Abstract
Key message
Sixteen major QTLs regulating maize kernel traits were mapped in multiple environments and one of them, qKW - 9.2 , was restricted to 630 Kb, harboring 28 putative gene models.
Abstract
To elucidate the genetic basis of kernel traits, a quantitative trait locus (QTL) analysis was conducted in a maize recombinant inbred line population derived from a cross between two diverse parents Zheng58 and SK, evaluated across eight environments. Construction of a high-density linkage map was based on 13,703 single-nucleotide polymorphism markers, covering 1860.9 cM of the whole genome. In total, 18, 26, 23, and 19 QTLs for kernel length, width, thickness, and 100-kernel weight, respectively, were detected on the basis of a single-environment analysis, and each QTL explained 3.2–23.7 % of the phenotypic variance. Sixteen major QTLs, which could explain greater than 10 % of the phenotypic variation, were mapped in multiple environments, implying that kernel traits might be controlled by many minor and multiple major QTLs. The major QTL qKW-9.2 with physical confidence interval of 1.68 Mbp, affecting kernel width, was then selected for fine mapping using heterogeneous inbred families. At final, the location of the underlying gene was narrowed down to 630 Kb, harboring 28 putative candidate-gene models. This information will enhance molecular breeding for kernel traits and simultaneously assist the gene cloning underlying this QTL, helping to reveal the genetic basis of kernel development in maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize is an important cereal grain crop worldwide and the most highly produced food staple (Statista 2014), playing a significant role in human and livestock nutrition (Blummel et al. 2013). Improving maize yield is the primary concern of crop production systems and is the most important goal of maize breeding (Prado et al. 2014). Maize grain yield can be explained as a function of several yield attributes among which kernel size, measured by kernel length (KL), width (KW), and thickness (KT), plays an important role in determining kernel weight and, therefore, grain yield (Gupta et al. 2006). Genetic studies of kernel size have been emphasized not merely because it is a component of yield but also because of its impact on end-use quality, grain filling, and seedling vigor, and as a domestication syndrome module (Brown et al. 2009; Gupta et al. 2006; Liu et al. 2011; Pozzi et al. 2004; Revilla et al. 1999). These grain qualities are classic quantitative traits, which harbor complex genetic mechanisms, are regulated by many genes, and are also influenced by non-genetic and environmental factors (Song and Ashikari 2008; Xing and Zhang 2010; Yan et al. 2014). Because, maize kernel size is quantitatively inherited, it is difficult for breeders to efficiently accomplish improvements by means of the traditional breeding methods although some significant progresses have already been made by the conventional approaches. Thus, the genetic mechanism of kernel-size variation must be known to competently use the accessible variation for the maize genetic improvement. The initial step of exploring such a genetic mechanism is the mapping of the underlying genetic factors known as quantitative trait loci (QTLs); the recent advent of high precision maize genome sequences, in addition to various molecular markers (http://www.maizegdb.org), has made QTL-based approaches readily accessible (Monaco et al. 2013).
A great number of QTLs have been dissected from diverse crop germplasm using the map-based cloning approaches (Bao 2014; Hong et al. 2014; Huang et al. 2013; Ramya et al. 2010). These achievements provide an opportunity to reveal molecular basis of grain development in cereal crops and suggest ways to improve respective grain yields. For instance, in rice, more than 400 QTLs coupled with grain traits have been explored, and 13 genes relevant to grain shape and weight have been isolated by the map-based cloning strategies (Huang et al. 2013). Rice researchers have reported several genes for grain width, GWs, and length, GLs or GSs, including GS3, a major QTL for grain length and weight, and a minor QTL for grain width and thickness (Fan et al. 2006); GW2 for grain width and also for grain weight (Song et al. 2007); GW5 for grain width (Shomura et al. 2008; Weng et al. 2008); GL3 for grain length, which was identified in three mapping populations (Hu et al. 2012; Qi et al. 2012; Zhang et al. 2012); GW8 for grain width, having a critical deletion polymorphism in the promoter region (Wang et al. 2012); and GS2 for grain length and width, which function in a dominant manner (Zhang et al. 2013). In addition to these cloned genes, ~167, 103, and 95 QTLs associated with thousand-grain weight, grain length, and grain width, respectively, have been mapped on chromosomes in independent studies (Huang et al. 2013). Nonetheless, in maize, several conserved QTLs have been identified that reveal the genetic basis of kernel-related traits, especially kernel weight (Guo et al. 2008; Li et al. 2010a; Liu et al. 2010, 2014; Tang et al. 2010; Zhang et al. 2014).
However, a very little work has been done to identify kernel-size QTLs in maize (Austin and Lee 1996; Nikolić et al. 2013; Peng et al. 2011; Li et al. 2009; Qi et al. 2009). Previously, a few attempts were made to isolate genes that govern kernel size and weight using maize mutants, such as rgf1, dek1, sh1/sh2, and incw2 (Borrás and Gambín 2010; Lid et al. 2002; Maitz et al. 2000; Thévenot et al. 2005). Recently, using an immortalized F2 population, Zhang et al. (2014) reported 42 main-effect QTLs for kernel-related traits, among which qKL3, qKWI6, and qKW10b for KL, KW, and weight, respectively, contributed 11.2–21.1 % of the total phenotypic variance. In addition, Liu et al. (2014) performed a QTL analysis for maize kernel-size traits and kernel weight in five environments, revealing 55 and 28 QTLs using a single-environment analysis and mixed linear model-based composite interval mapping (CIM) for joint analysis, respectively. In addition, Nikolic et al. (2013) reported five QTLs for grain yield per plant along with 18 QTLs for four yield components (KL, KW, KT, and weight) under the drought conditions. Moreover, ZmGS3 (GRMZM2G139878), a maize ortholog of the rice gene GS3, was isolated and contains five exons and a common domain with the rice gene that appears to be associated with KL, but not KW (Li et al. 2010c). Likewise, two maize homologs of the rice gene GW2, namely ZmGW2-CHR4 (GRMZM2G170088) and ZmGW2-CHR5 (GRMZM2G007288), have conserved functional protein domains with no non-synonymous polymorphisms. In addition, there is a negative correlation in the expression level of ZmGW2-CHR4 with KW (Li et al. 2010b). Very recently, Liu et al. (2015) identified a maize ortholog, ZmGS5 (GRMZM2G123815), of rice GS5 that contributes to the maize kernel development. In spite of these findings, there are very few efforts to address the genetic analysis of kernel size in maize.
To fine map and subsequently clone a defined QTL, constructing a mapping population in which the expected QTL behaves as a single classical Mendelian factor is a prerequisite. A recombinant inbred line (RIL) population, having the advantage of repeatability, is suitable for QTL mapping, as trials can be repeated over time and in different environments. The residual heterozygosity that exists in an RIL population, known as a heterogeneous inbred family (HIF), that segregates with the genomic region of the mapped QTL is the ideal material for QTL fine mapping and cloning (Coles 2009). Therefore, QTL mapping of maize kernel size and kernel weight using RILs developed from a cross between Zheng58, a widely used elite line with regular kernels, and SK, a selected line from tropical landrace with small kernels, was undertaken. A detected major QTL, qKW-9.2, was selected for fine mapping using an HIF to narrow the genomic region.
Materials and methods
Plant materials and field experiments
In total, 204 RILs developed using the single-seed descent method (Poehlman and Sleper 1995) to the F6 generation from the cross between two inbred lines, SK and Zheng58, which have distinct small and regular kernel morphologies, respectively (Fig. 1), were used in this experiment. The parent Zheng58 is a derivative of the inbred Y478, which has been used as a parent to breed a series of novel hybrids that widely cultivated in China. The Zheng58 inbred line possesses superior agronomic traits related to drought tolerance, disease resistance, and yield compared with the foundational genotype Y478. This line representing the predominant heterotic group (similar group with B73) is the common parent of the commercial hybrid ZD958 (Zheng58 × Chang 7-2 type combination) that is currently the most widely grown maize hybrid in China (Lai et al. 2010). In contrast, the SK parent is an inbred line selected from tropical landraces (Yang et al. 2011), possessing intermediate maturity and a black flint grain type with a very small kernel size, which provides a huge contrasting phenotypic difference to its counterpart Zheng58.
In 2011 and 2012, the kernel trait values of the RIL populations and the parental lines were evaluated for QTL mapping in the Hainan, Henan, Hubei, Yunnan, and Chongqing Provinces of China having diverse climatic attributes (Supplementary Table 1). Among the five locations listed, four were included in each year, for instance, in 2011, Chongqing (11CQ), Yunnan (11YN), Henan (11HN), and Hainan (11DHN), while Chongqing (12CQ), Yunnan (12YN), Henan (12HN), and Hubei (12HB) were considered for the year 2012. The experiment was conducted following a randomized complete block design with one random-block replication per location, and standard intercultural operations were followed throughout the entire growing season and over all of the locations. Simultaneously, self-pollination was carried out for each line in each generation to maintain the lines. On the basis of the QTL-mapping results, HIFs were derived from three RILs (KQ9-HZAU-1271-1, KQ9-HZAU-1382-2, and KQ9-HZAU-1341-1) that contained recombination breakpoints within the target QTL region, and near-isogenic lines (NILs) were constructed by repeated self-pollination of HIFs-driven progenies. To map the candidate gene, in the winter of 2013 in Hainan, the resultant RIL-derived HIF progenies were planted and genotyped at the qKW-9.2 locus using eight simple sequence repeat (SSR) markers, and thereafter, two subsequent recombinant families, HN-hap5 and HN-hap7, recovered in Hainan in 2013 were selected for the further fine mapping.
Phenotypic measurements
At least five well-pollinated ears in each row were harvested for phenotypic measurements of four kernel traits by standard procedures. Some RILs had abnormal ear development and they were discarded from the analysis. Four kernel-related traits, KL, KW, KT, and 100-kernel weight (HKW), were examined for each individual by randomly selecting kernels from the middle position of the respective cob, and five even ears were chosen for each line. Ten kernels for each individual were measured for the traits KL (mm), KW (mm), and KT (mm) using an electronic digital caliper (Guanglu Measuring Instrument Co. Ltd., China) with a precision of 0.1 mm (Yang et al. 2014). The average KLs, KWs, and KTs of five ears (10 kernels per ear) were used as the phenotypic values. Kernel weight in grams was based on 100 kernels (the mixed kernels of five ears were measured three times for each line) using a digital electrical balance. To reduce the influence of environmental effects on phenotypic variation (ANOVA; Supplementary Table 2), a best linear unbiased predictor (BLUP) value for each line was computed across all of the environments with the R package lem4 (Team 2014), and the BLUP values were then combined to reduce the prediction bias caused by the unbalanced data. In addition, the descriptive statistics (ANOVA, mean, correlation) of the populations were analyzed using the statistical software SPSS version 11.5 (SPSS 1999).
Heritability estimation
The broad-sense heritability (h 2 b ) for each trait was estimated according to the formula: h 2 b = σ 2 g /(σ 2 g + σ 2 e ), where σ 2 g is the between genotype variance component and σ 2 e is the within error variance component. Using a random effects model (model 2), one way ANOVA, according to the model γ ij = μ + g i + e ij , the total phenotypic variance for each trait was partitioned and the mean square of between groups and the mean square of within groups were obtained. Concurrently, difference between these two was divided by ‘n’ to estimate σ 2 g , where n is the number of replications.
Linkage map construction and QTL mapping
The Zheng58/SK RIL population was characterized with 56,110 single-nucleotide polymorphisms by Illumina MaizeSNP50 BeadChip, and polymorphisms were explored in 13,703 single-nucleotide polymorphisms. A very high-density genetic map for the RIL population was constructed (Pan et al. 2015) with 2486 genetic bins (a genomic region in which no recombination exists). On the basis of the established genetic map covering 1860.9 cM of mapping distance along with the data for the four kernel traits in the RIL population, QTLs controlling the traits were analyzed using the CIM method (Zeng 1994) in the Windows QTL Cartographer version 2.5 software (Wang et al. 2010) with a default setting of 0.5-cM scanning interval between markers and putative QTLs. The log10 of odds (LOD) ratio threshold value for QTL significance was determined by 1000 random permutations at an experiment-wise significance level of P = 0.05 (Doerge and Churchill 1996). A forward stepwise regression program was exploited to select the cofactors used for CIM calculations, and simultaneously, all of the linkage groups were scanned with a view to create an LOD curve. The QTLs with LOD values larger than the threshold value (threshold = 2.5 LOD) were considered, and concurrently, the position, genetic effects, and percentage of phenotypic variation explained by the identified QTL were estimated at the peak region of the significant LOD value. QTLs that were firmly identified from multiple environments for a defined trait with evidently similar positions (overlapping one LOD confidence interval) were predicted to be identical. In addition, QTLs, which were mapped in different environments and explained more than 10 % of the phenotypic variance, were considered major QTLs.
DNA extraction, PCR protocol, and SSR marker design
Total genomic DNA was extracted from the leaf tissue following the cetyl-trimethyl-ammonium bromide method as described by Murray and Thompson (1980) with minor modifications. Thereafter, the extracted DNA was dissolved in double-distilled water, separated on 1.5 % agarose gel stained with ethidium bromide and examined under UV light. DNA was quantified by a comparison of the sample DNA with the lambda DNA standards (TakaRa, Dalian, China) run at the same time. Finally, for the SSR study, the stock DNA solution was diluted to 20 ng/µl and PCR amplification was performed using a basic 15-µl reaction mixture with an amplification profile that consisted of an initial 94 °C for 3 min, followed by 35 cycles of 30 s at 94 °C, 30 s at 58–62 °C, and 10 min at 72 °C, with a 4-min extension at 25 °C. Denatured amplified products were then separated on 6 % polyacrylamide gels and visualized using the silver-staining method as described by Sanguinetti et al. (1994). For the SSR marker development, SSR sites were identified from the publicly available gramene database (http://www.gramene.org/). SSR primers were then designed using the online software of the NCBI primer designing tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) to produce a 125–500-bp PCR fragment, and polymorphisms were detected using the template DNA of the parents SK and Zheng58. Primer sequences and physical positions in the genome of the newly developed SSR markers are presented in Supplementary Table 3.
Fine mapping of qKW-9.2
Three HIFs derived from three RILs, KQ9-HZAU-1271-1, KQ9-HZAU-1382-2, and KQ9-HZAU-1341-1, which were heterozygous within the candidate region, were screened with SSR markers for the qKW-9.2 validation. Previous studies reported that this region was located in the recombination hot spot (Pan et al. 2015). Thereafter, 1200 individuals resulting from the self-pollination of KQ9-HZAU-1341-1 were evaluated to narrow down the location of the qKW-9.2 locus. In doing so, we searched for recombinant in each family member using SSR markers, and after an independent haplotype analysis of each family, we compared the important recombinants within the family along with their homozygous genotypes (NILs) at the qKW-9.2 locus. Finally, if there was a significant difference between the recombinants and homozygous NIL; then, we declared that specific recombinant breakpoint define the boundary (left or right ended) of a QTL. Progeny test relying on family-based ANOVA (t test: two sample assuming equal variance) was considered for statistical interpretation of the phenotypic difference between two haplotypes. For the significance test, the phenotypic value of a family was regarded as the arithmetic mean of the corresponding family in the QTL analysis.
Results
Phenotypic variation in kernel-related traits
The phenotypic performance and variation of kernel traits in the RIL population and the parents in 2011 and 2012 over eight locations are presented in Table 1 and Fig. 1. Highly significant differences in all of the four kernel traits studied were exhibited by the two parents. The parent Zheng58 showed a typical kernel size of 32.05 g per 100 kernels when grown in Yunnan in the year 2011, with a KL of 10.94 mm, a KW of 9.45 mm, and a KT of 5.39 mm, whereas its counterpart SK had an extremely small kernel size of only 5.63 g per 100 kernels, with correspondingly lower trait values of 7.17, 4.09, and 3.51 mm for KL, KW, and KT, respectively, in the same environment and year. A similar trend was also observed between the two parents regarding all of the trait values for all of the environments in the years 2011 and 2012 (Table 1). In 2011, the highest mean value for KL in the RIL population was 8.90 mm, with a range from 7.32 to 10.82 mm, recorded in the population grown in Hainan. Subsequently, in 2012, the RIL population grown in Yunnan exhibited the highest mean value for KL as 8.79, ranging from 7.13 to 10.96 mm. Nonetheless, the phenotypic affinity for KW and HKW was almost identical in 2011 and 2012 (Table 1), signifying that the phenotypic performance was fairly stable over the two consecutive years for these two traits (Supplementary Table 4). The KL, KW, and HKW revealed a pattern of continuous and approximately normal distributions across 2011 and 2012, indicating a quantitative inheritance of the characteristics studied (Fig. 2). Meanwhile, the frequency distribution of KT ranged from 3.6 to 5.7 mm, demonstrating a bimodal pattern with a KT of 4.5 mm as the boundary in the year 2012 (Fig. 2), and the transgressive segregation of KT could be observed in the RIL population for both years.
Heritability and correlation
High broad-sense heritabilities of 91.1, 96.2, 90.7, and 94.1 % were estimated for KL, KW, KT, and HKW, respectively (Table 1), which were rather similar to earlier reports of maize kernel traits (Zhang et al. 2014). Highly significant positive correlations were detected among the four kernel traits for both years in the RIL population (Fig. 3), except for the relationship between KL and KT in the year 2012, in which the Pearson correlation coefficient between these traits was 0.14 and non significant. Moreover, correlations within the traits KL, KW, and KT were comparatively smaller (0.14 < r < 0.55) in relation to their individual effects on HKW (0.55 < r < 0.83), suggesting the vital role of kernel weight in the maize kernel development (Fig. 3). In addition, heatmaps (Supplementary Fig. 1) using the phenotypic data for the eight environments also expressed high correlations for the four different traits, signifying that the BLUP data of the four traits were suitable for use in the QTL mapping. Among the different traits compared, KW and HKW were highly correlated, indicating that KW was an important trait affecting maize yield.
Genotypic and phenotypic correlations among four traits. The italic and bold numbers represent Pearson correlation coefficients between the traits in 2011 and 2012, respectively; Double asterisk significant at the 0.01 level. Total numbers of QTLs identified at eight environments in 2 years are shown within parentheses
QTL mapping of kernel traits in RIL populations
KL
In total, 18 QTLs for KL were detected across all of the environments in the two consecutive years, and 12 QTLs were detected in at least two environments (Supplementary Table 5). Moreover, for a single environment, Hainan was the best, resolving nine QTLs in 2011 (Supplementary Table 5). The maximum number of QTLs detected on a chromosome was three each on chromosome 1 (qKL1.1, qKL1.2, and qKL1.3), 3 (qKL3.1, qKL3.2, and qKL3.3), and 9 (qKL9.1, qKL9.2, and qKL91.3), from which qKL1.3 and qKL9.1 were ascertained in four environments. In addition, qKL7.1 also overlapped in four environments (11YN, 11DHN, 12CQ, and 12YN) elucidating 10.03, 4.61, 10.03, and 5.21 % of the phenotypic variance, respectively. In addition, the QTL qKL6.1 on chromosome 6 was repeatedly identified in all of the locations, except Chongqing and Yunnan in 2011, explaining 4.65–11.53 % of the total phenotypic variance. Furthermore, a major QTL, qKL8.2, flanking the marker interval PZE-108100624_PZE-108117143 was revealed over multiple environments, 11YN, 11DHN, 12CQ, 12YN, and 12HB, and explained 9.43, 7.20, 12.54, 12.15, and 10.43 %, respectively, of the total phenotypic variance. The additive effect was positive (alleles increased the KL from Zheng58) for all of the identified QTLs, except two, designated qKL1.1 and qKL1.3, on chromosome 1.
KW
In total, 26 QTLs were detected for KW during 2011 and 2012 over the eight environments (Supplementary Table 5), whereas the number QTLs revealed in multiple environments was 14. The highest number of QTLs, estimated as 11, was exposed when the RIL population was grown at Chongqing in 2012, followed by Henan and Hainan in 2011, each of which revealed 10 QTLs. Each of chromosomes 1, 3, and 8 had the four QTLs, and of these 12 QTLs, 8 (excluding qKW1.2, qKW1.3, qKW1.4, and qKW3.4) were detected in more than one environment. The lowest number of QTLs was detected on chromosomes 6, 7, 9, and 10, whereas no KW-related QTL was identified on chromosome 2. A major QTL, qKW1.1, bound by the marker interval PZE-101033801_PZE-101056681 was constantly resolved in all eight environments, explaining 5.49–23.69 % of the phenotypic variance. Moreover, another major QTL, qKW3.2, persisted over the environments, 11HN, 12CQ, 12HN, and 12HB, elucidating 12.81, 12.68, 13.30, and 10.80 %, respectively, of the KW phenotypic variance. Heat map with the BLUP data of eight environments (Fig. 4) also demonstrated the similar strong detection effect for chromosome 1 and 3 as well as the non detection of KW QTLs for chromosome 2. In addition, QTLs qKW3.3, qKW5.1, qKW5.3, qKW8.4, and qKW9.2 were also frequently identified in the same genomic region of their respective chromosomes over four different environments and contributed 3.42–12.32 % to the phenotypic variance.
KT
In total, 23 QTLs were identified for KT in 2011 and 2012 over the eight environments and only 10 (qKT1.1, qKT3.1, qKT3.3, qKT3.4, qKT3.6, qKT5.2, qKT7.2, qKT7.4, qKT8.1, and qKT10.1) of them were persistent in multiple locations (Supplementary Table 5). In a single environment, the maximum number of QTLs was detected in Henan in 2012. The highest number of QTLs detected was seven on chromosome 3, followed by four on chromosome 7. Single QTLs qKT4.1, qKT9.1, and qKT10.1 were identified on chromosomes 4, 9, and 10, respectively, whereas only qKT10.1 was found in multiple environments. Moreover, two QTLs were positioned on chromosomes 2 and 5, whereas no KT-related QTL was detected on chromosome 6. Furthermore, qKT3.3 was repeatedly identified over the locations 11YN, 11HN, and 12YN, explaining 10.84, 7.31, and 6.57 %, respectively, of the phenotypic variance. Likewise, qKT7.2 was also commonly found in 11YN, 11HN, and 12HB, and contributed 7.04, 10.92, and 5.86 %, respectively, to the phenotypic variance. In addition, a major QTL, qKT3.7, was revealed in 11DHN flanked by the marker interval PZE-103133167_PZE-103140275, with an LOD score of 10.87 and explained 17.93 % of the total phenotypic variance of KT.
HKW
In total, 19 QTLs were discovered for HKW in all of the environments and only seven (qHKW1.1, qHKW3.1, qHKW3.2, qHKW3.3, qHKW7.3, qHKW8.2, and qHKW9.1) overlapped in multiple environments, the lowest number of detected overlapping QTLs existed for four traits (Supplementary Table 5). In a single environment, Henan in 2012 provided the highest number, eight, QTLs for HKW. Four QTLs were detected on chromosome 1, and three each were found on chromosomes 3 and 7. Only a single QTL, qHKW6.1, was detected on chromosome 6, which elucidated 7.36 % of the phenotypic variance in 12HB, whereas no QTLs were identified on chromosomes 4 and 10 for this trait. Three major QTLs, qHKW1.1, qHKW7.3, and qHKW9.1, were constantly identified over seven different environments and explained 4.61–17.89, 5.43–11.24, and 3.48–14.13 %, respectively, of the total phenotypic variance. Similarly, qHKW3.2 and qHKW8.2 were also repeatedly identified in five and six environments, respectively, and these two QTLs elucidated 4.07–13.82 and 3.92–9.23, respectively, of the phenotypic variance. Furthermore, a major QTL, qHKW1.2, was revealed to have an LOD score of 9.47 in a single environment (11DHN) and explained 16.04 % of the total phenotypic variance. A positive additive effect was exposed for all of the QTLs except qHKW1.4, which was detected on chromosome 1 in 2011 in Henan and had a −0.73 effect.
QTL detection using BLUP data
In total, 34 QTLs were detected for all of the kernel traits studied using the BLUP data (Fig. 4; Supplementary Fig. 2) and 19 of those clustered into seven genomic regions on chromosomes 1, 3, 7, 8, and 9 (Fig. 4; Supplementary Table 6). Of these, 10, 9, 7, and 8 QTLs were identified for KL, KW, KT, and HKW, respectively. Of the kernel-related traits, a maximum of six QTLs were detected on chromosomes 7 and 8, followed by chromosome 1, which had five QTLs. Four QTLs were identified on chromosomes 3 and 9, and two QTLs were found on chromosomes 4 and 10. Only one QTL was detected on each of chromosomes 2 and 6, which were responsible for the trait KL. Three major QTLs, qKL-6, qKL-8, and qKL-9a, with LOD scores 6.33, 9.19, and 7.14, respectively, explained 9.20, 14.03, and 10.35 %, respectively, of the phenotypic variance of KL (Table 2). Likewise, for KW, qKW-1, qKW-3, qKW-5a, and qKW-9, with LOD scores of 9.39, 9.71, 7.02, and 5.81, respectively, contributed 12.68, 13.52, 9.37, and 6.45 %, respectively, to the total phenotypic variance. Meanwhile, only one major QTL, qKT-3, delimited by the markers PZE-103101073_PZE-103107669 for KT elucidated 13.32 % of the variance, having an LOD value of 8.46. Three major QTLs, qHKW-1b, qHKW-7b, and qHKW-9, were uncovered for HKW, with LOD values of 12.77, 8.64, and 6.05, respectively, and these three QTLs collectively explained 38.23 % of the total phenotypic variance. Moreover, qKL-9a, qKW-9, and qHKW-9 were located in the recombination hot spot of chromosome 9, and qKW-9 and qHKW-9 were flanked by the same marker interval between PZE-109109569 and SYN8851.
Validation and fine mapping of qKW-9.2
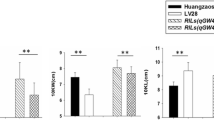
qKW-9.2 was detected for KW over four locations along with the BLUP data (qKW-9)and mapped to chromosome 9 (Fig. 5a; Table 2; Supplementary Table 5). The identified QTL, commonly designated as qKW-9.2, was consistent in populations grown across 11HN, 12CQ, 12HN, and 12HB, contributing 11.26, 4.64, 5.88, and 5.23 % (Supplementary Table 5), respectively, of the total variance in the four environments. To validate the identified QTL, three HIF lines, having heterozygous loci within the candidate region, were selected from the RIL (KQ9-HZAU-1271-1, KQ9-HZAU-1382-2, and KQ9-HZAU-1341-1) populations (Fig. 5b) and genotyped using eight SSR polymorphic markers. The segregation pattern of HIF from KQ9-HZAU-1341-1 represented three genotypic classes, SK homozygous (1/1), Zheng58 homozygous (2/2), and heterozygous (1/2), which had 25, 23, and 12 individuals in each respective class. After a progeny test, significant differences in KW were revealed within the HIF derived from KQ9-HZAU-1341-1 (Fig. 5c), whereas no differences were observed in the remaining two HIF derivatives of KQ9-HZAU-1271-1 and KQ9-HZAU-1382-2. This result indicated that qKW-9.2 was in the region between the markers SSR1 and MSR10.
Map-based fine mapping of the qKW-9.2 locus. a QTL mapping of Zheng58/SK RILs. An additive effect QTL was detected for kernel width over four locations along with the best linear unbiased predictor (BLUP) data and mapped to a 122.2–128.6-cM genomic interval of maize chromosome 9 (Physical confidence interval as according to maize B73 reference genome APGv2). b RIL selection within the candidate region for QTL validation. Three RILs, KQ9-HZAU-1271-1, KQ9-HZAU-1382-2, and KQ9-HZAU-1341-1, having heterozygous loci within the candidate region were screened for the qKW-9.2 validation. c QTL validation in RIL-derived heterogeneous inbred families (HIFs) with progeny test relying on family-based ANOVA (t test: two sample assuming equal variance). Three HIFs derived from RILs that were genotyped using eight SSR polymorphic markers revealed significant differences within the HIF derived only from KQ9-HZAU-1341-1. d Fine mapping of the qKW-9.2 locus using HIFs. In total, 1200 individuals resulting from the self-pollination of KQ9-HZAU-1341-1-driven HIFs were evaluated, and the location of qKW-9.2 was narrowed down to a genomic region of ~630 kb flanked by the FSR6 and MSR36 markers
To fine map qKW-9.2, a segregating population of 1200 individuals originating from the RIL (KQ9-HZAU-1341-1)-derived HIF with a heterozygous region around the locus qKW-9.2 was grown in Hainan in 2013 to identify the recombination events between qKW-9.2 and tightly linked markers. Accordingly, two markers, SSR1 and MSR10, were used for recombination screening, and 51 recombinants were sorted out. Among these recombinants, 31 were identified between SSR1 and qKW-9.2, along with another 20 between MSR10 and qKW-9.2 (Fig. 5d). Subsequently, another six SSR markers (SK15, MSR24, FSR6, MSR4, FSR27, and MSR36) were used to promote the resolution of the qKW-9.2 local linkage map. After the inclusion of these six markers, 22 recombinants (HN-hap4) were revealed between MSR24 and qKW-9.2 (Fig. 5d), in addition to 9 (HN-hap5) and 7 (HN-hap7) recombinants between FSR6 and qKW-9.2, and MSR36 and qKW-9.2, respectively. The genotypes of the recombinants, revealed by eight polymorphic SSR markers, are shown in Fig. 5d. After the progeny test, each recombinant family was found to contain an identical larger KW marker resulting in a significantly larger KW than the homozygous SK. Consequently, no difference was observed with homozygous Zheng58, indicating the identity of each recombinant progeny group. Thus, the location of qKW-9.2 was narrowed down to a genomic region of approximately 630 kb flanked by the FSR6 and MSR36 markers (Fig. 5d) that harbored 28 putative gene models (Supplementary Table 7).
Discussion
A QTL analysis using the Zheng58/SK RIL revealed 86 QTLs for four kernel traits, and this total QTL number is higher than found in the previous studies (Liu et al. 2014; Peng et al. 2011; Zhang et al. 2014). QTL mapping results explored the numbers of significant QTLs for each trait, ranging from 18 for KL to 26 for KW over the eight environments, with an asymmetric and clustered allocation in genomic regions that exposed the complex genetic architecture of the kernel traits. The reasons of such huge QTLs detection may reside in the origin and domestication process of the used parents. For instance, the Zheng58 is a regular inbred line with normal kernel size, while SK is near landrace with very small kernel size, as shown in Fig. 1, and thus, they may have more diverse genes or genomic differences that predominantly regulate the kernel traits. Moreover, the high heritability (>0.90) of the kernel traits resulting from the precise estimates of phenotypes at eight environments, and the ultrahigh-density linkage map may increase the power to detect QTLs. Two major QTLs, qKL1.3 and qKL8.2, regulated the major phenotypic variance for KL. Likewise, the QTL qKW1.1 for KW was continually identified over all of the environments, explaining the highest percentage of variance, and was similar to qKW1-2 reported by Liu et al. (2014). Moreover, the QTL mapping results for KL and KW demonstrated that in each case, chromosome 3 was included in the maximum QTL-revealing group, and this corroborated previously reviewed rice results (Bao 2014), which concluded that chromosome 3 harbors more QTLs for the GL and GW genes. Some major QTLs were steadily identified over seven environments, although the phenotypic variation explained by such stable QTLs diverged in magnitude among the different environments. This variation may be due to QTL–environment interactions (Xu 2010). Moreover, environment-specific QTLs, having a large phenotypic variation, were exposed for all of the studied traits, and context-dependent effects, as well as regulation of minor polygenes, may be responsible for such phenomena (Mackay et al. 2009). QTL analyses in single environments cannot precisely predict the positions and stability of QTLs (Messmer et al. 2009), and thus, the direction, as well as the magnitude, of QTL effects are often missed. Conversely, QTLs with apparent genetic effects could be selectively discovered in diverse environments. Here, in the Zheng58/SK RIL population, 16 QTLs significantly contributed more than 10 % to phenotypic variance, with fewer contributing over 20 %, and were mapped in multiple environments. In addition, a great portion of the detected QTLs was environmentally specific, and some of them also explained huge phenotypic variations, signifying a complex genetic architecture with a few major and many minor effects that might regulate the maize kernel development.
What morphological and physiological functions are possibilities for these QTLs to be affecting to cause the trait impacts that they do? The current data set may be fairly inadequate to resolve this query. Mostly, the use of different parental lines and populations with diverse morphological features results in different consequences, regarding the number of QTLs and their effects. Moreover, the tightly correlated traits, for instance, length of the female inflorescence or ear shoot, a notable yield component in maize and kernel-related traits, may also have huge possibilities to do so (Ross et al. 2006). Phenotying the related traits and performing the QTL mapping in the same population will help answer these questions; thus enhancing the understanding of plant development.
Mapping QTLs is often hindered by the limited resolution and lack of recombination events within the QTL region in a finite population size (Tanksley 1988; Yu and Buckler 2006). A fine-mapping approach can be employed to narrow the genomic interval through the interactions of novel recombination events within the targeted QTL region. Nonetheless, identical genetic backgrounds in the mapping population that place QTLs into typical Mendelian traits greatly enhance the efficiency of fine mapping (Alonso-Blanco and Koornneef 2000) and hence, an advanced population, such as an RIL-derived HIF, that minimizes the background noise is a great option for the QTL map-based cloning. In this study, to develop a HIF, we only genotyped a single individual from each RIL and selfed each of them to represent the whole RIL population, providing us a good opportunity to discover the HIFs for each QTL of interest. Selfed RILs were chosen as the primary mapping populations if they were segregating in the QTL region of chromosome 9 known as qKW-9.2, which represents an HIF of nearly isogenic individuals. HIF-based NIL development is much more consistent, as well as more swift, in identifying either major or minor QTLs as compared with the traditional approaches (Bai et al. 2010). The analysis of HIFs is convenient for identifying links between markers and QTLs, and is useful in complementing the RIL population, which allows for the quick confirmation of individual QTLs. Each segregating HIF population is characterized by its independence, possessing unique recombination events in the genomic regions of interest flanked by the QTL (Tuinstra et al. 1997), and these recombination events can explain the genetic interval containing the defined QTL (Paterson et al. 1990). In this study, a KW-related QTL detected over four unique locations (qKW-9.2), commonly designated as qKW-9.2, and having BLUP data, was chosen for fine mapping using HIFs. With one generation of mapping, the location of the underlying gene was narrowed down to 630 kb, harboring 28 putative gene models. Moreover, our synchronized effort is now aimed at distinguishing the genomic region to the gene level, which will help to explain the genetic basis of complex quantitative traits. In addition, as the different cereal species diverged from a common ancestor and underwent a parallel selection for domestication, it may be possible to draw a comparative map among the cereals for QTLs-governing complex traits, such as grain shapes in rice, wheat, and maize (Paterson et al. 1995). Unlike in maize, several QTLs-governing rice GW have been fine mapped, including GW2, qSW5, GS5, and GS2 that mapped to 8.2, 49.7, 11.6, and 33.2 kb, respectively (Li et al. 2011; Song et al. 2007; Wan et al. 2008; Zhang et al. 2013). GW2 encodes a RING-type protein with E3 ubiquitin ligase activity, and mutant alleles of GW2 render spikelet hull cell division and the increase of the grain-milk filling rate, resulting in an expansion of GW (Song et al. 2007). Two homologs of GW2 in maize, ZmGW2-CHR4 and ZmGW2-CHR5, are reported to have high amino acid sequence identities (81 %) with rice GW2 (Li et al. 2010b). Therefore, incorporating the QTL fine-mapping results of maize obtained here with previously reviewed rice results may provide insight into the molecular organization of the maize kernel development.
Conclusions
We performed a QTL analysis for maize kernel size using an RIL population derived from two parents having distinct variations in kernel shape and weight, and we identified several major QTLs for kernel weight or size, including KL, KW, KT, and HKW. Major QTLs detected in this study may be utilized in breeding programs following marker-assisted selection which were also good candidates for isolating the underlying genes using map-based cloning strategies. We validated the QTL qKW-9.2, which controls the trait KW on chromosome 9, and it was simultaneously fine mapped using an RIL-derived HIF. Because, few studies have been conducted to resolve the genetic basis underlying maize KW, the detection of novel genetic loci governing KW, followed by the simultaneous characterization of their corresponding genes, will facilitate our ability to discover the secrets of the maize kernel development.
Author contribution statement
J.Y. designed and supervised the study. M.S.R., J.L., J.H., H.G., and Q.P. performed the experiments. M.S.R. analyzed the data. M.S.R. and J.Y. prepared the manuscript and all authors read, and approved the manuscript.
References
Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5:22–29
Austin D, Lee M (1996) Comparative mapping in F2:3 and F6:7 generations of quantitative trait loci for grain yield and yield components in maize. Theor Appl Genet 92:817–826
Bai X, Luo L, Yan W, Kovi MR, Zhan W, Xing Y (2010) Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet 11:16
Bao J (2014) Genes and QTLs for rice grain quality improvement. In: Yan WG, Bao JS (ed) Rice-germplasm genetics and improvement, chap 9. InTech, pp 239–278
Blummel M, Grings E, Erenstein O (2013) Potential for dual-purpose maize varieties to meet changing maize demands: synthesis. Field Crop Res 153:107–112
Borrás L, Gambín BL (2010) Trait dissection of maize kernel weight: towards integrating hierarchical scales using a plant growth approach. Field Crop Res 118:1–12
Brown TA, Jones MK, Powell W, Allaby RG (2009) The complex origins of domesticated crops in the Fertile Crescent. Trends Ecol Evol 24:103–109
Coles ND (2009) The genetic architecture of maize photoperiod sensitivity revealed by recombinant inbred line, backcross, and heterogeneous inbred family populations. Ph.D. thesis, Department of Crop Science, North Carolina State University, Raleigh. http://www.lib.ncsu.edu/resolver/1840.16/4750
Doerge RW, Churchill GA (1996) Permutation tests for multiple loci affecting a quantitative character. Genetics 142:285–294
Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112:1164–1171
Guo J, Su G, Zhang J, Wang G (2008) Genetic analysis and QTL mapping of maize yield and associate agronomic traits under semi-arid land condition. Afr J Biotechnol 7:12
Gupta PK, Rustgi S, Kumar N (2006) Genetic and molecular basis of grain size and grain number and its relevance to grain productivity in higher plants. Genome 49:565–571
Hong Y, Chen L, Du LP, Su Z, Wang J, Ye X, Qi L, Zhang Z (2014) Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct Integr Genomics 14:341–349
Hu Z, He H, Zhang S, Sun F, Xin X, Wang W, Qian X, Yang J, Luo X (2012) A Kelch motif-containing serine/threonine protein phosphatase determines the large grain QTL trait in rice. J Integr Plant Biol 54:979–990
Huang R, Jiang L, Zheng J, Wang T, Wang H, Huang Y, Hong Z (2013) Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci 18:218–226
Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42:1027–1030
Li Y, Wang Y, Shi Y, Song Y, Wand T, Li Y (2009) Correlation analysis and QTL mapping for traits of kernel structure and yield components in maize. Sci Agric Sin 42:408–418
Li M, Guo X, Zhang M, Wang X, Zhang G, Tian Y, Wang Z (2010a) Mapping QTLs for grain yield and yield components under high and low phosphorus treatments in maize (Zea mays L.). Plant Sci 178:454–462
Li Q, Li L, Yang X, Warburton ML, Bai G, Dai J, Li J, Yan J (2010b) Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol 10:143
Li Q, Yang X, Bai G, Warburton ML, Mahuku G, Gore M, Dai J, Li J, Yan J (2010c) Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theor Appl Genet 120:753–763
Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43:1266–1269
Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen OA (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci 99:5460–5465
Liu XH, Zheng P, Tan ZB, Li Z, He C (2010) Quantitative trait locus (QTL) mapping for 100-kernel weight of maize (Zea mays L.) under different nitrogen regimes. Afr J Biotechnol 49:8283–8289
Liu Z, Ji H, Cui Z, Wu X, Duan L, Feng X, Tang J (2011) QTL detected for grain-filling rate in maize using a RIL population. Mol Breed 27:25–36
Liu Y, Wang L, Sun C, Zhang Z, Zheng Y, Qiu F (2014) Genetic analysis and major QTL detection for maize kernel size and weight in multi-environments. Theor Appl Genet 127:1019–1037
Liu J, Deng M, Guo H, Raihan S, Luo J, Xu Y, Dong X, Yan J (2015) Maize orthologs of rice GS5 and their trans-regulator are associated with kernel development. J Integr Plant Biol. doi:10.1111/jipb.12421
Mackay TF, Stone EA, Ayroles JF (2009) The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10:565–577
Maitz M, Santandrea G, Zhang Z, Lal S, Hannah LC, Salamini F, Thompson RD (2000) rgf1, a mutation reducing grain filling in maize through effects on basal endosperm and pedicel development. Plant J 23:29–42
Messmer R, Fracheboud Y, Bänziger M, Vargas M, Stamp P, Ribaut JM (2009) Drought stress and tropical maize: QTL-by-environment interactions and stability of QTLs across environments for yield components and secondary traits. Theorl Appl Genet 119:913–930
Monaco MK, Sen TZ, Dharmawardhana PD, Ren L, Schaeffer M, Naithani S, Amarasinghe V, Thomason J, Harper L, Gardiner J, Cannon EK (2013) Maize metabolic network construction and transcriptome analysis. Plant Genome 6:1–12
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nikolić A, Anđelković V, Dodig D, Mladenović-Drinić S, Kravić N, Ignjatović-Micić D (2013) Identification of QTL-s for drought tolerance in maize, II: yield and yield components. Genetika 45:341–350
Pan Q, Li L, Yang X, Tong H, Xu S, Li Z, Li W, Muehlbauer GJ, Li J, Yan J (2015) Genome wide recombination dynamics are associated with phenotypic variation in maize. New Phytol. doi:10.1111/nph.13810
Paterson AH, De Verna JW, Lanini B, Tanksley SD (1990) Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics 124:735–742
Paterson AH, Lin YR, Li Z, Schertz KF, Doebley JF, Pinson SR, Liu SC, Stansel JW, Irvine JE (1995) Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269:1714–1718
Peng B, Li Y, Wang Y, Liu C, Liu Z, Tan W, Zhang Y, Wang D, Shi Y, Sun B (2011) QTL analysis for yield components and kernel-related traits in maize across multi-environments. Theor Appl Genet 122:1305–1320
Poehlman JM, Sleper DA (1995) Breeding soybean. In: Breeding field crops, 4th edn. Iowa State University Press, Ames, pp 300–318
Pozzi C, Rossini L, Vecchietti A, Salamini F (2004) Gene and genome changes during domestication of cereals. In: Gupta PK, Varshney RK et al (eds) Cereal genomics. Springer, pp 165–198
Prado SA, César G, López M, Senior L, Borrás L (2014) The genetic architecture of maize (Zea mays L.) kernel weight determination. G3 (Bethesda) 4:1611–1621
Qi X, Zhao Y, Jiang L, Cui Y, Wang Y, Liu B (2009) QTL analysis of kernel soluble sugar content in super sweet corn. Afr J Biotechnol 8:6913–6917
Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang L, Gao JP, Lin HX (2012) The novel quantitative trait locus GL3. 1 controls rice grain size and yield by regulating Cyclin-T1; 3. Cell Res 22:1666–1680
Ramya P, Chaubal A, Kulkarni K, Gupta L, Kadoo N, Dhaliwal H, Chhuneja P, Lagu M, Gupt V (2010) QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.). J Appl Genet 51:421–429
Revilla P, Butrón A, Malvar R, Ordás R (1999) Relationship among kernel weight, early vigor, and growth in maize. Crop Sci 39:654–658
Ross AJ, Hallauer AR, Lee M (2006) Genetic analysis of traits correlated with maize ear length. Maydica 151(2):301
Sanguinetti CJ, Dias Neto E, Simpson AJG (1994) Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 17:915–919
Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40:1023–1028
Song XJ, Ashikari M (2008) Toward an optimum return from crop plants. Rice 1:135–143
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39:623–630
SPSS Inc. (1999) SPSS base 10.0 for Windows user’s guide. SPSS Inc., Chicago
Statista (2014) Worldwide production of grain in 2013, by type. http://www.statista.com/statistics/263977/world-grain-production-by-type/. Accessed 20 Aug 2015
Tang J, Yan J, Ma X, Teng W, Wu W, Dai J, Dhillon BS, Melchinger AE, Li J (2010) Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor Appl Genet 120:333–340
Tanksley SD (1988) Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335:6170
Team RC (2014) A language and environment for statistical computing 2012. R Foundation for Statistical Computing, Vienna. ISBN 3-900051-07-0
Thévenot C, Simond-Côte E, Reyss A, Manicacci D, Trouverie J, Le Guilloux M, Ginhoux V, Sidicina F, Prioul J-L (2005) QTLs for enzyme activities and soluble carbohydrates involved in starch accumulation during grain filling in maize. J Exp Bot 56:945–958
Tuinstra M, Ejeta G, Goldsbrough P (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95:1005–1011
Wan X, Weng J, Zhai H, Wang J, Lei C, Liu X, Guo T, Jiang L, Su N, Wan J (2008) Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on chromosome 5. Genetics 179:2239–2252
Wang S, Basten CJ, Zeng ZB (2010) Windows QTL cartographer 2.5. Department of statistics. North Carolina State University, Raleigh. http://www.statgen.ncsu.edu/qtlcart/WQTLCart.htm
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954
Weng J, Gu S, Wan X, Gao H, Guo T, Su N, Lei C, Zhang X, Cheng Z, Guo X (2008) Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res 18:1199–1209
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Xu Y (2010) Molecular plant breeding. CAB International, Wallingford
Yan B, Liu R, Li Y, Wang Y, Gao G, Zhang Q, Liu X, Jiang G, He Y (2014) QTL analysis on rice grain appearance quality, as exemplifying the typical events of transgenic or backcrossing breeding. Breed Sci 64:231
Yang X, Gao S, Xu S, Zhang Z, Prasanna BM, Li L, Li J, Yan J (2011) Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breed 28:511–526
Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, Fang W, Feng H, Xie W, Lian X, Wang G (2014) Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5:5087
Yu J, Buckler ES (2006) Genetic association mapping and genome organization of maize. Curr Opin Biotechnol 17:155–160
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang X, Wang J, Huang J, Lan H, Wang C, Yin C, Wu Y, Tang H, Qian Q, Li J (2012) Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc Natl Acad Sci 109:21534–21539
Zhang W, Sun P, He Q, Shu F, Wang J, Deng H (2013) Fine mapping of GS2, a dominant gene for big grain rice. Crop J 1:160–165
Zhang Z, Liu Z, Hu Y, Li W, Fu Z, Ding D, Li H, Qiao M, Tang J (2014) QTL analysis of kernel-related traits in maize using an immortalized F2 population. PLoS One 9:e89645
Acknowledgments
We thankfully acknowledge and greatly appreciate Mr. Xiongbing Yan for his admirable field work. This research was supported by the Genetically Modified Organisms Breeding Major Projects (2014ZX0800944B) and the National Natural Science Foundation of China (31222041).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Communicated by N. de Leon.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Raihan, M.S., Liu, J., Huang, J. et al. Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58 × SK maize population. Theor Appl Genet 129, 1465–1477 (2016). https://doi.org/10.1007/s00122-016-2717-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2717-z