Abstract
Key message
Mapping studies confirm that resistance to Ug99 race of stem rust pathogen in Aegilops tauschii accession Clae 25 is conditioned by Sr46 and markers linked to the gene were developed for marker-assisted selection.
Abstract
The race TTKSK (Ug99) of Puccinia graminis f. sp. tritici, the causal pathogen for wheat stem rust, is considered as a major threat to global wheat production. To address this threat, researchers across the world have been devoted to identifying TTKSK-resistant genes. Here, we report the identification and mapping of a stem rust resistance gene in Aegilops tauschii accession CIae 25 that confers resistance to TTKSK and the development of molecular markers for the gene. An F2 population of 710 plants from an Ae. tauschii cross CIae 25 × AL8/78 were first evaluated against race TPMKC. A set of 14 resistant and 116 susceptible F2:3 families from the F2 plants were then evaluated for their reactions to TTKSK. Based on the tests, 179 homozygous susceptible F2 plants were selected as the mapping population to identify the simple sequence repeat (SSR) and sequence tagged site (STS) markers linked to the gene by bulk segregant analysis. A dominant stem rust resistance gene was identified and mapped with 16 SSR and five new STS markers to the deletion bin 2DS5-0.47-1.00 of chromosome arm 2DS in which Sr46 was located. Molecular marker and stem rust tests on CIae 25 and two Ae. tauschii accessions carrying Sr46 confirmed that the gene in CIae 25 is Sr46. This study also demonstrated that Sr46 is temperature-sensitive being less effective at low temperatures. The marker validation indicated that two closely linked markers Xgwm210 and Xwmc111 can be used for marker-assisted selection of Sr46 in wheat breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) stem rust, caused by Puccinia graminis Pers.:Pers. f. sp. tritici Eriks. & E. Henn. (abbreviated as Pgt), is one of the most devastating diseases of wheat. P. graminis f. sp. tritici race TTKSK (known as Ug99), which was detected in Uganda in 1998 (Pretorius et al. 2000) and has virulence to most stem rust resistance (Sr) genes previously deployed in wheat cultivars, is a major threat to global wheat production (Singh et al. 2008, 2011). Since 2007, an enormous effort, under the coordination of the Borlaug Global Rust Initiative (http://www.globalrust.org), has been undertaken to identify TTKSK-effective Sr genes from wheat and its relatives and deploy them into wheat cultivars in the targeted regions. So far, at least 23 Sr genes cataloged in wheat have been identified as effective against TTKSK (Singh et al. 2011; McIntosh et al. 2013). However, a majority of these genes, such as Sr24, Sr25, Sr26, Sr32, Sr36, Sr37, Sr39, Sr40, Sr43, Sr44, Sr47, Sr50, Sr51, Sr52, and Sr53, were transferred from related species in the secondary and tertiary gene pools of wheat (Dundas et al. 2007; Anugrahwati et al. 2008; Liu et al. 2011a, b, 2013; Niu et al. 2011, 2014; Qi et al. 2011; Klindworth et al. 2012; Mago et al. 2013; McIntosh et al. 2013).
Among these TTKSK-effective alien genes, only four, including Sr24, Sr25, and Sr26 derived from tall wheatgrasses (Thinopyrum ponticum (Podp.) Barkworth & D.R. Dewey) and Sr36 from Triticum timopheevii (Zhuk.) Zhuk. have been deployed in wheat cultivars (McIntosh et al. 1995; Jin and Singh 2006; Tsilo et al. 2008). Two new Ug99 variant races TTKST and TTTSK, which are virulent to Sr24 and Sr36, respectively, were identified in Kenya in 2006 and 2007 (Jin et al. 2008, 2009). Recently, alien chromosome segments carrying five other genes, including Sr32 (Mago et al. 2013), Sr39 (Niu et al. 2011), and Sr47 (Klindworth et al. 2012) from goatgrass species Aegilops speltiodes, Sr43 (Niu et al. 2014) from Th. ponticum, and Sr50 (Anugrahwati et al. 2008) from rye (Secale cereale L.) have been substantially reduced by chromosome engineering. However, the effects of residual chromatin from Ae. speltoides, Th. ponticum and rye on wheat yield and quality have not been evaluated. For deploying Sr genes into new wheat cultivars, it is recognized that pyramiding several Sr genes can lead to increased durability of resistance (Singh et al. 2006, 2011). However, stacking several alien genes into one genotype will increase the total amount of alien chromatin, which may result in potential negative effects on cultivar yield stability and end-use quality.
Contrary to the genes derived from the secondary and tertiary gene pools, the useful genes from the primary gene pool of wheat are rarely associated with the deleterious linkage drags. Therefore, TTKSK-effective Sr genes from the wheat primary gene pool are highly desirable for accelerating development of TTKSK-resistant cultivars through gene pyramiding. So far, only 14 Sr genes cataloged in wheat (McIntosh et al. 2013), including Sr2, Sr13, Sr21, Sr22, Sr28, Sr33, Sr35, Sr42, Sr45, Sr46, Sr55, Sr56, Sr57, and SrWeb derived from primary gene pool, have been identified to be resistant to TTKSK and/or its variant races (Jin et al. 2007; Hiebert et al. 2010, 2011; Rouse et al. 2011a, 2012; Rouse and Jin 2011; Simons et al. 2011; Singh et al. 2011, 2013; Ghazvini et al. 2012; McIntosh et al. 2013; Bansal et al. 2014; Herrera-Foessel et al. 2014). Some of these genes such as Sr2, Sr55, Sr56, and Sr57 have been characterized to confer adult plant resistance (Singh et al. 2013; Bansal et al. 2014; Herrera-Foessel et al. 2014). They are effective only at the adult plant stage or confer inadequate resistance to stem rust when they are present in the cultivars alone (Singh et al. 2011; Rouse et al. 2014). The utility of Sr21, Sr28, Sr42, Sr45, and SrWeb should be limited to gene pyramids because these genes condition resistance to TTKSK, but are ineffective to many other races (Rouse et al. 2011a, 2012; Rouse and Jin 2011; Ghazvini et al. 2012). Therefore, the TTKSK-effective Sr genes from the primary gene pool are very limited. Further efforts are needed to identify more TTKSK-resistant Sr genes from untapped germplasm collections in the primary gene pool.
Among the various wheat-related species in the primary gene pool, Aegilops tauschii Cosson (2n = 2x = 14, DD), the D-genome donor of hexaploid wheat, is an excellent source of unique genes for resistance to many biotic and abiotic stresses. Numerous genes for resistance to the major wheat diseases and insects identified in Ae. tauschii have been transferred into common wheat through production of synthetic hexaploid wheat (SHW) or by direct hybridization (see reviews by Ogbonnaya et al. 2013). For resistance to stem rust, genes Sr33 (Kerber and Dyck 1979), Sr45 (Marais et al. 1998), and Sr46 (Evans Lagudah, unpublished; McIntosh et al. 2008) were identified in Ae. tauschii and transferred into hexaploid wheat. These three genes are all effective against TTKSK (Jin et al. 2007; Olson et al. 2013a; Periyannan et al. 2013). Both Sr33 and Sr45 have been mapped to chromosome arm 1DS at approximately, 10 cM from each other (Sambasivam et al. 2008). Recently, Sr33 was isolated by map-based cloning and the gene was found to encode a coiled-coil, nucleotide-binding, leucine-rich repeat protein (Periyannan et al. 2013).
The result from recent evaluation of 456 unique Ae. tauschii accessions for stem rust resistance showed that 22.2 % of accessions were resistant to TTKSK with diverse TTKSK infection types and diverse infection type patterns to other races (Rouse et al. 2011a), suggesting that there are more unique genes conferring resistance to TTKSK in the Ae. tauschii germplasm collections. The three new Sr genes, designated as SrTA1662, SrTA10187, and SrTA10171 have been identified in three TTKSK-resistant Ae. tauschii accessions, including TA 1662, TA 10187, and TA 10171, respectively, through molecular mapping (Olson et al. 2013a, b). SrTA1662 was mapped to the same position as Sr33, but race specificity suggested that SrTA1662 is different from Sr33 (Olson et al. 2013a). SrTA10187 and SrTA10171 are located on chromosome 6DS and 7DS, respectively (Olson et al. 2013b).
Among the TTKSK-resistant accessions identified by Rouse et al. (2011a), CIae 25 has a moderate level of resistance to TTKSK and several other races. This accession and its derived SHW line have been extensively used in our program to map the Hessian fly-resistant gene H26 (Yu et al. 2009). A large F2 population was developed from a cross between Ae. tauschii accessions CIae 25 and AL8/78. The objectives of this study were to identify any Sr gene(s) conferring the resistance to TTKSK in CIae 25 and to develop molecular markers for marker-assisted selection for the gene(s) in CIae 25.
Materials and methods
Plant materials
Ae. tauschii accession CIae 25 is maintained in USDA-ARS National Plant Germplasm System (http://www.ars-grin.gov/npgs/). The accession was previously identified as resistant to TTKSK and several other races including TRTTF, QTHJC, and TPMKC (Rouse et al. 2011a) while Ae. tauschii accession AL8/78 was susceptible to TTKSK, TRTTF and all other races that have been tested (Friesen et al. 2008; Zhang 2013). A large F2 population was developed from a cross between CIae 25 and AL8/78 to map the Sr gene(s) in CIae 25.
Stem rust resistance evaluation
The parental accessions CIae 25 and AL8/78 were first tested against three locally-maintained US Pgt races, including TPMKC (isolate TNMKsp1), QCCJB (isolate QCC-2, Zhong et al. 2009), and JCMNC (isolate gb-121; Sun and Steffenson 2005). The large F2 population (710 plants) was then tested against TPMKC. The evaluations of parental lines for resistance to the three local races and the F2 population to TPMKC were performed at the USDA-ARS, Northern Crop Science Laboratory (NCSL), Fargo, ND using similar procedures as described by Niu et al. (2011). Two seeds were sown in a super-cell cone (Stuewe and Sons, Inc., Corvallis, OR) filled with Sunshine SB100 mix (Sun Gro Horticulture Distribution Inc., Bellevue, WA) with an application of Osmocote Plus 15-19-12 fertilizer (Scotts Sierra Horticultural Product Company, Marysville, OH). The seedlings were grown in the greenhouse at 20–23 °C with 16/8 h (day/night) photoperiod. Ten days after sowing, seedlings were inoculated with urediniospores of P. graminis f. sp. tritici. Infection types were recorded for each plant at 14 days after the inoculation, using the infection type (IT) scales described by Stakman et al. (1962), where 0 = immune,; = necrotic flecks, 1 = small necrotic pustules, 2 = small to medium-sized chlorotic pustules with green island, 3 = medium-sized chlorotic pustules, and 4 = large pustules without chlorosis. The seedling plants of the F2 population with IT 2 or lower were considered as resistant while the plants scored with IT 3 and higher were considered susceptible. After the F2 plants were scored, they were moved to a vernalization chamber at 2–3 °C for about 6 weeks and then grown to maturity in the greenhouse at 20–23 °C with 16/8 h (day/night) photoperiod.
The parental accessions and 116 F2:3 families with adequate seeds harvested from the TPMKC-susceptible F2 plants and a subset of 14 F2:3 families from the TPMKC-resistant F2 plants were evaluated for their resistance to race TTKSK (isolate 04KEN156/04) at the USDA-ARS Cereal Disease Laboratory (CDL), St. Paul, MN. The evaluation of the parental accessions and their F2:3 families against TTKSK were performed according to the procedure described by Rouse et al. (2011b). For F2:3 family tests, 20 seeds per family were planted in trays filled with vermiculite. Plants were inoculated with urediniospores 8 days after sowing and were grown in a greenhouse at 22 ± 2 and 18 ± 2 °C (day and night, respectively) with supplemental lighting for a photoperiod of 16 h. Plants were scored at 14 days after inoculation using the Stakman et al. (1962) IT scales as described above. Chi-square (χ 2) goodness-of-fit tests were performed on segregating families.
SSR marker analysis
Leaf tissue was sampled from the parental accessions and each susceptible F2 plant. DNA was extracted from freeze-dried leaf samples using the procedure of Dellaporta et al. (1983). Bulked segregant analysis (BSA) was used to identify SSR markers linked to the stem rust resistance gene. Two bulks of DNA were made by pooling equal amounts of DNA from eight homozygous susceptible plants and the eight most resistant plants with the lowest IT scores. After the SSR markers linked to the Sr gene were identified, they were further analyzed on a subpopulation composed of 179 susceptible F2 plants. The polymerase chain reaction (PCR) amplification was carried out as described by Yu et al. (2009, 2010). The PCR products were separated on 6 % poly-acrylamide gels using the procedure of Yu et al. (2009). The gels were stained with Gel-Red, and then scanned with a Typhoon 9410 imager (Molecular Dynamics, Ithaca, NY, USA). The band sizes in the gel images were analyzed using Kodak 1D Image Analysis Software version 3.6.5 K2 (Eastman Kodak, Rochester, NY).
Sequence tagged site (STS) marker development
After the Sr gene was mapped to the distal region of chromosome 2D short arm (2DS5-0.47-1.00) based on the genetic linkage analysis with SSR markers, a total of 194 expressed sequence tags (ESTs) (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi) were used to develop sequence tagged site (STS) markers. The genomic sequences of the 2DS collinear region in the Brachypodium (http://www.phytozome.org/cgi-bin/gbrowse/brachy/) and rice genomes (http://www.phytozome.org/cgi-bin/gbrowse/rice/) were also employed for STS marker development. The program Primers3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3) (Koressaar and Remm 2007; Untergrasser et al. 2012) was used to design STS primers. The STS marker analysis was conducted in the same way as for SSR markers.
Genetic linkage analysis and identity confirmation of the stem rust resistance gene
A total of 21 polymorphic SSR and STS markers around the stem rust resistance gene were analyzed on the 179 F2 plants susceptible to TPMKC. In this homozygous susceptible subpopulation, the F2 plants carrying the marker alleles from the resistant parent CIae 25 were identified as recombinants. The recombination frequencies were calculated using the program Map Manager QTX (Manly et al. 2001) with the algorithm for an F2 population. The genetic linkage map was constructed using the program Map Manager QTX (Manly et al. 2001). A probability of 0.001 (equivalent to LOD = 3) was used for genetic linkage map construction. The Kosambi mapping function (Kosambi 1944) was used to calculate the genetic distance. The map was drawn using the program Mapchart 2.2 (Voorrips 2002).
The genetic linkage analysis showed that the Sr gene in CIae 25 was located in the same chromosomal region as Sr46, which was previously identified from Ae. tauschii accession AUS 18913 by Evans Lagudah (see McIntosh et al. 2008). AUS 18913 is an accession maintained in the Australian Winter Cereals Collection, Tamworth, New South Wales, Australia (http://www2.dpi.qld.gov.au/extra/asp/AusPGRIS/Centres.asp). To confirm that the Sr gene in CIae 25 was Sr46, we surveyed the origin of CIae 25 and AUS 18913 by examining accession details in the USDA-ARS National Plant Germplasm System (http://www.ars-grin.gov/cgi-bin/npgs/acc/display.pl?1000898) and AusPGRIS—Australian Plant Genetic Resource Information Service (http://www2.dpi.qld.gov.au/extra/asp/auspgris/), respectively. We also tested CIae 25, AUS 18913, and TA 1703 (synonymous with AUS 18913) for their seedling reactions to six Pgt races and genotyped them with the SSR markers previously mapped to seven D-genome chromosomes. The original seeds of AUS 18913 were obtained from the Commonwealth Scientific and Industrial Research Organization (Canberra, Australia) and the seed sample used in this study was currently maintained in the CDL. The seeds of TA 1703 were provided by Jon Raupp, Wheat Genetics and Genomics Resource Center (Manhattan, Kansas, USA).
The two Ae. tauschii accessions AUS 18913 and TA 1703 carrying Sr46 previously showed different seedling infection types to races QTHJC and RKQQC tested in different locations (Rouse et al. 2011a). To determine if the variation can be attributed to different environments, we tested the reaction of lines with Sr46 to Pgt races in three environments. The three Ae. tauschii accessions and checks were tested independently for reactions to six Pgt races RKQQC (isolate 99KS76A-1), QTHJC (isolate 75ND717C), QFCSC (isolate 06ND76C), MCCFC (isolate 59KS19), TPMKC (isolate 74MN1409), and TTTTF (isolate 01MN84A-1-2) at the CDL and five races RKQQC (isolate CRL-3, Zhong et al. 2009), QTHJC [isolate 64E(1)sp1], QFCSC (isolate 370C), MCCFC (isolate A-5), and TPMKC (isolate TNMKsp1) at the NCSL. At each location, the three Ae. tauschii accessions and checks were evaluated in the greenhouse and at two temperature (low and high) conditions in growth chambers. The greenhouses were maintained at 21 °C at the NSCL and 22 °C during the day and 19 °C during the night at the CDL with supplemental lighting for a photoperiod of 16 h. The growth chambers at both locations were set to 15 and 18 °C in the dark (8 h) and light (16 h), respectively, for the experiments at low temperature and to 20 and 24 °C in the dark (8 h) and light (16 h), respectively, for high temperature. The common wheat cultivar ‘Chinese Spring’, Ae. tauschii AL8/78, and the durum line Rusty (PI 639869) (Klindworth et al. 2006) were used as susceptible checks. The durum cultivar ‘Langdon’ (CItr 13165), which is known to have Sr13 (Simons et al. 2011), was used as a resistant check. Two SHW lines SW8 (PI 639730) (Xu et al. 2006) and SW73 derived from the crosses of Langdon and Rusty with CIae 25 were also included. In addition, common wheat line LMPG-6 was included as a susceptible check in the experiments conducted at the CDL. The stem rust inoculation and scoring in the experiments were performed using the same procedures described above except that the plants at high temperature conditions were scored at 11 and 13 days after the inoculation at the CDL and the NCSL, respectively.
For marker genotyping analysis, the three Ae. tauschii accessions CIae 25, AUS 18913, and TA 1703, along with checks (AL8/78, SW8, SW73, Langdon, and Rusty), were genotyped using 18 SSR markers that were previously mapped to chromosomes 1D through 7D (Somers et al. 2004) and one STS marker closely linked to the Sr gene in CIae 25 developed in this study. The 18 SSR markers (Xwmc222, Xwmc336, and Xwmc432 on chromosome 1D, Xgwm210 and Xwmc111 on 2D, Xgdm99 and Xgwm191 on 3D, Xgwm213 and Xwmc182 on 4D, Xcfa2141, Xgwm16, Xwmc289, and Xwmc405 on 5D, Xcfd49, Xcfd75, and Xgdm132 on 6D, Xcfa2040, Xcfd21, and Xwmc273 on 7D, and Xgwm133 on 4D and 6D) were selected based on evaluation of 29 markers for their polymorphism between CIae 25 and AL8/78. The procedure for marker genotyping was the same as described above.
Molecular marker validation for marker-assisted selection
A set of 31 common wheat cultivars and a breeding line were used to validate the markers closely-linked to the stem rust resistance gene for marker-assisted selection (MAS). Among them, seven are eastern Chinese winter wheat cultivars, 23 are the hard red spring wheat cultivars developed in the states of Minnesota, North Dakota, and South Dakota in the U.S., and two are the hard red winter wheat cultivar (‘Newton’) and breeding line (IL06-14262) from Kansas and Illinois, respectively. Ae. tauschii accessions CIae 25, AUS 18913, and TA 1703 and two SHW lines SW8 and SW73 derived from CIae 25 were included as positive checks and Ae. tauschii AL8/78 and durum wheat Langdon and Rusty were used as negative checks, The DNA extraction and marker analysis were performed using the procedures described above.
Results
Resistance of Ae. tauschii accession CIae 25 to TTKSK and other P. graminis f. sp. tritici races
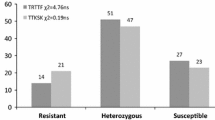
Evaluation of the two parental Ae. tauschii accessions CIae 25 and AL8/78 showed that CIae 25 was resistant to races TTKSK (IT 2), TPMKC (IT 1−), QCCJB (IT 1+), and JCMNC (IT 1+) while AL8/78 showed susceptible reactions to these races (IT 3−, 3+, 3, and 3+, respectively). Because CIae 25 and AL8/78 exhibited a stronger differential reaction to race TPMKC than the other races (Fig. 1), we used this race to evaluate 710 F2 plants from the cross between CIae 25 and AL8/78. From the 710 F2 plants, we initially identified 180 susceptible plants. To verify if the resistance gene detected in the F2 population using race TPMKC was the same gene for resistance to race TTKSK, 116 F2:3 families with adequate seeds derived from the F2 plants susceptible to TPMKC and a sample set of 14 F2:3 families from the F2 plants resistant to TPMKC were evaluated for their reactions to TTKSK. Among the 14 F2:3 families derived from TPMKC-resistant F2 plants, 10 families (16–20 plants per family) were homozygous for resistance to TTKSK (IT 2) and four families segregated for resistant (IT 2) and susceptible (ITs 3 to 3+) plants (Table 1). The segregation of resistant and susceptible plants in the four families fit a 3:1 ratio (Table 1), indicating that the stem rust resistance in CIae 25 is controlled by a single dominant gene. Among the 116 F2:3 families derived from the race TPKMC-susceptible F2 plants, 104 families were homozygous for susceptibility to race TTKSK (IT 3–3+), seven families showed all the plants with intermediate ITs (ITs 2+3 or 32+), four families showed a mixture of intermediate (IT 2+3) and susceptible reactions, and one family segregated for resistant and susceptible reactions. The family segregating for resistant and susceptible plants should be derived from a heterozygous F2 plant which was misclassified as susceptible to TPMKC. The F2:3 family tests substantiated that the Sr gene in CIae 25 confers the resistance to both TPMKC and TTKSK.
Molecular mapping of the stem rust resistance gene in Ae. tauschii CIae 25
Molecular mapping was performed using a population of 179 F2 plants that were homozygous susceptible to race TPMKC (the F2 plant which was misclassified as susceptible to TPMKC was excluded). Since stem rust resistance genes are frequently identified on chromosome 2B of wheat and group 2 chromosomes of wild relative species such as Ae. speltoides, we started BSA with the SSR markers previously mapped to chromosome 2D. The result showed that 11 SSR markers in the distal region of the short arm of chromosome 2D were polymorphic between the two parents as well as between the resistant and susceptible bulks. The 11 SSR markers were then analyzed on the 179 F2 susceptible plants used as a mapping population. The initial linkage analysis showed that the Sr gene was proximal to Xbarc124 and Xgwm210, and distal to Xcfd36, Xgwm455, and Xgwm296, which are all mapped to the distal deletion bin 2DS5-0.47-1.00 (Sourdille et al. 2004), indicating that the gene is located within this deletion bin.
Based on the preliminary linkage analysis, we used the sequences of the ESTs mapped to the distal deletion bin (2DS5-0.47-1.00) and co-linearity with Brachypodium or rice to develop new markers linked to the Sr gene in CIae 25. Five co-dominant STS markers were developed and designated as Xrwgs33 to Xrwgs37 (Table 2). Xrwgs36 amplified two DNA fragments in estimated sizes of 1,200 and 1,240 bp, respectively, while the other four STS markers, including Xrwgs33, Xrwgs34, Xrwgs35, and Xrwgs37, all amplified polymorphic bands of less than 500 bp (Fig. 2). Four STS markers, including Xrwgs33, Xrwgs35, Xrwgs36, and Xrwgs37, amplified highly intense bands. Two STS markers Xrwgs33 and Xrwgs34 amplified the bands with a large size difference between the parents (Fig. 2). To determine the relationship between Sr46 and Sr6 on chromosome arm 2DS (Tsilo et al. 2010), the SSR markers around Sr6 were also screened for polymorphism between CIae 25 and AL8/78 and additional five polymorphic SSR markers in the region were mapped with the mapping population.
Gel images of five new codominant sequence-tagged site (STS) markers linked to the stem rust resistance gene in Ae. tauschii accession CIae 25. Amplicons of five codominant STS markers were separated on polyacrylamide gels and generated polymorphic bands distinguishing the CIae 25 (lane 1) and AL8/78 (lane 2) parents of the CIae 25 × AL8/78 F2 population. Lanes 3 through 8 contain samples from the population with the parental marker genotypes. The numbers on the left and right indicate size of polymorphic bands and DNA ladder (lane L) in base pair (bp), respectively
For determining the identities of CIae 25 and AUS 18913 in which Sr46 was originally identified, we found that CIae 25 was originally collected from Gilan, Iran in 1955 with original accession number 2147 designated by Kyoto University, Kyoto, Japan (USDA-ARS, National Genetic Resources Program. Available: http://www.ars-grin.gov/cgi-bin/npgs/acc/display.pl?1000898. Accessed on November 23 2014). In the AusPGRIS, AUS 18913 has additional names listed as CIae 25 and 2147 (Supplementary material 1), clearly indicating that AUS 18913 and CIae 25 are the same accession that was assigned different names in the two germplasm systems in Australia and the United States, respectively. The genotyping analysis revealed that CIae 25, AUS 18913, and TA 1703 produced identical amplicons from the STS marker Xrwgs33 linked to the Sr gene in CIae 25 and 18 SSR markers that were polymorphic between CIae 25 and AL8/78. As shown in Fig. 3, four illustrative SSR markers Xwmc432, Xgwm210, Xwmc405, and Xcfa2040 produced bands with sizes of 217 and 238, 178, 217, and 284 and 316 bp, respectively, from the three Ae. tauschii accessions TA 1703, AUS 18913, and CIae 25 and the two SHW lines SW8 and SW73. The results from the stem rust evaluation showed that CIae 25 had the same or similar reactions to the six Pgt races as AUS 18913 and TA 1703 when they were tested at the various environmental conditions (Table 3; Fig. 4; Supplementary material 2–6). Therefore, marker genotyping and stem rust responses confirmed that the CIae 25 has the same genotype as AUS 18913 and TA 1703 and that the Sr gene in CIae 25 is the gene Sr46 identified in AUS 18913.
Gel images of four Ae. tauschii accessions and controls analyzed with four illustrative simple sequence repeat (SSR) markers. The numbers on the top of each image indicate the samples for Ae. tauschii accessions AL8/78 (lane 1), TA 1703 (lane 2), AUS 18913 (lane 3), and CIae 25 (lane 4), synthetic hexaploid wheat lines SW8 and SW73 having pedigrees of Langdon/CIae 25 (lane 5) and Rusty/CIae 25 (lane 6), respectively, and durum wheat line Rusty (lane 7) and cultivar Langdon (lane 8). The numbers on the left of each image indicate size of polymorphic bands between AL8/78 and the other three Ae. tauschii accessions (TA 1703, AUS 18913, and CIae 25) in base pair (bp). The numbers on the right of each image indicate size of DNA ladder (lane L). The four SSR markers Xwmc432, Xgwm210, Xwmc405, and Xcfa2040 were previously mapped onto chromosome 1D, 2D, 5D, and 7D, respectively (Somers et al. 2004). They produced identical amplicons from the three Ae. tauschii accessions TA 1703, AUS 18913, and CIae 25
Seedling reactions of four Ae. tauschii accessions AL8/78, CIae 25, TA 1703, and AUS 18913, common wheat cultivar ‘Chinese Spring’ (susceptible check), and durum wheat cultivar ‘Langdon’ (resistant check) to races RKQQC and QTHJC of P. graminis f. sp. tritici in greenhouse experiment. The three Ae. tauschii accessions CIae 25, TA 1703, and AUS 18913 showed the similar reactions to the two races
Stem rust tests at different environmental conditions showed that Sr46 is a temperature-sensitive gene. In the greenhouse experiments, the three Ae. tauschii accessions exhibited similar moderate or high levels of resistance to races RKQQC, QTHJC, QFCSC, TPMKC, and TTTTF and intermediate reactions to race MCCFC (Table 3; Fig. 4). At the low temperature environment (15/18 °C), the accessions displayed susceptible or intermediate reactions to races RKQQC, QTHJC, and MCCFC and they also had susceptible reactions or decreased levels of resistance to races QFCSC and TTTTF (Supplementary material 2–6). At the high temperature environment (20/24 °C), the three Ae. tauschii accessions had susceptible reactions only to RKQQC and MCCFC in the CDL test. They had intermediate or susceptible reactions to RKQQC at the NCSL and TTTTF at the CDL. The data suggest Sr46 was less effective at the low temperature environment compared to the high temperature environment and that Sr46 was most effective in the greenhouse tests at both the CDL and the NCSL. Infection type data also showed that the SHW line SW8 was resistant to all six races but SW73 was susceptible to five of the six races except for TPMKC in all the experiments (Table 3; Supplementary material 2–6), suggesting that the resistance of Sr46 is suppressed in the SHW having Rusty background. Because SW8 had a similar level of resistance compared to its durum parent Langdon in all the experiments, the resistance in SW8 may be due to the presence of Sr13 or other genes from Langdon.
The linkage map of Sr46 consists of 16 SSR and five STS markers and its total length was 64.0 cM (Fig. 5). The average genetic distance between two markers was 3.2 cM. Among the 16 SSR markers, six, including Xbarc124, Xgwm210, Xcfd36, Xgwm455, Xgwm296, and Xcfd43, were previously mapped to deletion bin 2DS5-0.47-1.00; marker Xgwm102 was mapped to deletion bin 2DS1-0.33-0.47; two SSR markers, Xgwm261 and Xgwm484, were mapped to deletion bin C-2DS1-0.33; and the other nine SSR markers are unassigned (Sourdille et al. 2004). Based on the genetic linkage map, several markers were identified to be closely linked to Sr46 in the distal region of chromosome arm 2DS (Fig. 5). On the distal side of the gene, there are two SSR markers, Xgwm210 and Xbarc124, linked to the gene at a distance of 3.9 and 7.7 cM, respectively (Fig. 5). On the proximal side of the gene, there are three SSR markers and one STS marker, including Xcfd36, Xwmc111, and Xgwm455, and Xrwgs33, linked to the gene at distances of 5.6, 5.9, 7.6, and 7.9, respectively.
Alignment of genetic and physical location of Sr46 and its linked markers on chromosome arm 2DS. a Genetic map of 2DS on CIae 25 × AL8/78 F2 population. b Physical map of 2DS of Chinese Spring, which was reproduced from Sourdille et al. (2004). Deletion bins are indicated by rectangle boxes
Validation of markers for marker-assisted selection
Three markers, including SSR markers Xgwm210 and Xwmc111 and STS marker Xrwgs33, which are closely linked to Sr46, were analyzed on a panel of 31 common wheat cultivars and a breeding line (Table 4; Supplementary material 7). Because flanking marker Xcfd36 produced multiple bands, it was not used for the validation. Marker Xgwm210 amplified a 178-bp band from CIae 25 and a 174-bp band from AL8/78 and all the wheat cultivars and line IL06-14262. This marker also amplified a 176-bp band from IL06-14262 and a 172-bp band from all the cultivars except for Jinan 177 from which a 173-bp band was produced. Marker Xwmc111 amplified two bands with sizes of 385 and 404 bp from CIae 25 and two bands with sizes of 394 and 410 bp from AL8/78. Among the 32 wheat cultivars and line, nine cultivars had the marker allele of AL8/78 (394 + 410 bp) and all other cultivars and the line had different amplicons from both CIae 25 and AL8/78. This result suggests that SSR markers Xgwm210 and Xwmc111 can be effectively used in deployment of Sr46 into diverse backgrounds of common wheat germplasm.
Marker Xrwgs33 generated a 226-bp band from CIae 25 and a 200-bp band from AL8/78. It produced 200-bp bands from six of the seven Chinese cultivars, a 224-bp band from three cultivars, and a 151-bp band from the remaining 22 cultivars and line IL06-14262 (Table 4; Supplementary material 7). Thus, Xrwgs33 produced amplicons with a large size difference (75 bp) between the marker allele in CIae 25 and the alleles of a majority of the wheat cultivars and line. However, its usefulness in marker-assisted selection for Sr46 may need to be further tested because it amplified two bands (approximately, 226 and 228 bp) from Rusty and Langdon and a band (225 bp) from the line IL06-14262 and 28 cultivars with the same or similar size as the 226-bp band from CIae 25.
Discussion
Stem rust evaluation results from Rouse et al. (2011a) and this study suggest that Ae. tauschii accession CIae 25 conferred moderate levels of resistance to races TTKSK, TRTTF, QTHJC, and RKQQC and high levels of resistance to TPMKC, QCCJB, and JCMNC. Phenotyping of the CIae 25 × AL8/78 F2 population indicated that a single gene in CIae 25 confers resistance to races TTKSK and TPMKC. Using a population of 179 susceptible F2 plants derived from the cross between CIae 25 and AL8/78, we mapped the Sr gene to the distal region of the deletion bin 2DS5-0.47-1.00 on the short arm of chromosome 2D with 16 SSR and five newly developed STS markers. The population of 179 susceptible F2 plants is genetically equivalent to 358 gametes or 358 doubled haploid lines. The genetic linkage map developed from such a population should be more accurate than the ones based on smaller populations. The order of most SSR markers on the map is consistent with that of the wheat SSR consensus map (Somers et al. 2004). However, several SSR markers are not in the same order as those on the consensus SSR map. The inconsistency between the Sr gene map and the SSR consensus map might be caused by difference in 2D chromosomes between bread wheat and Ae. tauschii because the SSR consensus map was developed primarily based on hexaploid wheat (Somers et al. 2004).
The map location of the Sr gene in CIae 25 identified from our study prompted us to examine its relationship with gene Sr46 previously assigned to chromosome arm 2DS from Ae. tauschii accession AUS 18913. Based on the survey on CIae 25 and AUS 18913 in the USDA-ARS National Plant Germplasm System and AusPGRIS, respectively, we found that AUS 18913 and CIae 25 are actually the same accession originally collected from Iran. In addition, evidences from cross-referenced marker and stem rust test information also support our conclusion that CIae 25 carries Sr46. The information in the Catalog of Gene Symbols for Wheat indicates that Sr46 co-segregated with RFLP marker Xpsr649 at both the diploid and hexaploid levels and a PCR-based marker, csSC46, was developed from a BAC clone containing Xpsr649 (Evans Lagudah, unpublished; McIntosh et al. 2013). Xpsr649 is 0.3 cM from another RFLP marker Xpsr908 on the RFLP map of chromosome 2D (Devos et al. 1993; http://wheat.pw.usda.gov/cgi-bin/cmap/viewer?mapMenu=1&featureMenu=1&corrMenu=1&displayMenu=1&advancedMenu=1&ref_map_accs=Ta-Gale-2D&sub=Draw+Selected+Maps&ref_map_set_acc=Ta-Gale-2D&data_source=GrainGenes), whereas Xpsr908 is closely linked to the SSR marker Xgwm210 on the wheat composite map of chromosome 2D (http://wheat.pw.usda.gov/cgi-bin/cmap/viewer?ref_map_set_acc=Wheat_Composite_2004&ref_map_accs=Wheat-Composite2004-2D&ref_species_acc=1&data_source=GrainGenes), suggesting that Xgwm210 is closely linked to Sr46 identified in AUS 18913. Xgwm210 is physically located in the deletion bin 2DS5-0.47-1.00 (Somers et al. 2004) and it is closely linked to the Sr gene in CIae 25, indicating that the Sr gene in CIae 25 and Sr46 are located in the same chromosome region on the short arm of chromosome 2D.
Rouse et al. (2011a) included two Ae. tauschii accessions (TA 1703 and AUS 18913) for Sr46 in the Ae. tauschii germplasm screening for reactions to multiple races of stem rust. The two accessions exhibited consistent reactions to TTKSK, TRTTF, TTTTF, and TPMKC, but they showed variations in reactions to races QTHJC and RKQQC. Rouse et al. (2011a) indicated that variable reactions of TA 1703 and AUS 18913 to QTHJC and RKQQC might be attributed to unstable intermediate reactions to the two races and/or the screening of two accessions in different experiments. The stem rust evaluations conducted in this study showed that the three Ae. tauschii accessions (CIae 25, AUS 18913, and TA 1703) had similar reactions to each of the six Pgt races including RKQQC and QTHJC when they were tested in the same experiments or in the different experiments with the same or similar environmental conditions. The results from genotyping analysis revealed that the three Ae. tauschii accessions had the same alleles at 19 marker loci distributed in seven D-genome chromosomes. Thus, molecular mapping, genotyping analysis, and the stem rust tests performed in this study confirmed that the resistance to stem rust in Clae 25 is conditioned by Sr46. The stem rust tests also showed that Sr46 is a temperature-sensitive gene that can become less resistant or susceptible to several races such as RKOOC, QTHJC, MCCFC, and TTTTF in both low- and high-temperature conditions. The decreased effectiveness of Sr46 in both the high and low temperature environments compared to the greenhouse environment may be due to the difference in light intensity in the growth chambers compared to the greenhouse. Unfortunately, our experimental design did not permit the evaluation of the effects of light intensity per se. Overall, Sr46 was most ineffective at the lower temperature.
Among the 57 known Sr genes (McIntosh et al. 2013), only Sr6 (Tsilo et al. 2009, 2010) and Sr46 (Evans Lagudah, unpublished, McIntosh et al. 2008) were previously mapped to chromosome arm 2DS. We therefore, examined the relationship between Sr6 and Sr46. The gene Sr6, which originated from common wheat ‘Red Egyptian’ (CItr 12345) (Loegering and Harmon 1969), exists in many Australian and North American wheat cultivars (McIntosh et al. 1995) and has been one of the most important Sr genes in North America due to its resistance against a wide range of races, including TPMKC (Tsilo et al. 2009). However, Sr6 is ineffective against TTKSK (Jin et al. 2007). Sr6 was mapped to the interval between SSR markers Xcfd43 and Xgwm102 on chromosome arm 2DS (Tsilo et al. 2010). On the current genetic linkage map, the Sr46 in CIae 25 is 48.6 cM away from marker Xcfd43 (Fig. 5), which is 1.2 and 1.5 cM away from Sr6 in two different populations (Tsilo et al. 2009, 2010). Thus, Sr46 is at least 48.6 cM away from Sr6. Such a genetic distance may allow pyramiding the two genes together into a cultivar.
The D genome of Ae. tauschii has been involved in the origin and evolution of hexaploid wheat. A previous study based on meiotic pairing showed that the D genome of Ae. tauschii remains largely unaltered and homologous to the D genome of common wheat (Kimber and Zhao 1983). Useful genes in Ae. tauschii could easily be incorporated into common wheat through homologous recombination. Therefore, Sr46 can be transferred through direct crossing between CIae 25 and a common wheat line similar to transfer of SrTA1662, SrTA10187, and SrTA10171, which were introduced into hard winter wheat lines from Ae. tauschii using direct crossing (Olson et al. 2013a, b). The two SSR markers closely linked to Sr46, including Xgwm210 and Xwmc111, which differentiate CIae 25 from all the 31 wheat cultivars and a breeding line genotyped in this study, can be used for marker-assisted selection for Sr46 in wheat breeding programs.
Author contribution statement
SSX, GY, and JBR initiated the project, designed the experiment, developed and evaluated the populations, marker development and analysis and prepared the manuscript; QZ performed stem rust test and genotyping for the three Ae. tauschii accessions and controls; SZ and TLF-assisted stem rust test using local races; MNR and YJ conducted stem rust test using TTKSK; ESL provided information on Sr46 originally identified in Ae. tauschii AUS 18913; all authors provided comments and revisions of the manuscript.
References
Anugrahwati DR, Shepherd KW, Verlin DC, Zhang P, Mirzaghaderi G, Walker E, Francki MG, Dundas IS (2008) Isolation of wheat-rye 1RS recombinants that break the linkage between the stem rust resistance gene SrR and secalin. Genome 51:341–349
Bansal U, Bariana H, Wong D, Randhawa M, Wicker T, Hayden M, Keller B (2014) Molecular mapping of an adult plant stem rust resistance gene Sr56 in winter wheat cultivar Arina. Theor Appl Genet 127:1441–1448
Dellaporta SL, Wood J, Hicks JB (1983) A plant molecular DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Devos KM, Millan T, Gale MD (1993) Comparative RFLP maps of the homoeologous group 2 chromosomes of wheat, rye and barley. Theor Appl Genet 85:784–792
Dundas IS, Anugrahwati DR, Verlin DC, Park RF, Bariana HS, Mago R, Islam AKMR (2007) New sources of rust resistance from alien species: meliorating linked defects and discovery. Aust J Agric Res 58:545–549
Friesen TL, Xu SS, Harris MO (2008) Stem rust, tan spot, Stagonospora nodorum blotch, and Hessian fly resistance in Langdon durum–Aegilops tauschii synthetic hexaploid wheat lines. Crop Sci 48:1062–1070
Ghazvini H, Hiebert CW, Zegeye T, Liu S, Dilawari M, Tsilo T, Anderson JA, Rouse MN, Jin Y, Fetch T (2012) Inheritance of resistance to Ug99 stem rust in wheat cultivar Norin 40 and genetic mapping of Sr42. Theor Appl Genet 125:817–824
Herrera-Foessel SA, Singh RP, Lillemo M, Huerta-Espino J, Bhavani S, Singh S, Lan C, Calvo-Salazar V, Lagudah ES (2014) Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor Appl Genet 127:781–789
Hiebert CW, Fetch T Jr, Zegeye T (2010) Genetics and mapping of stem rust resistance to Ug99 in the wheat cultivar Webster. Theor Appl Genet 121:65–69
Hiebert CW, Fetch TG, Zegeye T, Thomas JB, Somers DJ, Humphreys DG, McCallum BD, Cloutier S, Singh D, Knott DR (2011) Genetics and mapping of seedling resistance to Ug99 stem rust in Canadian wheat cultivars ‘Peace’ and ‘AC Cadillac’. Theor Appl Genet 122:143–149
Jin Y, Singh RP (2006) Resistance in US wheat to recent eastern African isolates of Puccinia graminis f. sp. tritici with virulence to resistance gene Sr31. Plant Dis 90:476–480
Jin Y, Singh RP, Ward RW, Wanyera R, Kinyua M, Njau P, Pretorius ZA, Yahyaoui A (2007) Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp tritici. Plant Dis 91:1096–1099
Jin Y, Szabo LJ, Pretorius ZA, Singh RP, Ward R, Fetch T Jr (2008) Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp tritici. Plant Dis 92:923–926
Jin Y, Szabo LJ, Rouse MN, Fetch T Jr, Pretorius ZA, Wanyera R, Njau P (2009) Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp tritici. Plant Dis 93:367–370
Kerber ER, Dyck PL (1979) Resistance to stem and leaf rust of wheat in Aegilops squarrosa and transfer of a gene for stem rust resistance to hexaploid wheat. In: Ramanujam S (ed) Proceeding of the 5th International Wheat Genetic Symposium. New Delhi, India, pp 358–364
Kimber G, Zhao YH (1983) The D genome of the Triticeae. Can J Genet Cytol 25:581–589
Klindworth DL, Miller JD, Xu SS (2006) Registration of rusty durum wheat. Crop Sci 46:1012–1013
Klindworth DL, Niu Z, Chao S, Friesen TL, Jin Y, Faris JD, Cai X, Xu SS (2012) Introgression and characterization of a goatgrass gene for a high level of resistance to Ug99 stem rust in tetraploid wheat. Genes Genom Genet 2:665–673
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Liu W, Jin Y, Rouse M, Friebe B, Gill B, Pumphrey MO (2011a) Development and characterization of wheat-Ae. searsii Robertsonian translocations and a recombinant chromosome conferring resistance to stem rust. Theor Appl Genet 122:1537–1545
Liu W, Rouse M, Friebe B, Jin Y, Gill B, Pumphrey MO (2011b) Discovery and molecular mapping of a new gene conferring resistance to stem rust, Sr53, derived from Aegilops geniculata and characterization of spontaneous translocation stocks with reduced alien chromatin. Chromosome Res 19:669–682
Liu W, Danilova TV, Rouse MN, Bowden RL, Friebe B, Gill BS, Pumphrey MO (2013) Development and characterization of a compensating wheat-Thinopyrum intermedium Robertsonian translocation with Sr44 resistance to stem rust (Ug99). Theor Appl Genet 126:1167–1177
Loegering WQ, Harmon DL (1969) Wheat lines near-isogenic for reaction to Puccinia graminis tritici. Phytopathology 59:456–460
Mago R, Verlin D, Zhang P, Bansal U, Bariana H, Jin Y, Ellis J, Hoxha S, Dundas I (2013) Development of wheat-Aegilops speltoides recombinants and simple PCR-based markers for Sr32 and a new stem rust resistance gene on the 2S#1 chromosome. Theor Appl Genet 126:2943–2955
Manly KF, Cudmore RH Jr, Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
Marais GF, Wessels WG, Horn M, du Toit F (1998) Association of a stem rust resistance gene (Sr45) and two Russian wheat aphid resistance genes (Dn5 and Dn7) with mapped structural loci in common wheat. S Afr J Plant Soil 15:67–71
McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. CSIRO Press, Victoria
McIntosh RA, Yamazaki Y, Dubcovsky J, Rogers J, Morris C, Somers DJ, Appels R, Devos KM (2008) Catalogue of gene symbols for wheat. (http://www.shigen.nig.ac.jp/wheat/komugi/genes/download.jsp. Accessed 11 Sept 2014). Committee for the National BioResource Project (NBRP)/KOMUGI, Japan
McIntosh RA, Yamazaki Y, Dubcovsky J, Rogers J, Morris C, Appels R, Xia XC (2013) Catalogue of gene symbols for wheat. (http://www.shigen.nig.ac.jp/wheat/komugi/genes/download.jsp. Accessed 3 April 2014). Committee for the National BioResource Project (NBRP)/KOMUGI, Japan
Niu Z, Klindworth DL, Friesen TL, Chao S, Jin Y, Cai X, Xu SS (2011) Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187:1011–1021
Niu Z, Klindworth DL, Yu G, Friesen TL, Chao S, Jin Y, Cai X, Ohm JB, Rasmussen JB, Xu SS (2014) Development and characterization of wheat lines carrying stem rust resistance gene Sr43 derived from Thinopyrum ponticum. Theor Appl Genet 127:969–980
Ogbonnaya FC, Abdalla O, Mujeeb-Kazi A, Kazi AG, Xu SS, Gosman N, Lagudah ES, Bonnett D, Sorrells ME, Tsujimoto H (2013) Synthetic hexaploids: harnessing species of the primary gene pool for wheat improvement. Plant Breed Rev 37:35–122
Olson EL, Rouse MN, Pumphrey MO, Bowden RL, Gill BS, Poland JA (2013a) Simultaneous transfer, introgression, and genomic localization of genes for resistance to stem rust race TTKSK (Ug99) from Aegilops tauschii to wheat. Theor Appl Genet 126:1179–1188
Olson EL, Rouse MN, Pumphrey MO, Bowden RL, Gill BS, Poland JA (2013b) Introgression of stem rust resistance genes SrTA10187 and SrTA10171 from Aegilops tauschii to wheat. Theor Appl Genet 126:2477–2484
Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, Deal K, Luo M, Kong X, Bariana H, Mago R, McIntosh R, Dodds P, Dvorak J, Lagudah E (2013) The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341:786–788
Pretorius ZA, Singh RP, Wagoire WW, Payne TS (2000) Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis 84:203
Qi LL, Pumphrey MO, Friebe B, Zhang P, Qian C, Bowden RL, Rouse MN, Jin Y, Gill BS (2011) A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor Appl Genet 123:159–167
Roelfs AP, Martens JW (1988) An international system of nomenclature for Puccinia graminis f. sp. tritici. Phytopathology 78:526–533
Rouse MN, Jin Y (2011) Stem rust resistance in A-genome diploid relatives of wheat. Plant Dis 95:941–944
Rouse MN, Olson EL, Gill BS, Pumphrey MO, Jin Y (2011a) Stem rust resistance in Aegilops tauschii germplasm. Crop Sci 51:2074–2078
Rouse MN, Wanyera R, Njau P, Jin Y (2011b) Sources of resistance to stem rust race Ug99 in spring wheat germplasm. Plant Dis 95:762–766
Rouse MN, Nava IC, Chao S, Anderson JA, Jin Y (2012) Identification of markers linked to the race Ug99 effective stem rust resistance gene Sr28 in wheat (Triticum aestivum L.). Theor Appl Genet 125:877–885
Rouse MN, Talbert LE, Singh D, Sherman JD (2014) Complementary epistasis involving Sr12 explains adult plant resistance to stem rust in Thatcher wheat (Triticum aestivum L.). Theor Appl Genet 127:1549–1559
Sambasivam PK, Bansal UK, Hayden MJ, Dvorak J, Lagudah ES, Bariana HS (2008) Identification of markers linked with stem rust resistance genes Sr33 and Sr45. In: Appels R, Eastwood R, Lagudah E, Langridge P, Mackay M, McIntyre L, Sharp P (eds) Proceedings of 11th international wheat genetics symposium. Sydney University Press, Sydney, pp 351–353
Simons K, Abate Z, Chao S, Zhang W, Rouse M, Jin Y, Elias E, Dubcovsky J (2011) Genetic mapping of stem rust resistance gene Sr13 in tetraploid wheat (Triticum turgidum ssp. durum L.). Theor Appl Genet 122:649–658
Singh RP, Hodson DP, Jin Y, Huerta-Espino J, Kinyua MG, Wanyera R, Njau P, Ward R (2006) Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour 1(54):1–13
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Njau P, Wanyera R, Herrera-Foessel SA, Ward RW (2008) Will stem rust destroy the world’s wheat crop? Adv Agron 98:271–309
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V (2011) The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 49:465–481
Singh A, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Campbell HL, Singh D, Bhavani S, Fetch T, Clarke F (2013) Identification and mapping in spring wheat of genetic factors controlling stem rust resistance and the study of their epistatic interactions across multiple environments. Theor Appl Genet 126:1951–1964
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genomics 4:12–25
Stakman EC, Stewart DM, Loegering WQ (1962) Identification of physiological races of Puccinia graminis var. tritici. United States Department of Agriculture, Agricultural Research Service E-617
Sun Y, Steffenson BJ (2005) Reaction of barley seedlings with different stem rust resistance genes to Puccinia graminis f. sp. tritici and Puccinia graminis f. sp. secalis. Can J Plant Pathol 27:80–89
Tsilo TJ, Jin Y, Anderson JA (2008) Diagnostic microsatellite markers for the detection of stem rust resistance gene Sr36 in diverse genetic backgrounds of wheat. Crop Sci 48:253–261
Tsilo TJ, Chao S, Jin Y, Anderson JA (2009) Identification and validation of SSR markers linked to the stem rust resistance gene Sr6 on the short arm of chromosome 2D in wheat. Theor Appl Genet 118:515–524
Tsilo TJ, Jin Y, Anderson JA (2010) Identification of flanking markers for the stem rust resistance gene Sr6 in wheat. Crop Sci 50:1967–1970
Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3: new capabilities and interfaces. Nucleic Acids Res 40:e115
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Xu SS, Cai X, Wang T, Harris MO, Friesen TL (2006) Registration of two synthetic hexaploid wheat germplasms resistant to Hessian fly. Crop Sci 46:1401–1402
Yu GT, Cai X, Harris MO, Gu YQ, Luo M-C, Xu SS (2009) Saturation and comparative mapping of the genomic region harboring Hessian fly resistance gene H26 in wheat. Theor Appl Genet 118:1589–1599
Yu GT, Zhang Q, Klindworth DL, Friesen TL, Knox R, Jin Y, Zhong S, Cai X, Xu SS (2010) Molecular and cytogenetic characterization of wheat introgression lines carrying the stem rust resistance gene Sr39. Crop Sci 50:1393–1400
Zhang QJ (2013) Development and characterization of wheat germplasm for resistance to stem rust Ug99 in wheat. PhD Dissertation. North Dakota State University, Fargo, North Dakota
Zhong S, Leng Y, Friesen TL, Faris JD, Szabo LJ (2009) Development and characterization of expressed sequence tag-derived microsatellite markers for the wheat stem rust fungus Puccinia graminis f. sp. tritici. Phytopathology 99:282–289
Acknowledgements
We thank Drs. Chao-Chien Jan and Lili Qi for critically reviewing the manuscript. The authors also thank Danielle Holmes and Daryl Klindworth for technical support. This research was supported in part by funds to S. S. X. provided through a grant from the Bill & Melinda Gates Foundation to Cornell University for the Borlaug Global Rust Initiative (BGRI) Durable Rust Resistance in Wheat (DRRW) Project and the USDA-ARS CRIS Project No. 5442-22000-037-00D. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Conflict of interest
All authors have no conflict of interest.
Ethical standards
The experiments were performed in compliment with the current laws of United States of America.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Susanne Dreisigacker.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, G., Zhang, Q., Friesen, T.L. et al. Identification and mapping of Sr46 from Aegilops tauschii accession CIae 25 conferring resistance to race TTKSK (Ug99) of wheat stem rust pathogen. Theor Appl Genet 128, 431–443 (2015). https://doi.org/10.1007/s00122-014-2442-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2442-4