Abstract
The ability to sense and recognize various classes of compounds is of particular importance for survival and reproduction of insects. Ionotropic receptor (IR), a sub-family of the ionotropic glutamate receptor family, has been identified as one of crucial chemoreceptor super-families, which mediates the sensing of odors and/or tastants, and serves as non-chemosensory functions. Yet, little is known about IR characteristics, evolution, and functions in Lepidoptera. Here, we identify the IR gene repertoire from a destructive polyphagous pest, Spodoptera litura. The exhaustive analyses with genome and transcriptome data lead to the identification of 45 IR genes, comprising 17 antennal IRs (A-IRs), 8 Lepidoptera-specific IRs (LS-IRs), and 20 divergent IRs (D-IRs). Phylogenetic analysis reveals that S. litura A-IRs generally retain a strict single copy within each orthologous group, and two lineage expansions are observed in the D-IR sub-family including IR100d-h and 100i-o, likely attributed to gene duplications. Results of gene structure analysis classify the SlitIRs into four types: I (intronless), II (1–3 introns), III (5–9 introns), and IV (10–18 introns). Extensive expression profiles demonstrate that the majority of SlitIRs (28/43) are enriched in adult antennae, and some are detected in gustatory-associated tissues like proboscises and legs as well as non-chemosensory organs like abdomens and reproductive tissues of both sexes. These results indicate that SlitIRs have diverse functional roles in olfaction, taste, and reproduction. Together, our study has complemented the information on chemoreceptor genes in S. litura, and meanwhile allows for target experiments to identify potential IR candidates for the control of this pest.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chemosensory systems, referring to olfactory and taste systems, are of particular importance for survival and reproduction of insects involved in various vital behaviors, such as host searching, partner recognition, predator avoidance, and food assessment (Dahanukar et al. 2005; Hallem et al. 2006; Hansson and Stensmyr 2011). The olfactory-related behaviors are driven primarily by volatile organic compounds (VOCs), including sex pheromones, and host or non-host odors (Arimura et al. 2009; Gomez-Diaz and Benton 2013). In addition, behaviors that rely on the sensing of taste are mediated by tastants such as sugars, carbon dioxide, and bitter compounds (Agnihotri et al. 2016; Isono and Morita 2010; Slone et al. 2007; Xu and Anderson 2015). These crucial chemosensory behaviors are associated with three distinctive super-families of chemoreceptors: odorant (ORs), gustatory (GRs), and ionotropic (IRs) receptors. These receptor proteins are expressed in specific chemosensory sensilla harboring one to five gustatory (GSNs) or olfactory (OSNs) sensory neurons that are tuned to various classes of compounds (Ihara et al. 2013; Su et al. 2009; Touhara and Vosshall 2009).
IRs, belonging to a variant sub-family of ionotropic glutamate receptors (iGluRs), were first discovered from Drosophila melanogaster genome using bioinformatics-based methods (Benton et al. 2009). Initially, this gene family was proposed to solely detect the VOCs, and thereby was defined as olfactory receptors. Except those IRs involved in olfaction, functions of the remaining IRs were still unknown due to no expression in various tissues (Benton et al. 2009). Accordingly, Drosophila IRs were distinguished into two sub-families: divergent IRs (D-IRs) and antennal IRs (A-IRs), based on sequence characteristics and phylogenetic analyses (Croset et al. 2010). More recently, a growing number of studies have improved our knowledge on IR functions with involvement in olfaction, gustation, hearing, temperature, and humidity (Chen and Amrein 2017; Enjin et al. 2016; Hussain et al. 2016; Knecht et al. 2017, 2016; Koh et al. 2014; Ni et al. 2016; Prieto-Godino et al. 2017; Senthilan et al. 2012; Silbering et al. 2011; Stewart et al. 2015; Tauber et al. 2017). For example, members of the IR20a clade in D. melanogaster, consisting of 35 IR genes, were identified as candidate taste and pheromone receptors (Koh et al. 2014). Moreover, three combinatorial patterns of IR21a/IR25a/IR93a, IR25a/IR40a/IR93a, and IR25a/IR68a/IR93a have been shown to mediate the sensing of cool-temperature avoidance, humidity, and moist, respectively (Knecht et al. 2016, 2017; Ni et al. 2016). Interestingly, two candidate co-receptor IRs, IR25a and IR76b, have dual roles, i.e., participating in the formation of hetero-complexes as co-receptors and sensing compounds or temperature as stimulus-specific IRs (Chen et al. 2015; Croset et al. 2016; Ganguly et al. 2017; Hussain et al. 2016; Knecht et al. 2017; Ni et al. 2016; Rytz et al. 2013; Silbering et al. 2011; Zhang et al. 2013).
Although the IR gene family has been intensively investigated since 2009, especially focusing on dipteran species including Drosophila species and Anopheles gambiae (Abuin et al. 2011; Benton et al. 2009; Croset et al. 2016; Ganguly et al. 2017; Grosjean et al. 2011; Hussain et al. 2016; Liu et al. 2010; Pitts et al. 2017; van Giesen and Garrity 2017), little information on its functions and evolution in other insect orders is available until now. Apart from the two chemosensory super-families of ORs and GRs, the IR gene repertoire is also important for lepidopteran chemosensory reception. More recently, a study on IR evolution has been conducted in Heliconius butterflies (van Schooten et al. 2016), which has enhanced our knowledge of lepidopteran IRs. However, the IR gene family has still been largely unexplored and poorly understood in moth species, which has largely restricted our ability to understand IR evolution and functions within lepidopteran species or across insects. With the increased availability of genome data in Lepidoptera, we are able to explore the problems of IR sequence characteristics, gene structures, gene expansions, phylogenetic relationships, expression profiles, and evolution.
In the current study, we integrated bioinformatics-based approaches and molecular biology strategies to exhaustively identify IR genes from the genome of Spodoptera litura (Cheng et al. 2017). This study greatly complements the information on chemoreceptors in S. litura and other lepidopteran species, and meanwhile provides reference data for further functional studies and evolution of IRs across insects.
Materials and methods
Insect rearing and tissue collection
The larvae of S. litura were reared on an artificial diet (Huang et al. 2002) in the laboratory of Southwest Forestry University, under the conditions of 14:10 h light/dark and 65 ± 5% relative humidity. Pupae were sexed and kept in separate cages until eclosion. The emerging adults were supplied with 10% honey solution.
Adult tissues were dissected between the 6th and 8th hours of the scotophase from 2-day-old moths of both sexes. These tissues included antennae, proboscises, heads without antennae and proboscises, thorax, abdomens, legs, wings, and reproductive systems (female: accessory gland, bursa copulatrix, ovary, spermatheca, and spermathecal gland; male: accessory gland, ejaculatory duct, seminal vesicle, and testis) of both sexes. Two independent samples of each tissue were collected as two templates.
Total RNA extraction and first-strand cDNA synthesis
Total RNA was extracted from collected tissues using TRIzol Reagent (Ambion, Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instruction. The quality and concentration of RNA were examined by using NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific, USA). Synthesis of first-strand cDNA was performed with 1 μg of total RNA using PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, Liaoning, China) according to the protocol. Briefly, genomic DNA was first removed by the treatment with gDNA Eraser at 42 °C for 2 min, and then cDNA template was synthesized at 37 °C for 15 min and 85 °C for 5 s. The prepared cDNA templates were stored at − 20 °C prior to use.
Gene identification
The genome assembly and transcriptomes of S. litura were downloaded from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/genome/?term=spodoptera+litura) (Cheng et al. 2017) and NCBI Sequence Read Archive (SRA) Databases (https://www.ncbi.nlm.nih.gov/genbank/tsa/), respectively. Candidate IR genes were identified from S. litura based on the stand-alone genomic and transcriptomic data, using the IRs of Bombyx mori, Helicoverpa armigera, Spodoptera frugiperda, and D. melanogaster (Croset et al. 2010; Gouin et al. 2017; Liu et al. 2014; Pearce et al. 2017; van Schooten et al. 2016) as initial queries in exhaustive TBLASTN and PSI-BLAST searches, with a E value cutoff of e−5. Further, all identified S. litura IRs were used as queries to screen the genome in an alternative method, with manual adjustments.
Gene nomenclature
Conserved S. litura IRs were named following the nomenclature conventions of B. mori, H. armigera, S. frugiperda, and D. melanogaster IRs (Croset et al. 2010; Gouin et al. 2017; Pearce et al. 2017), together with the orthology. In addition, other IRs were designated as IR100 appended with a lower-case “a-p”-like SlitIR100a to SlitIR100p. Notably, SlitIR100a was specific to moths and had no orthology to D. melanogaster IR100a. S. litura iGluRs were named with the Arabic numerals from 1 to 12, based on the orthology to H. armigera iGluRs (Liu et al. 2014; Pearce et al. 2017) (Table S1 and Additional file 1).
Sequence and phylogenetic analysis
Transmembrane segments and signal peptides of candidate IRs were predicted using TMHMM Server v2.0 (Krogh et al. 2001) and SignalP 4.1 Server (Petersen et al. 2011), respectively. Multiple alignments of IR protein sequences were generated by MAFFT v7.308 (Katoh and Standley 2013) or MUSCLE (Edgar 2004), with the default parameters and necessary manual adjustments. Aligned sequences were edited using JalView 2.8 (Waterhouse et al. 2009). Exon and intron boundaries were predicted and analyzed using GeneWise with the synchronous model and GT-AG rule (Birney et al. 2004). Gene models were generated with Exon-Intron Graphic Marker (http://www.wormweb.org/exonintron). For comparison of IR gene numbers across insects, S. litura and ten other species were selected including B. mori, Danaus plexippus, H. armigera, Heliconius melpomene, Manduca sexta, S. frugiperda, Acyrthosiphon pisum, Apis mellifera, D. melanogaster, and Tribolium castaneum. Gene gains and losses were analyzed between the above-mentioned lepidopteran species.
In the phylogenetic tree, the IRs were selected from B. mori, H. armigera, S. frugiperda, and S. litura as well as a model insect D. melanogaster (Croset et al. 2010; Gouin et al. 2017; Pearce et al. 2017; van Schooten et al. 2016). S. litura iGluRs were used as an outgroup to root the tree. The tree was constructed using PhyML 3.2 (Guindon et al. 2010) based on the aligned protein sequences, under Jones-Taylor-Thornton (JTT) model with Nearest-Neighbor Interchange (NII). Support values were estimated by approximate likelihood ratio test (Chi2). The tree was viewed and edited using FigTree 1.4.3.
Gene expression profile analysis
Expression profiles of candidate S. litura IR genes in various adult tissues and internal reproductive systems of both sexes, were examined using reverse transcription PCR (RT-PCR). Gene-specific primers (Table S2) were designed by Primer Premier 6.0 (PREMIER Biosoft International, CA, USA), with an expected product size of about 600 bp, containing at least one intron if possible. RT-PCR was conducted with rTaq DNA polymerase (TaKaRa, Dalian, Liaoning, China) under the following conditions: 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 40 s, and final extension for 5 min at 72 °C. Ribosomal protein L10 (RPL10) gene (Lu et al. 2013) was used as the reference gene to check the quality and integrity of cDNA templates. Negative control was set using sterile water as the template. For some IR genes, two biological replicates were carried out to acquire accurate data.
Results
Identification of candidate IRs in S. litura
Based on the genome sequences of S. litura, coupled with published transcriptomes, 45 genes encoding IRs were identified by combining bioinformatics-based approaches and manual efforts. They contained two putative pseudogenes (IR2 and IR2.1) and one partial sequence (IR68a). The remaining IRs were full-length sequences with various sizes containing 543 to 919 amino acids (AAs). Apart from the IR family, we also identified two α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), two N-methyl-d-aspartate (NMDA), and eight Kainate receptors. Of these, iGluR1–iGluR9 were full-length sequences varying from 871 to 985 AAs while iGluR10–iGluR12 were partial sequences (Table S1 and Additional file 1).
In comparison with most of the SlitIRs, the three other iGluR sub-families had longer full-length sequences (871–985 AAs). However, we noticed that for two co-receptors (IR8a and IR25a), the predicted genes encoded protein sequences of 899 AAs and 919 AAs, respectively. By contrast, the third co-receptor, IR76b, showed the shortest open reading frame, encoding 543 AAs. Seven IR genes encoded over 650 AAs, including IR21a, IR40a, IR60a, IR93a, IR100a, IR100b, and IR100b.1. The remaining IR genes (79%) encoded 580 to 643 AAs (Table S1 and Additional file 1).
Comparison of IRs between Lepidoptera and other insect orders
To compare the differences of IR gene numbers among various lepidopteran species as well as between Lepidoptera and other insect orders, we examined seven lepidopteran species and four non-lepidopteran species. By comparison, the number of IRs in Lepidoptera was larger than in A. mellifera, A. pisum, and T. castaneum but smaller than in D. melanogaster (Fig. 1a). To further unravel gene gains or losses in Lepidoptera, another six species were selected. In seven IR orthologous groups (LS-IRs: IR1.2, IR2.1, IR100b, and IR100b.1; A-IRs: IR60a and IR75p; D-IRs: IR7d.4), the orthologous analysis revealed an absence of IRs in some lepidopteran species. Of these, IR2.1 and IR100b.1 were found only in noctuid moths, and IR100b was present in moths rather than in butterflies. In the A-IR sub-family, IR60a was not identified from B. mori and M. sexta, and IR75p was present only in moths. For the D-IR sub-family, IR7d.4 was not detected in B. mori. The remaining 23 orthologous groups were highly conserved among lepidopteran species (Fig. 1b).
Lepidoptera IR gene family. a Numbers of ionotropic receptor genes across insect species. b Presence and absence of IR orthologous groups within seven lepidopteran species. Numbers in colored boxes represent gene copies identified from the genome. “P” and “F” mean pseudogene and fragment, respectively. Blank space means gene losses in the corresponding species. A-IRs, antennal IRs; LS-IRs, Lepidoptera-specific IRs; D-IRs, divergent IRs
Sequence and phylogenetic analyses of candidate IRs in S. litura
To explore three key amino acid residues involved in ligand binding, the protein sequences of 12 iGluRs and 43 IRs (except IR2 and IR2.1) were aligned and analyzed. Only seven iGluRs (iGluR2, iGluR3, iGluR4, iGluR6, iGluR7, iGluR10, and iGluR12) retained the conserved pattern of R-T-D/E, whereas this conserved pattern was lacking in all IRs. The threonine in the second position was not present in all IRs, and ten IRs exhibited a conserved aspartate or glutamate in the third position (Fig. 2).
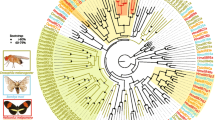
Our phylogenetic analysis revealed that twelve A-IR groups and one D-IR group were homologous to D. melanogaster IRs with each clade showing a strict single copy. In Lepidoptera, two large lineage expansions were observed in IR7d and IR75 clades. In addition, two expansions were observed in the D-IR sub-family, representing IR100d-h and IR100i-o. These expansions are possibly derived from gene duplications, further supported by IR gene physical location on a single chromosome and relative high identities among the paralogs (Figs. 3 and 4a). Two candidate co-receptors IR8a and IR25a were phylogenetically clustered together with iGluRs. Interestingly, IR60a and IR87a members belonging to the A-IR and LS-IR sub-families showed a close phylogenetic relationship to the D-IR family (Fig. 3).
Phylogenetic relationship of insect IRs. The tree was constructed using PhyML 3.2 under the JTT model of substitution with NNIs, based on the aligned amino acid sequences by MAFFT v7.308. Branch support value was estimated using an approximate likelihood ratio test (Chi2) and indicated (circles < 0.90). The IRs were grouped into various small clades with species-specific color patterns. Three sub-families (A-IRs, LS-IRs, and D-IRs) were distinguished based on the classification of Drosophila IRs. iGluRs was used as the outgroup to root the tree
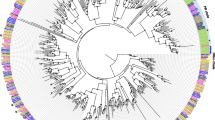
Candidate IR gene family in S. litura. a Possible duplication events of candidate IRs in S. litura. Transcriptional orientation of genes is indicated by red arrows. Protein sequence identities between the adjacent IRs are shown, and the identities in the brackets represent a mean value among the paralogs. b Phylogenetic tree and sequence lengths of S. litura iGulRs and IRs. The tree was constructed using PhyML 3.2, with estimated branch support values (circles < 0.90) using an approximate likelihood ratio test (Chi2). Color-coded branches represent intron numbers of IR genes, of which intron numbers of incomplete IRs are unknown and indicated in black. At the bottom of the tree, ORF length of each gene is presented. c Models of candidate IRs in S. litura. Gene structures of some representative IRs in S. litura are shown. Various colors mean different functional domains and transmembrane segments (S1 and S2, purple; P, green; M1, M2, and M3, red). Scale bars represent a size of 500 bases. Models of the remaining IRs of S. litura are shown in Fig. S1
Structure and organization of candidate IRs in S. litura
We mapped all the IRs onto the genome of S. litura by TBLASTN searches. These IR genes were oriented onto fifteen chromosomes, three scaffolds, and two contigs, respectively. In particular, six possible gene duplication events were detected including IR7d.1/7d.2, IR75p/75p.2, IR75q.1/75q.2, IR100b/100b.1, IR100d-h, and IR100i-o. In each duplication event, these IR genes were tandemly located on one chromosome. Moreover, except IR75p and IR75p.2 presenting an opposite transcriptional orientation, the IR genes of other five events were closely located on a single chromosome with the same transcriptional orientation (Table S1; Fig. 4a).
To compare exon and intron structures of IR genes in S. litura, we first analyzed their intron numbers. Based on the analysis of intron numbers, four types of IR genes could be defined: type I—intronless: the D-IR sub-family (except IR85a), and IR60a and IR87a from the A-IR and LS-IR sub-families, respectively; type II—one to three introns: IR85a/100a/100b/100b.1 from the LS-IR and D-IR sub-families; type III—five to nine introns: five A-IR groups of IR21a, IR31a, IR41a, IR64a, and IR76b; and type IV—ten to eighteen introns: the remaining IRs including IR1.1, IR1.2, IR8a, IR25a, IR40a, IR93a, and members of IR75 groups (Fig. 4b, c).
Intron size and position were analyzed based on genome and cDNA sequences. Several typical characteristics could be summarized. For example, the type I IRs had similar positions of functional domains and transmembrane segments (one ion channel pore (P), two lobes of ligand-binding domains (S1 and S2), three transmembrane domains (M1, M2, and M3)) due to their similar sequence lengths (Fig. S1). For the type II IRs, intron lengths were usually short, varying from 60 to 444 bp. Notably, IR100b and IR100b.1 shared the same intron numbers, similar intron lengths, and similar positions of functional domains and transmembrane segments. Unlike the type I and type II IRs, two other types of IRs possessed variable intron lengths. Of these, the shortest intron was 67 bp (intron 3 of IR1.1 and intron 9 of IR75q.2), while the longest intron was 7559 bp (intron 9 of IR25a). In addition, we found that members of IR75 groups had similar positions of functional domains and transmembrane segments (Table S1; Fig. S1; Fig. 4c).
Expression profile of candidate IRs in S. litura
To explore possible functional roles of S. litura IRs, we investigated global expression of 43 IRs (except two IR pseudogenes) and 12 iGluRs in different tissues of both sexes. In total, 13 IR genes belonging to the D-IR sub-family were not detected in all tested tissues, which was consistent with the expression of Drosophila D-IRs (Benton et al. 2009; Rytz et al. 2013). Overall, 12 iGluR genes presented a broad tissue distribution, and a similar expression profile was also observed in seven D-IR members (IR7d.1, 7d.2, 7d.3, 100k, 100l, 100o, and 100p), four A-IRs (IR25a, IR60a, IR64a, and IR76b) including two co-receptors and one LS-IR member (IR100a) (Fig. 5a, b). As expected, the majority of IRs were expressed predominantly in adult antennae. Of these, IR1.2, IR8a, IR31a, and IR87a were specifically expressed in the antennae. Over half of the IRs (proboscis, 21/30 and leg, 17/30) were detected in gustatory tissues with IR7d.4 and IR85a being specific to proboscises of both sexes, although some displayed extremely low expression. Notably, one A-IR member IR68a was abundant in female abdomens, but it had low expression in male abdomens. In addition, some IRs were present in non-chemosensory tissues including the thorax, abdomens, and wings. For example, IR7d.1, IR7d.2, IR7d.3, IR25a, IR76b, and IR100a were expressed in all three non-chemosensory tissues. IR60a had expression in thorax and wings (Fig. 5b).
Expression profile of candidate S. litura IRs in different tissues of both sexes. a Expression of candidate SlitiGluRs. b Expression of candidate SlitIRs. S. litura RPL10 gene was used as quality and quantity control for all cDNA templates. Negative control (NC) is set using sterile water as the template. An, antennae; Pro, proboscises; He, heads without antennae and proboscises; T, thorax; Ab, abdomens; Le, legs; Wi, wings
To unravel potential reproductive-related roles of S. litura IRs, we further investigated the expression profiles of 43 IRs and 12 iGluRs in reproductive systems of male and female moths, consisting of the accessory gland, ejaculatory duct, seminal vesicle, and testis of male adults as well as accessory gland, bursa copulatrix, ovary, spermathecal gland, and spermatheca of female adults (Fig. 6a). Results showed that 26 IRs and seven iGluRs were detected in at least one reproductive-related tissue. Of these, iGluR6, iGluR7, and iGluR9 as well as IR7d.1, IR7d.2, IR7d.3, IR25a, IR64a, IR76b, and IR100a were broadly expressed in tested tissues. Intriguingly, IR68a and IR75q.2 exhibited relatively high expression in ovary and accessory gland of females, respectively (Fig. 6b). The expression of IR68a was enriched in the ovary, which was consistent with its female abdomen-biased expression characteristics (Fig. 6b). Additionally, IR76b showed abundant expression in bursa copulatrix and ovary, and IR100o was highly expressed in male accessory gland (Fig. 6b).
Expression profile of candidate S. litura IRs in reproductive tissues of both sexes. a Reproductive systems of male (left) and female (right) S. litura. The photos were taken by light microscopy. ED, ejaculatory ducts; MAG, male accessory glands; SV, seminal vesicles; Te, testes; BC, bursa copulatrix; FAG, female accessory glands; Ov, ovaries; Sp + SG, spermathecae and spermathecal glands. b Expression of candidate SlitiGluRs and IRs. S. litura RPL10 gene was used as quality and quantity control for all cDNA templates. Negative control (NC) is set using sterile water as the template. The abbreviation of tissues is showed in (a)
Discussion
Outside dipteran species (mainly D. melanogaster and A. gambiae), our ability to understand IR characteristics, evolution, and functions in other insects has been restricted to the studies on gene identification, gene numbers, phylogenetic analyses, or expression profiles (Dong et al. 2016; Feng et al. 2015; Xu et al. 2015). Little is known about IR functions and evolution in Lepidoptera and other insect orders (Gouin et al. 2017; Harrison et al. 2018; Liu et al. 2014; Olivier et al. 2011; Pearce et al. 2017; Wang et al. 2018). In the current study, we have identified and characterized the IR gene repertoire from S. litura, which has been earmarked as potential molecular targets for pest control, in parallel with two other multigene families of ORs and GRs. Together, our work greatly complements the information on the chemoreceptor super-families in S. litura (Feng et al. 2015) or other lepidopteran species (Olivier et al. 2011; van Schooten et al. 2016).
The number of S. litura IRs (45) is larger compared with that of A. mellifera (10), A. pisum (11), B. mori (30), D. plexippus (32), H. melpomene (33), H. armigera (39), H. zea (39), M. sexta (34), and T. castaneum (23), equal to that of A. gambiae (46) and S. frugiperda (45), but smaller than that of Aedes aegypti (95) and D. melanogaster (66) (Croset et al. 2010; Gouin et al. 2017; Pearce et al. 2017; Rytz et al. 2013; van Schooten et al. 2016). Although the species-specific D-IR sub-family generally exhibits relatively low protein sequence identities across insects (Croset et al. 2010; Gouin et al. 2017; Pearce et al. 2017), a typical characteristic of gene structures, i.e., intronless, makes the identification of this sub-family easier. Based on the analyses of gene structures, several typical characteristics such as intron insertion sites, intron numbers, sequence lengths, gene organization on chromosomes, and conserved positions of functional domains, greatly assist in completing manual curation. Notably, we notice that some prior studies, showing IR identification, sequence, and phylogenetic characteristics, have some errors in full-length sequences or conclusion (Bengtsson et al. 2012; Dong et al. 2016; Feng et al. 2015; Koenig et al. 2015; Liu et al. 2014; Xu et al. 2015). Thus, our work would be helpful for the improvement of previous IR work and subsequent IR identification and annotation in other lepidopteran species, especially in moths.
In comparison with the classification of Drosophila IRs (Croset et al. 2010), lepidopteran species possess a specific sub-family (namely LS-IRs), described in this study and previous studies in Lepidoptera (Liu et al. 2014; Olivier et al. 2011; van Schooten et al. 2016). Of these, IR100a is Lepidoptera specific, whereas IR100b is present only in moths and IR100b.1 is restricted to noctuid moths (van Schooten et al. 2016). Exon and intron structural analyses indicate that the LS-IR (except IR1.1 and IR1.2) and D-IR sub-families have fewer introns relative to conserved A-IRs. Sequence alignment analysis reveals that S. litura IRs possess diverse amino acids in three conserved positions, together with diversities of expression profiles, suggesting IR diverse functional roles in this species. Our phylogenetic results show the relationships between three sub-families and intron numbers. For example, the D-IR (except IR85a) sub-family is intronless, while the A-IR sub-family in general has multiple introns, similar to Drosophila IRs (Croset et al. 2010). Interestingly, two groups of IR60a and IR87a that have no introns and respectively belong to the A-IR and LS-IR sub-families are close to the D-IR sub-family in phylogeny, but they show considerable expression in antennae deserving further functional studies.
The broad expression profiles of S. litura IRs provide insights into their functional diversities. Most of the A-IR sub-family (28/43) display antennal expression at a high level and some (IR1.1, IR8a, IR31a, IR75d, and IR87a) are specifically expressed in the antennae, indicating their potential olfactory roles (Abuin et al. 2011; Benton et al. 2009; Croset et al. 2010). The result is consistent with IR tissue-expression characteristics in the noctuid moths of H. armigera and Spodoptera littoralis (Liu et al. 2014; Olivier et al. 2011). Of three co-receptors (IR8a, IR25a, and IR76b), IR8a expression is restricted to the antennae, possibly indicating that it has extremely low or no expression in other tissues. The two other co-receptors exhibit broad expression, consistent with their designated co-receptor roles. Similar results have been observed in H. armigera IR8a, IR25a, and IR76b (Liu et al. 2014). Intriguingly, some IRs from the LS-IR and D-IR sub-families are also detected in the antennae, possibly guiding olfactory-related behaviors.
Outside olfactory roles driven by antennae, the sensing of taste is mediated primarily by gustatory-associated tissues like proboscis, leg, mouthpart, labellum, pharynx, or wing margin as previously indicated in A. aegypti, Culex quinquefasciatus, D. melanogaster, H. armigera, H. melpomene, and S. littoralis (Koh et al. 2014; Leal et al. 2013; Liu et al. 2014; Olivier et al. 2011; Sparks et al. 2014; van Schooten et al. 2016). Our current analysis reveals that over half of SlitIRs (22/43) are expressed in proboscises or legs with involvement in taste detection. Notably, IR7d.4 and IR85a are detected only in the proboscises of both sexes, possibly indicating their involvement in feeding behaviors of adults. In addition to chemosensory tissues, SlitIRs are present in non-chemosensory organs including the thorax, abdomens, wings, and reproductive-related tissues, similar to H. armigera and S. littoralis IRs (our unpublished data; Liu et al. 2014; Olivier et al. 2011). In D. melanogaster, IRs have been demonstrated to participate in hearing, and also humidity and temperature sensing (Enjin et al. 2016; Knecht et al. 2016; Senthilan et al. 2012). For example, DmelIR68a expressed in antenna and head mediates moist sensing (Knecht et al. 2017). DmelIR76b is required for the detection of polyamines that are attractive egg-laying substrates and increase reproductive success of Drosophila species (Hussain et al. 2016). In our study, an interesting result is the high expression of S. litura IR68a in female ovary, as a novel candidate for female reproductive regulation. Likewise, IR75q.2 and IR76b are likely to be involved in female reproductive behaviors, while IR100o may mediate male reproduction. Together, these results suggest that SlitIRs may have additional functions other than chemoreception, especially reproductive-associated behaviors.
References
Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R (2011) Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69(1):44–60. https://doi.org/10.1016/j.neuron.2010.11.042
Agnihotri AR, Roy AA, Joshi RS (2016) Gustatory receptors in Lepidoptera: chemosensation and beyond. Insect Mol Biol 25(5):519–529. https://doi.org/10.1111/imb.12246
Arimura G, Matsui K, Takabayashi J (2009) Chemical and molecular ecology of herbivore-induced plant volatiles: proximate factors and their ultimate functions. Plant Cell Physiol 50(5):911–923. https://doi.org/10.1093/pcp/pcp030
Bengtsson JM, Trona F, Montagne N, Anfora G, Ignell R, Witzgall P, Jacquin-Joly E (2012) Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS One 7(2):e31620. https://doi.org/10.1371/journal.pone.0031620
Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB (2009) Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136(1):149–162. https://doi.org/10.1016/j.cell.2008.12.001
Birney E, Clamp M, Durbin R (2004) GeneWise and Genomewise. Genome Res 14(5):988–995. https://doi.org/10.1101/gr.1865504
Chen Y, Amrein H (2017) Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Curr Biol 27(18):2741–2750. https://doi.org/10.1016/j.cub.2017.08.003
Chen C, Buhl E, Xu M, Croset V, Rees JS, Lilley KS, Benton R, Hodge JJ, Stanewsky R (2015) Drosophila ionotropic receptor 25a mediates circadian clock resetting by temperature. Nature 527(7579):516–520. https://doi.org/10.1038/nature16148
Cheng T, Wu J, Wu Y, Chilukuri RV, Huang L, Yamamoto K, Feng L, Li W, Chen Z, Guo H, Liu J, Li S, Wang X, Peng L, Liu D, Guo Y, Fu B, Li Z, Liu C, Chen Y, Tomar A, Hilliou F, Montagne N, Jacquin-Joly E, d'Alencon E, Seth RK, Bhatnagar RK, Jouraku A, Shiotsuki T, Kadono-Okuda K, Promboon A, Smagghe G, Arunkumar KP, Kishino H, Goldsmith MR, Feng Q, Xia Q, Mita K (2017) Genomic adaptation to polyphagy and insecticides in a major east Asian noctuid pest. Nat Ecol Evol 1(11):1747–1756. https://doi.org/10.1038/s41559-017-0314-4
Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R (2010) Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet 6(8):e1001064. https://doi.org/10.1371/journal.pgen.1001064
Croset V, Schleyer M, Arguello JR, Gerber B, Benton R (2016) A molecular and neuronal basis for amino acid sensing in the Drosophila larva. Sci Rep 6:34871. https://doi.org/10.1038/srep34871
Dahanukar A, Hallem EA, Carlson JR (2005) Insect chemoreception. Curr Opin Neurobiol 15(4):423–430. https://doi.org/10.1016/j.conb.2005.06.001
Dong J, Song Y, Li W, Shi J, Wang Z (2016) Identification of putative chemosensory receptor genes from the Athetis dissimilis antennal transcriptome. PLoS One 11(1):e0147768. https://doi.org/10.1371/journal.pone.0147768
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Enjin A, Zaharieva EE, Frank DD, Mansourian S, Suh GS, Gallio M, Stensmyr MC (2016) Humidity sensing in Drosophila. Curr Biol 26(10):1352–1358. https://doi.org/10.1016/j.cub.2016.03.049
Feng B, Lin X, Zheng K, Qian K, Chang Y, Du Y (2015) Transcriptome and expression profiling analysis link patterns of gene expression to antennal responses in Spodoptera litura. BMC Genomics 16(1):269. https://doi.org/10.1186/s12864-015-1375-x
Ganguly A, Pang L, Duong VK, Lee A, Schoniger H, Varady E, Dahanukar A (2017) A molecular and cellular context-dependent role for Ir76b in detection of amino acid taste. Cell Rep 18(3):737–750. https://doi.org/10.1016/j.celrep.2016.12.071
Gomez-Diaz C, Benton R (2013) The joy of sex pheromones. EMBO Rep 14(10):874–883. https://doi.org/10.1038/embor.2013.140
Gouin A, Bretaudeau A, Nam K, Gimenez S, Aury JM, Duvic B, Hilliou F, Durand N, Montagne N, Darboux I, Kuwar S, Chertemps T, Siaussat D, Bretschneider A, Mone Y, Ahn SJ, Hanniger S, Grenet AG, Neunemann D, Maumus F, Luyten I, Labadie K, Xu W, Koutroumpa F, Escoubas JM, Llopis A, Maibeche-Coisne M, Salasc F, Tomar A, Anderson AR, Khan SA, Dumas P, Orsucci M, Guy J, Belser C, Alberti A, Noel B, Couloux A, Mercier J, Nidelet S, Dubois E, Liu NY, Boulogne I, Mirabeau O, Le Goff G, Gordon K, Oakeshott J, Consoli FL, Volkoff AN, Fescemyer HW, Marden JH, Luthe DS, Herrero S, Heckel DG, Wincker P, Kergoat GJ, Amselem J, Quesneville H, Groot AT, Jacquin-Joly E, Negre N, Lemaitre C, Legeai F, d'Alencon E, Fournier P (2017) Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci Rep 7(1):11816. https://doi.org/10.1038/s41598-017-10461-4
Grosjean Y, Rytz R, Farine JP, Abuin L, Cortot J, Jefferis GS, Benton R (2011) An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature 478(7368):236–240. https://doi.org/10.1038/nature10428
Guindon S, J-Fo D, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321. https://doi.org/10.1093/sysbio/syq010
Hallem EA, Dahanukar A, Carlson JR (2006) Insect odor and taste receptors. Annu Rev Entomol 51:113–135. https://doi.org/10.1146/annurev.ento.51.051705.113646
Hansson BS, Stensmyr MC (2011) Evolution of insect olfaction. Neuron 72(5):698–711. https://doi.org/10.1016/j.neuron.2011.11.003
Harrison MC, Jongepier E, Robertson HM, Arning N, Bitard-Feildel T, Chao H, Childers CP, Dinh H, Doddapaneni H, Dugan S, Gowin J, Greiner C, Han Y, Hu H, Hughes DST, Huylmans A-K, Kemena C, Kremer LPM, Lee SL, Lopez-Ezquerra A, Mallet L, Monroy-Kuhn JM, Moser A, Murali SC, Muzny DM, Otani S, Piulachs M-D, Poelchau M, Qu J, Schaub F, Wada-Katsumata A, Worley KC, Xie Q, Ylla G, Poulsen M, Gibbs RA, Schal C, Richards S, Belles X, Korb J, Bornberg-Bauer E (2018) Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat Ecol Evol 2(3):557–566. https://doi.org/10.1038/s41559-017-0459-1
Huang CX, Zhu LM, Ni JP, Chao XY (2002) A method of rearing the beet armyworm Spodoptera exigua. Entomol Knowl 39:229–231
Hussain A, Zhang M, Ucpunar HK, Svensson T, Quillery E, Gompel N, Ignell R, Grunwald Kadow IC (2016) Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol 14(5):e1002454. https://doi.org/10.1371/journal.pbio.1002454
Ihara S, Yoshikawa K, Touhara K (2013) Chemosensory signals and their receptors in the olfactory neural system. Neuroscience 254(7):45–60. https://doi.org/10.1016/j.neuroscience.2013.08.063
Isono K, Morita H (2010) Molecular and cellular designs of insect taste receptor system. Front Cell Neurosci 4(2):20. https://doi.org/10.3389/fncel.2010.00020
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Knecht ZA, Silbering AF, Ni L, Klein M, Budelli G, Bell R, Abuin L, Ferrer AJ, Samuel AD, Benton R, Garrity PA (2016) Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. eLife 5:e17879. https://doi.org/10.7554/eLife.17879
Knecht ZA, Silbering AF, Cruz J, Yang L, Croset V, Benton R, Garrity PA (2017) Ionotropic receptor-dependent moist and dry cells control hygrosensation in Drosophila. eLife 6:e26654. https://doi.org/10.7554/eLife.26654
Koenig C, Hirsh A, Bucks S, Klinner C, Vogel H, Shukla A, Mansfield JH, Morton B, Hansson BS, Grosse-Wilde E (2015) A reference gene set for chemosensory receptor genes of Manduca sexta. Insect Biochem Mol Biol 66:51–63. https://doi.org/10.1016/j.ibmb.2015.09.007
Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, Carlson JR (2014) The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83(4):850–865. https://doi.org/10.1016/j.neuron.2014.07.012
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580. https://doi.org/10.1006/jmbi.2000.4315
Leal WS, Choo YM, Xu P, da Silva CS, Ueira-Vieira C (2013) Differential expression of olfactory genes in the southern house mosquito and insights into unique odorant receptor gene isoforms. Proc Natl Acad Sci U S A 110(46):18704–18709. https://doi.org/10.1073/pnas.1316059110
Liu C, Pitts RJ, Bohbot JD, Jones PL, Wang G, Zwiebel LJ (2010) Distinct olfactory signaling mechanisms in the malaria vector mosquito Anopheles gambiae. PLoS Biol 8(8):e1000467. https://doi.org/10.1371/journal.pbio.1000467
Liu NY, Xu W, Papanicolaou A, Dong SL, Anderson A (2014) Identification and characterization of three chemosensory receptor families in the cotton bollworm Helicoverpa armigera. BMC Genomics 15(1):597. https://doi.org/10.1186/1471-2164-15-597
Lu Y, Yuan M, Gao X, Kang T, Zhan S, Wan H, Li J (2013) Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One 8(7):e68059. https://doi.org/10.1371/journal.pone.0068059
Ni L, Klein M, Svec KV, Budelli G, Chang EC, Ferrer AJ, Benton R, Samuel AD, Garrity PA (2016) The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. eLife 5:e13254. https://doi.org/10.7554/eLife.13254
Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E (2011) Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol 20(2):189–199. https://doi.org/10.1111/j.1365-2583.2010.01057.x
Pearce SL, Clarke DF, East PD, Elfekih S, Gordon KHJ, Jermiin LS, McGaughran A, Oakeshott JG, Papanikolaou A, Perera OP, Rane RV, Richards S, Tay WT, Walsh TK, Anderson A, Anderson CJ, Asgari S, Board PG, Bretschneider A, Campbell PM, Chertemps T, Christeller JT, Coppin CW, Downes SJ, Duan G, Farnsworth CA, Good RT, Han LB, Han YC, Hatje K, Horne I, Huang YP, Hughes DST, Jacquin-Joly E, James W, Jhangiani S, Kollmar M, Kuwar SS, Li S, Liu NY, Maibeche MT, Miller JR, Montagne N, Perry T, Qu J, Song SV, Sutton GG, Vogel H, Walenz BP, Xu W, Zhang HJ, Zou Z, Batterham P, Edwards OR, Feyereisen R, Gibbs RA, Heckel DG, McGrath A, Robin C, Scherer SE, Worley KC, Wu YD (2017) Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol 15(1):63. https://doi.org/10.1186/s12915-017-0402-6
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth 8(10):785–786. https://doi.org/10.1038/nmeth.1701
Pitts RJ, Derryberry SL, Zhang Z, Zwiebel LJ (2017) Variant ionotropic receptors in the malaria vector mosquito Anopheles gambiae tuned to amines and carboxylic acids. Sci Rep 7:40297. https://doi.org/10.1038/srep40297
Prieto-Godino LL, Rytz R, Cruchet S, Bargeton B, Abuin L, Silbering AF, Ruta V, Dal Peraro M, Benton R (2017) Evolution of acid-sensing olfactory circuits in Drosophilids. Neuron 93(3):661–676.e6. https://doi.org/10.1016/j.neuron.2016.12.024
Rytz R, Croset V, Benton R (2013) Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem Mol Biol 43(9):888–897. https://doi.org/10.1016/j.ibmb.2013.02.007
Senthilan PR, Piepenbrock D, Ovezmyradov G, Nadrowski B, Bechstedt S, Pauls S, Winkler M, Möbius W, Howard J, Göpfert MC (2012) Drosophila auditory organ genes and genetic hearing defects. Cell 150(5):1042–1054. https://doi.org/10.1016/j.cell.2012.06.043
Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS, Benton R (2011) Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci 31(38):13357–13375. https://doi.org/10.1523/JNEUROSCI.2360-11.2011
Slone J, Daniels J, Amrein H (2007) Sugar receptors in Drosophila. Curr Biol 17(20):1809–1816. https://doi.org/10.1016/j.cub.2007.09.027
Sparks JT, Bohbot JD, Dickens JC (2014) The genetics of chemoreception in the labella and tarsi of Aedes aegypti. Insect Biochem Mol Biol 48:8–16. https://doi.org/10.1016/j.ibmb.2014.02.004
Stewart S, Koh TW, Ghosh AC, Carlson JR (2015) Candidate ionotropic taste receptors in the Drosophila larva. Proc Natl Acad Sci U S A 112(14):4195–4201. https://doi.org/10.1073/pnas.1503292112
Su CY, Menuz K, Carlson JR (2009) Olfactory perception: receptors, cells, and circuits. Cell 139(1):45–59. https://doi.org/10.1016/j.cell.2009.09.015
Tauber JM, Brown EB, Li Y, Yurgel ME, Masek P, Keene AC (2017) A subset of sweet-sensing neurons identified by IR56d are necessary and sufficient for fatty acid taste. PLoS Genet 13(11):e1007059. https://doi.org/10.1371/journal.pgen.1007059
Touhara K, Vosshall LB (2009) Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol 71:307–332. https://doi.org/10.1146/annurev.physiol.010908.163209
van Giesen L, Garrity PA (2017) More than meets the IR: the expanding roles of variant ionotropic glutamate receptors in sensing odor, taste, temperature and moisture. F1000Research 6:1753. https://doi.org/10.12688/f1000research.12013.1
van Schooten B, Jiggins CD, Briscoe AD, Papa R (2016) Genome-wide analysis of ionotropic receptors provides insight into their evolution in Heliconius butterflies. BMC Genomics 17(1):1–15. https://doi.org/10.1186/s12864-016-2572-y
Wang TT, Si FL, He ZB, Chen B (2018) Genome-wide identification, characterization and classification of ionotropic glutamate receptor genes (iGluRs) in the malaria vector Anopheles sinensis (Diptera: Culicidae). Parasit Vectors 11:34. https://doi.org/10.1186/s13071-017-2610-x
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Xu W, Anderson A (2015) Carbon dioxide receptor genes in cotton bollworm Helicoverpa armigera. Naturwissenschaften 102(3–4):1260. https://doi.org/10.1007/s00114-015-1260-0
Xu W, Papanicolaou A, Liu NY, Dong SL, Anderson A (2015) Chemosensory receptor genes in the oriental tobacco budworm Helicoverpa assulta. Insect Mol Biol 24(2):253–263. https://doi.org/10.1111/imb.12153
Zhang YV, Ni J, Montell C (2013) The molecular basis for attractive salt-taste coding in Drosophila. Science 340(6138):1334–1338. https://doi.org/10.1126/science.1234133
Acknowledgements
We thank Miss Mei-Yan Zheng (College of Plant Protection, Nanjing Agricultural University) for providing S. litura pupae.
Funding
This work was supported by the National Natural Science Foundation of China (31601647).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Sven Thatje
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhu, JY., Xu, ZW., Zhang, XM. et al. Genome-based identification and analysis of ionotropic receptors in Spodoptera litura. Sci Nat 105, 38 (2018). https://doi.org/10.1007/s00114-018-1563-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00114-018-1563-z