeled as F 1103SS1187SS2 ) and 285 individuals harboring the same genotype as Williams 82 (labeled as F 1103WW1187WW2 ). All the F 1103SS1187SS2 individuals were self-pollinated to produce F 1103SS1187SS3 population, from which 100 randomly selected seeds were planted and separately inoculated by SC6-N and SC7-N. All tested plants showed a resistant phenotype same as that of Suweon 97. Also, when 100 randomly selected F 1103WW1187WW3 seeds were planted and inoculated by SMVs, 47 out 48 SC6-N inoculated plants showed mosaic symptoms, and 46 out 50 plants were successfully infected by SC7-N. These data supported that no other locus except for Rsv1 confers resistance to SC6-N or SC7-N in Suweon 97 (Chi-test, P < .05), and the Rsv1-h is flanked by SSR markers 13_1103 and 13_1187 or closely linked to these two markers.
Recombinant F 2:3 lines mapped the Rsv1-h gene between SSR markers 13_1114 and 13_1115
During screening of the F 2 individuals using markers 13_1103 and 13_1187, we also identified 529 F 1103SW1187SW2 individuals that showed the same genotype as that of the F 1 hybrids and 74 recombinant individuals that had cross-over points between the two markers. Twenty additional SSR markers were used to determine genotypes of the 74 recombinants and for each F 2 recombinant, the cross-over point was roughly inferred (Fig. 2). These recombinants were then divided into three groups. The first group included 25 F 2 recombinants that had cross-over points between markers 13_1103 and 13_1124 (group I1103–1124). Similarly, group II1124–1140 and group III1140–1187 included 19 and 30 F 2 recombinants, respectively (Fig. 2). Next, three representatives from each group were randomly chosen and their F 2:3 lines were inoculated by SMVs. Figure 2a shows that the F 2-442 had a cross-over between markers 13_1103 and 13_1109, and its F 2:3 lines were all mosaic upon either SC6-N or SC7-N infection, thereby suggesting that the Rsv1-h gene is located at the right of the cross-over point (because the markers at the right are same as that of Williams 82). Also, the F 2-620 had a cross-over that occurred between 13_1122 and 13_1124, and the phenotypes of its F 2:3 lines segregated upon SMV inoculations, thereby suggesting that the Rsv1-h gene is located left side of the cross-over point (because the markers at the left are heterozygotes). Similar reasoning was applied to other seven representative F 2 recombinants (Fig. 2a), and the obtained results showed no contradiction, which together supported that the Rsv1-h gene is located between the markers 13_1103 and 13_1124.
A physical map showing the genotypes of recombinant F 2 individuals and the disease reactions of corresponding F 2:3 recombinant lines. a Left, the 74 recombinant F 2 individuals were separated into three groups: group I1103–1124 (25), group II1124–1140 (19) and group III1140–1187 (30); middle three representatives from each group are shown for their genotypes on 22 SSR markers; right, the phenotypes of corresponding F 2:3 lines upon inoculation by SMV strains SC6-N and SC7-N were shown. b The 25 recombinant F 2 individuals belonging to group I1103–1124 were assorted into 12 kinds according to their genotypes, and the phenotypes of corresponding F 2:3 lines (numbers in bold) upon inoculation by SMV strains were also shown. All M, all tested plants showed mosaic disease symptoms; all R, all tested plants showed no disease symptoms (resistant); Seg, some tested plants showed mosaic disease symptoms and some tested plants (more than half) showed no disease symptoms
Next, the 25 F 2 recombinants in group I1103–1124 were assorted into 12 kinds of genotypes (Fig. 2b), and for each genotype, F 2:3 lines of one to three representatives were inoculated by SMVs. The data obtained for F 2-382, F 2-87, F 2-457, F 2-216, F 2-358, and F 2-377 together supported that the Rsv1-h gene is located between the markers 13_1114 and 13_1115, and the data of remaining F 2 recombinants showed no contradiction (Fig. 2b). Thus, by determining the cross-over points for each of the 74 F 2 recombinants and inoculating representative F 2:3 lines with SC6-N and SC7-N to deduce their genotypes on Rsv1-h, we were able to draw a conclusion that 13_1114 and 13_1115 are the two closest SSR markers that flank the Rsv1-h gene in Suweon 97.
The results of mapping is further corroborated by data on the F 3:4 homozygotes
We further planted 13 F 2:3 lines to select certain F 3:4 lines, which were homozygotes at both sides of the cross-over points but one side shared the same genotype as that of Suweon 97 and the other side was same as that of Williams 82 (Fig. 3). After inoculation by SC6-N and SC7-N, the phenotypes of these F 3:4 lines were all consistent with the previous mapping results. For example, the homozygotic F 3:4-382 lines are all resistant to SC6-N or SC7-N, showing that the Rsv1-h gene is located at the right side of marker 13_1114. On the other hand, the data of homozygotic F 3:4-128 supported that the Rsv1-h gene is located on the left side of marker 13_1119 (Fig. 3). No contradictory results were observed for other lines.
Markers 13_1114 and 13_1115 delimit a 97.5-kb region in the soybean reference genome that contains eight genes, including two NBS-LRR genes
By examining the soybean reference genome sequences of Williams 82 (Schmutz et al. 2010) in Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html), we found that the SSR markers 13_1114 and 13_1115 together define a 97.5-kb region in the Rsv1 locus. Eight genes were present in this block (Table 2), namely, Glyma.13g184400, Glyma.13g184500, Glyma.13g184600, Glyma.13g184700, Glyma.13g184800, Glyma.13g184900, Glyma.13g185000, and Glyma.13g185100. Among these, Glyma.13g184800, and Glyma.13g184900 encode two coiled-coil-NBS-LRR (CNL) type R genes that represent the most likely candidates for Rsv1-h. However, it should be noted that in this defined block, the presence or absence of polymorphisms of these genes might occur between Suweon 97 and Williams 82, especially for the NBS-LRR type R genes, which exhibited a fast and dynamic evolutionary pattern under the pressure of pathogen.
Discussion
Among the three SMV-resistant loci (Rsv1, Rsv3 and Rsv4) reported in soybean, the Rsv1 locus represents the most complicated one, with up to 10 different alleles reported (Buss et al. 1989; Chen et al. 1991, 2001, 2002; Kiihl and Hartwig 1979; Ma et al. 1995, 2003; Silva et al. 2004; Tucker et al. 2009). However, our understanding of the molecular nature of these different alleles is still limited. One of the difficulties is that Rsv1 is located in a genomic region that is rich in R genes that confer resistance to various pathogens such as Rpg1 to bacterial blight (Ashfield et al. 1998, 2004), Rps3 to Phytophthora sojae (Diers et al. 1992) and Rpv1 to peanut mottle virus (Gore et al. 2002). Therefore, studies focusing on this region should be of a broader scope that addresses a series of questions such as: What are the relationships of different NBS-LRR genes in this region? How did these genes change over time? And most importantly, how did the diversified pathogen-resistance functions evolve? Answering these questions, undoubtedly, is a long-lasting task and requires knowledge from multiple aspects.
By examining the soybean reference genome of Williams 82, we were able to identify a mixed cluster of 14 nonTIR-NBS-LRR genes (nTNL) and 5 TIR-NBS-LRR (TNL) genes in this region (for details, see Table S1 in Shao et al. 2014). All these 19 NBS-LRR genes were flanked by SSR markers 13_1103 and 13_1187. Among these, the four nTNL genes flanked by SSR markers 13_1133 and 13_1136 were of special interests because two of these, 13g190000 (alias name, 13g25920) and 13g190800 (alias name, 13g26000), were respective alleles of the 5gG3 and 3gG2 genes, which have been reported to be tightly linked to the Rsv1 allele in cultivar PI 96983 (Gore et al. 2002; Hayes et al. 2004). A recombinant line inheriting the 3gG2 (but not 5gG3) gene from PI 96983 showed similar responses to certain SMV strains as cultivars Marshall and Ogden did, thereby raising the hypothesis that different Rsv1 alleles in certain cultivars reflect various recombination results among a few critical NBS-LRR genes (Hayes et al. 2004). A recent study by Yang et al. (2013) further demonstrated that in PI 96983, two different genes likely mediate resistance to different SMV strains. Utilizing four Chinese SMV strains (SC3, SC6, SC7, and SC17), the authors mapped two candidate SMV resistance genes in the Rsv1 locus, with one located between SSR makers 13_1128 and 13_1136 for resistance to strains SC3, SC6 and SC17, and the other located between SSR markers 13_1140 and 13_1155 for resistance to strain SC7 (Yang et al. 2013).
To shed more light on the molecular nature of different Rsv1 alleles, in the present study, we chose Suweon 97 as the resistant cultivar to map its Rsv1-h allele. Chen et al. (2002) earlier reported that Suweon 97 contains a single dominant gene (Rsv1-h) that confers resistance to US SMV strains G1-G7. However, in a recent study, Jeong and Jeong reported that in cultivar Hwangkeum, which is a later-released cultivar for Suweon 97, at least two genes at the Rsv1 locus and also a possible Rsv3 gene together confer resistances to G1 and/or G7 (Jeong and Jeong 2014). Because different SMV strains were used in our study, we first verified that there is only one dominant locus that confers resistance to SMV strains SC6-N and SC7-N in Suweon 97. Table 1 shows that inoculation with either SC6-N or SC7-N resulted in resistant F 2 individuals and sensitive individuals that follow a 3:1 inheritance ratio (P > .05), thereby supporting a single-locus hypothesis. Furthermore, by screening a population of 1150 F 2 individuals for markers 13_1103 and 13_1187, a total of 74 recombinants that had cross-overs occurring between the two markers were identified. Utilizing 20 more SSR markers within the Rsv1 region, we were able to genotype the 74 recombinants and estimate their cross-over points. About 23 recombinant F 2:3 lines were inoculated by two Chinese SMV strains SC6-N and SC7-N to infer the location of Rsv1-h (Fig. 2). The obtained data consistently supported that the Rsv1-h is located between the markers 13_1114 and 13_1115, which defines a 97.5-kb region in the Williams 82 reference genome. Finally, to preclude any possible phenotype recording errors that might have occurred with heterozygotes, we further developed 13 F 3:4 homozygote lines. Again, the obtained results were in agreement with our findings (Fig. 3). Therefore, in addition to the previously mapped Rsv1 loci from PI 96983, in the present study, Rsv1-h from Suweon 97 was fine mapped.
An explanation for the inconsistent results between our study and Jeong and Jeong (2014) is that the soybean material of Suweon 97 used in this study could be genetically diverged from Hwangkeum. The dynamic and rapid evolution of the R genes among plant accessions or crop cultivars has been well realized. Even for a cultivar with the same name (e.g., Kwanggyo tested by Zhou et al. 2015), the material kept in the Chinese soybean germplasm collection and in the US showed different phenotypes upon inoculation with multiple SMV strains, thereby suggesting that the responsible resistance genes were different. Our finding that Suweon 97 does not have Rsv3 is consistent to Chen et al. (2002). They used G7 to inoculate F 2 plants from the cross L29 × Suweon 97 to test whether Suweon 97 contained a resistance allele on the Rsv3 locus. The results showed that 4 out of 102 plants were sensitive to G7 inoculation, thereby supporting that Suweon 97 possessed only an Rsv1-h resistance gene and no Rsv3 resistance gene. Although the same SMV strain G7 was used in both previous studies, the difference in results suggests that the materials used by Chen et al. and Jeong and Jeong are not exactly the same.
Compared to results of previous mapping work conducted on the cultivar PI 96983 (Gore et al. 2002; Hayes et al. 2004; Yang et al. 2013), the Rsv1-h gene was mapped to a different interval between markers 13_1114 and 13-1115. Examination of this 97.5-kb region in the Williams 82 reference genome identified eight protein-coding genes, which include two expected NBS-LRR genes, two MYB transcription factors, two membrane proteins, and two proteins with unknown function. Although it is not entirely impossible that Rsv1-h is one of the other six genes in this region, more attention should be given to the two characteristic NBS-LRR genes, 13g184800 (alias 13g25420) and 13g184900 (alias 13g25440). Phylogenetically, gene 13g184800 was assigned in subfamily 11 and gene 13g184900 was placed in subfamily 3 of legume nTNL-45 family according to our previous study (Shao et al. 2014). Shi et al. (2008) had utilized a pair of Rsv1-f/r primers to amplify the 3gG2 gene in 97 soybean cultivars. Only 51 showed positive fragments. Hence, NBS-LRR-type R genes such as 3gG2 may show presence/absence polymorphisms among soybean cultivars. Next, it is imperative to confirm the presence of NBS-LRR genes in Suweon 97 within the mapped region reported in this study and to verify their roles in resistance as Williams 82 does not have SMV resistance. The sequence variation in the other six genes between Suweon 97 and Williams 82 should be also examined.
In summary, after tedious work in performing crosses, screening genotypes, and scoring phenotypes upon SMV inoculations, we present evidence that supports that the Rsv1-h allele in Suweon 97 is flanked by SSR markers 13_1114 and 13_1115, a position that is revealed for the first time within Rsv1 locus. Such results help us better understand the functional diversification process of the soybean Rsv1 locus in response to different SMV strains.
Author contribution statement
J.Q.C. and B.W. conceived and designed the project; F.F.M., X.Y.W., Z.Q.S., P.W., M.W., Y.J.Y., D.X.L., and B.W. collected plant materials; F.F.M., X.Y.W., Y.X.C., Y.N.L., C.C.L. and W.P.W. performed most of the molecular assays, F.F.M., X.Y.W., and B.W. analyzed the data and all other authors joined in the discussion; F.F.M. X.Y.W. and B.W. wrote the paper; J.Q.C. supervised the analysis and writing.
References
Ashfield T, Danzer J, Held D, Clayton K, Keim P, Maroof MS, Webb D, Innes R (1998) Rpg1, a soybean gene effective against races of bacterial blight, maps to a cluster of previously identified disease resistance genes. Theor Appl Genet 96:1013–1021
Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16:309–318
Buss G, Roane C, Tolin S, Chen P (1989) Inheritance of reaction to soybean mosaic virus in two soybean cultivars. Crop Sci 29:1439–1441
Buss G, Ma G, Chen P, Tolin S (1997) Registration of V94-5152 soybean germplasm resistant to soybean mosaic potyvirus. Crop Sci 37:1987–1988
Buzzell RI, Tu JC (1989) Inheritance of a soybean stem-tip necrosis reaction to soybean mosaic virus. J Hered 80:400–401
Chen P, Buss GR, Roane CW, Tolin SA (1991) Allelism among genes for resistance to soybean mosaic-virus in strain-differential soybean cultivars. Crop Sci 31:305–309
Chen P, Ma G, Buss GR, Gunduz I, Roane CW, Tolin SA (2001) Inheritance and allelism tests of Raiden soybean for resistance to soybean mosaic virus. J Hered 92:51–55
Chen PY, Buss GR, Tolin SA, Gunduz I, Cicek M (2002) A valuable gene in Suweon 97 soybean for resistance to soybean mosaic virus. Crop Sci 42:333–337
Cho EK, Goodman RM (1979) Strains of soybean mosaic-virus: classification based on virulence in resistant soybean cultivars. Phytopathology 69:467–470
Cho EK, Goodman RM (1982) Evaluation of resistance in soybeans to soybean mosaic-virus strains. Crop Sci 22:1133–1136
Diers B, Mansur L, Imsande J, Shoemaker R (1992) Mapping Phytophthora resistance loci in soybean with restriction fragment length polymorphism markers. Crop Sci 32:377–383
Gore M, Hayes A, Jeong S, Yue Y, Buss G, Maroof MS (2002) Mapping tightly linked genes controlling potyvirus infection at the Rsv1 and Rpv1 region in soybean. Genome 45:592–599
Guo D, Zhi H, Wang Y (2005) Identification and distribution of soybean mosaic virus strains in Middle and Northern Huang Huai Region of China. Chin J Oil Crop Sci 27:64–68
Hayes AJ, Jeong SC, Gore MA, Yu YG, Buss GR, Tolin SA, Maroof MA (2004) Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 166:493–503
Hill JH, Bailey TB, Benner HI, Tachibana H, Durand DP (1987) soybean mosaic virus: effects of primary disease incidence on yield and seed quality. Plant Dis 71:237–239
Jeong N, Jeong SC (2014) Multiple genes confer resistance to soybean mosaic virus in the soybean cultivar Hwangkeum. Plant Genet Resour 12:S41–S44
Jeong S, Hayes A, Biyashev R, Maroof MS (2001) Diversity and evolution of a non-TIR-NBS sequence family that clusters to a chromosomal “hotspot” for disease resistance genes in soybean. Theor Appl Genet 103:406–414
Kiihl RA, Hartwig E (1979) Inheritance of reaction to soybean mosaic virus in soybeans. Crop Sci 19:372–375
Li K, Yang QH, Zhi HJ, Gai JY (2010) Identification and distribution of soybean mosaic virus strains in Southern China. Plant Dis 94:351–357
Ma G, Chen P, Buss GR, Tolin SA (1995) Genetic characteristics of two genes for resistance to soybean mosaic virus in PI486355 soybean. TAG Theor Appl Genet 91:907–914
Ma G, Chen P, Buss G, Tolin S (2003) Genetic study of a lethal necrosis to soybean mosaic virus in PI 507389 soybean. J Hered 94:205–211
Ren Q, Pfeiffer TW, Ghabrial SA (1997) Soybean mosaic virus incidence level and infection time: interaction effects on soybean. Crop Sci 37:1706–1711
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Shao ZQ, Zhang YM, Hang YY, Xue JY, Zhou GC, Wu P, Wu XY, Wu XZ, Wang Q, Wang B, Chen JQ (2014) Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol 166:217–234
Shi A, Chen P, Zheng C, Hou A, Zhang B (2008) A PCR-based marker for the Rsv1 locus conferring resistance to soybean mosaic virus. Crop Sci 48:262–268
Silva MF, Kiihl RA, Almeida AM (2004) Inheritance of resistance to Soybean mosaic virus in FT-10 soybean. Euphytica 135:339–343
Song Q, Jia G, Zhu Y, Grant D, Nelson RT, Hwang E-Y, Hyten DL, Cregan PB (2010) Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci 50:1950–1960
Tucker DM, Maroof MAS, Jeong SC, Buss GR, Tolin SA (2009) Validation and interaction of the soybean mosaic virus lethal necrosis Allele, Rsv1-n, in PI 507389. Crop Sci 49:1277–1283
Wang X, Gai J, Pu Z (2003) Classification and distribution of strain groups of soybean mosaic virus in middle and lower Huang-Huai and Changjiang valleys. Soybean Sci 22:102–107
Whitham SA, Qi M, Innes RW, Ma W, Lopes-Caitar V, Hewezi T (2016) Molecular soybean–pathogen interactions. Ann Rev Phytopathol 54:443–468
Yang Y, Gong J, Li H, Li C, Wang D, Li K, Zhi H (2011) Identification of a novel Soybean mosaic virus isolate in China that contains a unique 5′ terminus sharing high sequence homology with Bean common mosaic virus. Virus Res 157:13–18
Yang Y, Zheng G, Han L, Dagang W, Yang X, Yuan Y, Huang S, Zhi H (2013) Genetic analysis and mapping of genes for resistance to multiple strains of Soybean mosaic virus in a single resistant soybean accession PI 96983. TAG Theor Appl Genet 126:1783–1791
Yang Y, Lin J, Zheng G, Zhang M, Zhi H (2014) Recombinant soybean mosaic virus is prevalent in Chinese soybean fields. Arch Virol 159:1793–1796
Yu YG, Maroof MAS, Buss GR, Maughan PJ, Tolin SA (1994) RFLP and microsatellite mapping of a gene for soybean mosaic virus resistance. Phytopathology 84:60–64
Zhan Y, Zhi H, Yu D, Gai J (2006) Identification and distribution of SMV strains in Huang-Huai valleys. Sci Agricult Sin 39:2009–2015
Zhou GC, Shao ZQ, Ma FF, Wu P, Wu XY, Xie ZY, Yu DY, Cheng H, Liu ZH, Jiang ZF, Chen QS, Wang B, Chen JQ (2015) The evolution of soybean mosaic virus: an updated analysis by obtaining 18 new genomic sequences of Chinese strains/isolates. Virus Res 208:189–198
Acknowledgments
The National Natural Science Foundation of China (31170210, 91231102, 31470327, and 31570217) supported this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Communicated by B. Diers.
F.-F. Ma and X.-Y. Wu have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, FF., Wu, XY., Chen, YX. et al. Fine mapping of the Rsv1-h gene in the soybean cultivar Suweon 97 that confers resistance to two Chinese strains of the soybean mosaic virus. Theor Appl Genet 129, 2227–2236 (2016). https://doi.org/10.1007/s00122-016-2769-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2769-0