Abstract
Base excision repair (BER) is a frontline repair mechanism that operates through the G1 phase of the cell cycle, which ensures the genome integrity by repairing thousands of DNA lesions due to endogenous and exogenous agents. Its correct functioning is fundamental for cell viability and the health of the organism. Uracil is one of the most prevalent lesions that appears in DNA arising by spontaneous or enzymatic deamination of cytosine or misincorporation of the deoxyuridine 5′-triphosphate nucleotide (dUTP) in place of deoxythymidine 5′-triphosphate (dTTP) during DNA replication. In the first pathway, the uracil will preferentially pair with adenine, leading to C:G → T:A transition. When uracil in DNA arises from misincorporation of dUTP instead of dTTP, this process will result in A:U pairs. Organisms counteract the mutagenic effects of uracil in DNA using the BER repair system, which is mediated by a member of the uracil-DNA glycosylase (UDG) superfamily. Several assays evaluating the in vitro BER enzyme activity have been described so far. Some of these measure the BER activity by an oligonucleotide incision assay using radiolabeled duplex oligo. Others use circular double-stranded DNA substrates containing a defined lesion. The novelty of our method resides in its rapidity and safety (radioactive free detection) as well as in the possibility of having a reliable quantitative determination of UDG activity in both cell and tissue extracts. We also demonstrated the effectiveness of our method in assessing UDG activity in cell lines with a reduced DNA repair capacity and in different kinds of tissues.
Key messages
• Base excision repair is a fundamental repair mechanism ensuring the genome integrity.
• Uracil is one of the most prevalent lesions that appears in DNA.
• The mutagenic effects of uracil in DNA are mitigated by the uracil-DNA glycosylase.
• Several assays evaluating the in vitro BER activity have been described so far.
• A safe and quantitative assay evaluating the in vitro UDG activity is required.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genome of a cell is continuously damaged. Inevitable DNA lesions arise from the intrinsic chemical instability of the DNA molecule in the cellular environment and the normal cellular processes, which determine base oxidations, deaminations, alkylations, hydrolytic loss of DNA bases, strand breaks, misincorporated nucleotides, and other chemical modifications, such as reactions with endogenous DNA reactive species [1,2,3]. It has been estimated that every single human proliferating cell can acquire approximately 10,000–20,000 lesions every day under physiological conditions [3, 4]. These lesions, if not repaired, can alter the base-pairing of DNA and interfere with the replication and transcription process, leading to mutation accumulation and related disease [3]. Among the cellular strategies to maintain the genome integrity, base excision repair (BER) is an essential mechanism that copes with DNA base alterations, especially due to reactive species generated by metabolism [2]. BER is the principal repair mechanism and operates through the G1 phase of the cell cycle, during which error-free DNA replication is ensured by removing DNA lesions [3]. It has been widely demonstrated that the BER pathway deficiency determines genomic instability and may trigger the development of many human diseases, such as sporadic cancer, neurodegeneration, and premature aging [3,4,5,6]. The importance of BER is also emphasized by the fact that the knockout of genes belonging to the BER pathway is lethal at the embryonic stage [3].
BER is initiated by a damage-specific DNA glycosylase that recognizes the incorrect or damaged base and cleaves the N-glycosylic bond that links the base to the sugar phosphate, creating an abasic intermediate site. This site is incised by an apurinic/apyrimidinic (AP) endonuclease or AP lyase. Then, a lyase removes the remaining sugar fragment, and a DNA polymerase fills the gap. Finally, a DNA ligase seals the nick [2, 3]. The first step in BER is generally the enzymatic excision of the incorrect or damaged base from duplex DNA by a DNA glycosylase. There are several DNA glycosylases, each of which possesses binding pockets that specifically receive a set of modified bases [2, 7, 8]. Mammals have evolved 11 DNA glycosylases that recognize specific base modifications in DNA and are subdivided into four structurally distinct superfamilies: the uracil-DNA glycosylases (UDG), the helix-hairpin-helix (HhH) glycosylases, the 3-methyl-purine glycosylases (MPG), and the endonuclease VIII-like (NEIL) glycosylases [9, 10].
Uracil is one of the most prevalent lesions that appears in DNA, arising by two different pathways. These include either spontaneous/enzymatic hydrolytic deamination of cytosine or misincorporation of deoxyuridine 5′-triphosphate nucleotide (dUTP) in place of thymidine 5′-triphosphate (dTTP) during DNA synthesis. In the first pathway, the uracil, if not repaired, will preferentially pair with adenine during replication, leading to C:G → T:A transition mutations [2, 11, 12]. It has been assessed that 70 to 200 genomic cytosine bases are converted to uracil in each human cell per day [11, 12]. On the other hand, when uracil in DNA arises from misincorporation of dUTP instead of dTTP, this process will result in A:U pairs. Although not mutagenic, this pairing can be lethal for cells when present at a high level in genome [12].
To counteract the mutagenic effects of uracil in DNA, organisms use the BER pathway, which is mediated by a member of the UDG superfamily proteins. Based on substrate specificity, four different UDG activities have been identified in humans, including the uracil-DNA glycosylase (UNG), with UNG1 involved in uracil removal from mitochondrial DNA and UNG2 acting in nuclear DNA, the single-strand-selective mono-functional uracil-DNA glycosylase 1 (SMUG1), the thymine DNA glycosylase (TDG), and the methyl-CpG-binding domain 4 (MBD4). These proteins can remove uracil from both single- and double-stranded DNA when located opposite G, A, or a CpG sequence [2, 13]. It has been suggested that the principal role of UNG2 may be to excise misincorporated uracil opposite A during DNA replication. On the other hand, SMUG1 has been showed to target uracil derived from cytosine deamination as well as TDG and MBD4 may preferentially excise uracil present at CpG sequences [14,15,16,17].
The development of assays able to evaluate the activity of key enzymes involved in BER is being studied by physicians and researchers since a defective DNA repair system could lead to deleterious effects for the whole body. Many protocols evaluating BER enzyme activity have been described so far. Some of these methods measure BER enzyme activity by an oligonucleotide incision assay using a radiolabeled duplex oligo [18, 19]. Others use circular double-stranded DNA substrates containing a defined lesion [20,21,22].
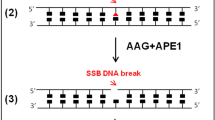
In this paper, we describe a new, simple, safe, and quantitative in vitro assay to assess the activity of UDG in both cell and fresh tissue extracts. Unlike the protocols proposed thus far, our method does not need technically demanding and time-consuming procedures. Our protocol requires the (i) preparation of cell or fresh tissue protein extracts, (ii) synthesis of a DNA substrate for UDG, (iii) assembly of an in vitro DNA incision reaction, and (iv) evaluation of UDG activity by qPCR (Fig. 1).
Illustrative diagram of method setup to test the activity of the UDG enzyme in cell and tissue extracts. Step 1: protein extraction from cell cultures and fresh tissues. Step 2: synthesis of DNA substrate for UDG. Step 3: assembly of in vitro DNA incision reaction and DNA precipitation. Step 4: evaluation of UDG activity by qPCR and data analysis
Materials and methods
Human bone marrow mesenchymal stromal cell cultures
Human bone marrow–derived mesenchymal stromal cells (MSCs) (ATCC® PCS-500-012™) were purchased from ATCC (Manassas, VA, USA). We seeded 5 × 103 cells/cm2 in alpha-minimum essential medium (Alpha-MEM) containing 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 3 ng/ml basic fibroblast growth factor (bFGF) (ThermoFisher, Waltham, MA, USA). These cells (passage 0) were further propagated to conduct experiments at passages 2–3. Cells were grown in monolayers and maintained at 37 °C under 5% CO2. All reagents were obtained from EuroClone S.p.A, Pero, Italy, unless otherwise specified.
SH-SY5Y cultures
SH-SY5Y neuroblastoma cells (ATCC® CRL-2266™) were purchased from ATCC (Manassas, VA, USA). We seeded 2 × 104 cells/cm2 in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS, 2 mM L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were grown in monolayers and maintained at 37 °C under 5% CO2. All reagents were obtained from EuroClone S.p.A., Pero, Italy.
MSC osteogenic differentiation and alizarin red staining
2 × 104 cells/cm2 were plated in osteogenic differentiation medium containing DMEM (EuroClone S.p.A., Pero, Italy) supplemented with 10% FBS (EuroClone S.p.A., Pero, Italy), 0.05 mM ascorbic acid (Sigma-Aldrich, Saint Louis, MO, USA), 10 mM β-glycerophosphate (Sigma-Aldrich, Saint Louis, MO, USA), and 100 nM dexamethasone (Sigma-Aldrich, Saint Louis, MO, USA) for 21 days, changing medium every 3 days. Thereafter, the differentiated MSCs were stained with Alizarin Red S (Sigma-Aldrich, Saint Louis, MO, USA) to visualize calcium sediments that were acquired by microscope [23].
SH-SY5Y neural differentiation
5 × 104 cells/cm2 were plated in neural differentiation medium containing DMEM/F12 (1:1) 10% FBS, and 1 μM retinoic acid (Sigma-Aldrich, Saint Louis, MO, USA) for 7 days, changing medium every 3 days. All reagents were obtained from EuroClone S.p.A., Pero, Italy, unless otherwise specified. Neural differentiation was evaluated by analyzing the mRNA expression of several neural-specific markers as already reported [24].
Protein extraction from cell cultures
After removing the culture medium, adherent cells were washed in PBS (EuroClone S.p.A., Pero, Italy). Subsequently, cells were washed in hypotonic buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 1 mM DTT) by scraping and gently resuspending them, followed by pelleting at 5000 rpm in a cooled centrifuge (Centrifuge 5417R Eppendorf Italia, Milan, Italy) for 5 min. The cellular pellet was suspended in 1 mL of hypotonic buffer in the presence of protease inhibitors (cOmplete™, EDTA-free Protease Inhibitor Cocktail-Roche, Basilea, Switzerland) and left on ice for 20 min with occasional agitation. Then, cells were homogenized in a tight-fitting glass homogenizer (Thomas Scientific, Swedesboro, NJ, USA) and mixed with 0.5 volume of high-salt buffer (50 mMTris-HCl pH 7.5, 1 M KCl, 2 mM EDTA, 1 mM DTT). The suspension was left on ice for 20 min with occasional agitation and then centrifuged at 5000 rpm for 10 min in a cooled centrifuge [25]. The Bradford assay was performed on the collected supernatant to determine protein concentration. All reagents were obtained from Sigma-Aldrich, Saint Louis, MO, USA, unless otherwise specified.
Protein extraction from tissues
Livers and muscles were dissected from C57BL/6 mice (3-month-old; male) (Jackson Laboratory, Bar Harbor, ME, USA). Mice were handled in compliance with protocols that were approved by the Animal Care and Use Committee of the University of Campania “Luigi Vanvitelli,” Naples, Italy. Tissue samples were washed with one volume of ice-cold PBS, immediately snap-frozen in liquid nitrogen, and pulverized with a porcelain mortar followed by resuspension in five fold by weight the amount of hypotonic buffer (10 mMTris-HCl pH 8.0, 1 mM EDTA, 1 mM DTT) in the presence of protease inhibitors (cOmplete™, EDTA-free Protease Inhibitor Cocktail-Roche, Basilea, Switzerland). Homogenate tissues were left on ice for 20 min, with occasional agitation, and processed as the cell culture samples.
Preparation of substrate for UDG
As substrate for UDG, we used a human DNA that was amplified by PCR with a primer pair encompassing a region of the GAPDH gene containing in its sequence several dTTPs (ID: 2597 of NCBI Gene bank; forward primer: 5′-GCATCCTGCACCACCAACTG-3′; reverse primer: 5′-GCCTGCTTCACCACCTTCTT-3′; amplicon size: 327 bp). DNA was obtained from a human blood sample with the DNeasy Blood & Tissue kit (Qiagen Italia, Milan, Italy) in compliance with the manufacturer’s instructions. Fifty nanograms of isolated DNA was amplified in 25 μL reaction mixtures containing 1X GoTaq® Flexi Buffer, 1.5 mM MgCl2, 100 μM of dNTP mix, 0.3 μM of each primer, and 1.25 U of GoTaq® Flexi DNA Polymerase. We utilized a dNTP mix containing 2.5 mM of A, C, and G deoxynucleotides, 0.25 mM of T deoxynucleotides, and 5 mM of U deoxynucleotides. All PCR reagents were obtained from Promega Italia, Milan, Italy [26]. A denaturation step at 94 °C for 3 min was followed by 32 amplification cycles (94 °C for 45 s, 55 °C for 45 s, 72 °C for 45 s). DNA was amplified on a Verity Thermal Cycler (ThermoFisher, Waltham, MA, USA). The obtained PCR products containing dUTP were used without purification as substrates for subsequent experiments.
Incubation of DNA substrates with cell and tissue extracts
Protein extracts obtained from cell cultures and fresh tissues contain active enzymes involved in DNA repair, including UDG. One hundred and fifty nanograms (ng) of DNA substrates (obtained as above reported) was incubated with 10 μg of cell/tissue extract in 20 μl of reaction buffer (70 mM HEPES-KOH pH 7.5, 1 mM EDTA, 1 mM DTT, 75 mM NaCl, 0,5% BSA) for 30 min at 37 °C [19]. In these conditions, the UDG activity of cell and tissue extracts might recognize and remove deoxyuracil from DNA substrates and decrease the subsequent PCR amplicon yield. DNA substrates incubated without protein extract represented the negative control (NC). As the positive control (PC), an equal amount of DNA substrate was treated with 0.1 U/μl AmpErase Uracil N-Glycosylase (ThermoFisher, Waltham, MA, USA) at 50 °C for 10 min, followed by enzyme inactivation at 95 °C for 5 min. To demonstrate the specificity of the UDG assay, we also treated an equal amount of DNA substrate that did not contain dUTP with cell and fresh tissue extracts. This template was obtained by PCR amplifying human DNA with a primer pair encompassing a region of the GAPDH gene. The reaction was carried out with canonical dNTP mix (dATP, dCTP, dGTP, dTTP).
All the above reactions were terminated adding 2 μl of 2 mg/ml Proteinase K and 0.8 μl of 10% SDS, followed by incubation at 55 °C for 15 min. Then, the DNA was precipitated by the addition of 2 volumes of absolute ethanol, 1 μg of glycogen, and 4 μl of 11 M ammonium acetate [19]. Samples were centrifuged at 16.000 rpm for 30 min in a cooled centrifuge (Centrifuge 5417R Eppendorf Italia, Milano, Italy). The DNA pellets were rinsed twice with 70% ethanol, dried at room temperature, and resuspended in 20 μl nuclease-free water. These samples represent the templates for the qPCR reaction described in the next paragraph.
UDG inhibition assay
For UDG inhibition assay, 0.1 U/μl uracil glycosylase inhibitor (UGI) (New England Biolabs, Ipswich, MA, USA) was incubated with 10 μg of each cell extracts (proliferating MSCs or proliferating SH-SY5Y) at 37 °C for 10 min before the addition of 150 ng of DNA substrate, followed by same procedures described above for the UDG activity measurement. As the positive control, an equal amount of DNA substrate was treated with 0.1 U/μl AmpErase Uracil N-Glycosylase (ThermoFisher, Waltham, MA, USA) and 0.1 U/μl UGI at 37 °C for 10 min, followed by same procedures described above.
UDG activity measurement with the standard curve method
Two microliters of each purified DNA template (obtained as reported above) was amplified by quantitative PCR (qPCR) in 20 μl reaction mixtures containing 1X iTaq universal SYBRGreen Supermix (Bio-Rad, Hercules, CA, USA) and 0.3 μM of each GAPDH primer. A denaturation step at 95 °C for 10 min was followed by 40 amplification cycles (95 °C for 15 s, 55 °C for 40 s) [27]. The DNA was amplified on a CFX96 Real-Time PCR detection System (Bio-Rad, Hercules, CA, USA). All reagents were obtained from Sigma-Aldrich, Saint Louis, MO, USA, unless otherwise specified.
We obtained an absolute quantification of PCR products using the standard curve method. Different known amounts (nanograms) of the DNA template were amplified by qPCR in the presence of canonical dNTPs to create a standard curve with threshold cycles (Ct) (Fig. 2a). The number of molecules of the known amounts was obtained using the following formula:
Panel aCalibration curve. The graph shows the obtained calibration curve to quantize the obtained GAPDH amplicons from analyzed samples. Different known amounts of DNA templates (without dUTP) were amplified by qPCR to create a standard curve with Ct. Panel bUDG activity in cell extracts. The graph reports the number of amplified molecules before (negative control—NC) and after the treatment of GAPDH substrates with the different cell extracts (± SD, n = 3). The asterisk(s) indicate the statistical difference between NC and each of the other experimental conditions (*p < 0.05, **p < 0.01). The hashtag indicates statistical difference between proliferating and differentiated SH-SY5Y cells (#p < 0.05). Panel cUDG assay specificity measured in cell extracts. The graphs report the number of amplified molecules before (negative control—NC) and after the cell extract treatments of GAPDH substrates that did not contain dUTP (± SD, n = 3). Panel dEffect of UDG on DNA templates containing dUTPs. DNA templates containing dUTP (as those in panel b) were incubated with AmpErase Uracil N-Glycosylase (ThermoFisher, Waltham, MA, USA) and then PCR amplified. This was a positive control (PC) of the UDG effect on DNA templates (± SD, n = 3)
This allowed us to obtain a relative measure of the amounts of amplicons in the samples treated with different protein extracts by comparing them to the standard curve and extrapolating the value. A lower quantity of obtained amplicon represents a higher UDG activity.
Statistical analysis
Statistical significance was evaluated using an analysis of variance, followed by Student’s t and Bonferroni’s tests. For data with continuous outcomes, we used the mixed-model variance analysis and, in any case, we analyzed all data with the GraphPad Prism, version 5.01 (GraphPad, La Jolla, CA, USA).
Results
Measurement of UDG activity in proliferating and differentiated cell lines
It is well known that the cell’s capacity for repairing DNA damage depends on the enzymatic activity of proteins involved in DNA repair mechanisms. Damage-specific DNA glycosylases are key enzymes in the BER pathway; they recognize and remove the incorrect or damaged base from DNA [2]. We set up a simple method to evaluate the UDG capacity to recognize and cleave the misincorporated bases.
We incubated protein extracts from human bone marrow MSCs or SH-SY5Y neuroblastoma cells with DNA templates containing dUTP. The UDG present in protein extracts recognizes and cleaves the misincorporated uracils, and the enzymatic activity is indirectly determined by calculating the PCR amplicon yield obtained following the incubation of templates with cell extracts. Therefore, a lower quantity of obtained amplicons will representa higher UDG activity.
We evaluated UDG activity both in normal and tumor proliferating cell lines, that is, MSCs and SH-SY5Y cells, respectively. We chose MSCs since they have a robust and effective DNA repair mechanism given their well-known key role in promoting the homeostasis of several tissues and organs [28,29,30,31,32]. Moreover, stem cells, such as those present in MSC population, persist for long periods in the body and undergo several rounds of intrinsic and extrinsic stresses. As a result, they present an effective DNA repair system in order to cope DNA damage and avoid mutation accumulation and related diseases [33]. Cancer cells should also present high DNA repair capacity due to their ability to remove therapy-induced DNA lesions and thus to develop therapy resistance [34]. Among cancer cell lines we chose to evaluate the UDG activity in SH-SY5Y cells, one of the most commonly used cell lines in cancer and neuroscience research [35].
In addition, we collected cellular extracts from MSCs and SH-SY5Y cells following induction of differentiation to assess the effectiveness of our method in experimental conditions showing a low DNA repair activity. Indeed, many terminally differentiated cells seem to have a downregulation of DNA repair systems. They do not need to replicate their DNA and may dispense the several repair systems to constantly control the genome integrity [36, 37].
MSCs contain multipotent cells able to differentiate in vitro into several cell types, such as bone, osteocytes, chondrocytes, and adipocytes [29, 38]. We chose to differentiate MSCs towards the osteocyte phenotype by treating cells with ascorbic acid, β-glycerophosphate, and dexamethasone (see “Materials and methods”). On the other hand, we induced neuronal differentiation of SH-SY5Y cells by using retinoic acid [39].
DNA substrates containing several dUTP in their sequence were incubated with cell lysates from proliferating and differentiated MSCs or SH-SY5Y cells. Subsequently, these substrates were used as templates for a qPCR reaction that amplifies a 327 bp fragment of GAPDH gene. In these conditions, the UDG activity of cell extracts might recognize and remove deoxyuracil from the DNA substrate and decrease the PCR amplicon yield. Different known amounts of GAPDH substrates (without dUTP) were amplified by qPCR to create a standard curve with threshold cycles (Ct) (Fig. 2a). This allowed us to obtain a relative measure of the amount of amplicon in the samples treated with different cell extracts by comparing unknowns to the standard curve and extrapolating the value of the sample under analysis. A lower quantity of obtained amplicon represents a higher UDG activity.
The in vitro incision assay showed that in both proliferating and differentiated cell lines, the BER pathway was active. In detail, a higher UDG activity was observed in proliferating conditions for both MSCs and SH-SY5Y cells with respect to their differentiated correspondents (Fig. 2b). Still, the UDG’s ability to remove the incorrect bases varies depending on the cell line analyzed; the SH-SY5Y cells in proliferating conditions had a greater ability to remove the misincorporated uracils compared to proliferating MSCs (Fig. 2b). The absence of interference by non-specific nuclease activity was demonstrated by the PCR amplicon yield obtained following the incubation in cell extracts of DNA templates that did not contain UTP. Indeed, in all these cases, the PCR amplicon yield was approximately the same as that obtained in the negative control (qPCR of DNA templates with no cell extract incubation) (Fig. 2c).
As a positive control, we treated an equal amount of DNA templates containing dUTP with AmpErase Uracil N-Glycosylase enzyme. Obviously, in this case, we obtained a negligible PCR amplicon yield (Fig. 2d).
Finally, to further confirm that observed incision activity was due to UDG rather than other components in cell extracts, we performed a UDG inhibition assay by pre-treating proliferating MSC and SH-SY5Y lysates with UGI (see “Materials and Methods”). The UGI is able to bind with UDG in 1:1 UDG:UGI stoichiometry by forming an irreversible complex [40, 41]. As shown in Fig. 3a, following UGI treatment, MSC and SH-SY5Y lysates lost the ability to remove the misincorporated bases. Indeed, for both cell lines, the PCR amplicon yield was comparable to that observed in the negative control sample (qPCR of DNA templates with no cell extract incubation) (Fig. 3a).
Panel aUDG inhibition assay in cell extracts. The graph reports the number of amplified molecules before (negative control—NC) and after the treatment of GAPDH substrates with the extracts from proliferating MSC or proliferating SH-SY5Y cells with or without (w/o) UGI treatment, respectively (± SD, n = 3). The asterisk(s) indicate the statistical difference between NC and each of the other experimental conditions (*p < 0.05, **p < 0.01). The hashtag indicates statistical difference between proliferating MSCs w/o UGI and proliferating MSCs with UGI (#p < 0.05). The degrees indicate statistical difference between proliferating SH-SY5Y cells w/o UGI and prolioferating SH-SY5Y cells with UGI (°°p < 0.01). Panel bEffect of UDG on DNA templates containing dUTPs. DNA templates containing dUTP (as those in panel a) were incubated with AmpErase Uracil N-Glycosylase (ThermoFisher, Waltham, MA, USA) or with AmpErase Uracil N-Glycosylase plus UGI and then PCR amplified, respectively. The latter was the positive control (PC) of the UGI effect on UDG inhibition (± SD, n = 3). The asterisks indicate the statistical difference between PC with UGI and PC w/o UGI (**p < 0.01)
As a positive control of the inhibition assay, an equal amount of DNA substrate was treated with AmpErase Uracil N-Glycosylase and UGI. As expected, also in this case, we observed a PCR amplicon yield similar to that of the negative control (Fig. 3a, b).
Measurement of UDG activity in tissue samples
For many studies, it is preferable to evaluate BER activity in the tissue of interest instead of using cell lines derived from that tissue [42, 43]. We also tested our assay to assess UDG activity in tissue samples. We measured UDG activity in the liver and muscle of adult mice using the same protocol described for the cell extracts in order to compare DNA repair in different tissue types. We chose two different tissues to demonstrate that our protocol was suitable for obtaining comparable total protein extracts from different kinds of samples. The liver and muscle tissues exhibit high structural and morphological differences; the former is made of very soft tissues encapsulated by a connective tissue unlike the latter, which is a hard tissue made of multiple bundles of cells (fibers) surrounded by connective tissue layers [44, 45].
Figure 4 shows that our method allowed us to detect UDG activity in both analyzed tissue samples. In detail, we observed no significant differences in the PCR amplicon yield following incubation with the different tissue extracts (Fig. 4a). As for the cell lines, the specificity of UDG activity in tissue extracts was demonstrated by the PCR amplicon yield obtained following the incubation of an equal amount of DNA templates (not containing dUTP) with the extracts from liver and muscle, respectively. In all the cases, the PCR amplicon yield was comparable to that observed in the negative control samples (DNA templates with no tissue extract) (Fig. 4b). As a positive control, we treated an equal amount of DNA templates containing dUTP with AmpErase Uracil N-Glycosylase enzyme (Fig. 4c).
Panel aUDG activity in tissue extracts. The graphs report the number of amplified molecules before (negative control—NC) and after the treatment of GAPDH substrates with the different tissue extracts (± SD, n = 3). The asterisk indicates the statistical difference between NC and each of the other experimental conditions (*p < 0.05). Panel bAssay specificity for UDG activity measured in tissue extracts. The graphs report the number of amplified molecules before (negative control—NC) and after the cell extract treatments of GAPDH substrates that did not contain dUTP(± SD, n = 3). Panel cEffect of UDG on DNA templates containing dUTPs. DNA templates containing dUTP (as those in panel a) were incubated with AmpErase Uracil N-Glycosylase (ThermoFisher, Waltham, MA, USA) and then PCR amplified. This was a positive control (PC) of the UDG effect on DNA templates (± SD, n = 3)
Discussion
BER is a frontline mechanism that operates through the G1 phase of the cell cycle, which ensures the genome integrity by repairing thousands of DNA lesions due to endogenous and exogenous mutagens or stressor agents. Thus, BER activity prevents many human diseases, such as cancer, neurodegeneration, and premature aging [3,4,5,6]. Its correct functioning is fundamental for cell viability and, therefore, for the health of the whole organism.
Uracil is one of the most prevalent lesions that appears in DNA, arising by spontaneous or enzymatic hydrolytic deamination of cytosine or misincorporation of dUTP in place of dTTP during DNA replication phase [2, 12]. Organisms counteract the mutagenic effects of uracil in DNA using the BER repair system, which is, in turn, mediated by a member of the UDG superfamily proteins [2, 13].
Several assays evaluating the BER activity in vitro using cell or tissue extracts have been described thus far [18,19,20,21,22, 46,47,48,49]. Some of these methods measure BER activity by an oligonucleotide incision assay using radiolabeled duplex oligo [18, 19]; others use circular double-stranded DNA substrates containing a defined lesion [20,21,22].
We described a rapid, safe, and quantitative in vitro assay to assess the enzyme activity of UDG in both cell and tissue extracts. Unlike several protocols proposed thus far, our method requires simple, not technically demanding and time-consuming procedures that are the (i) preparation of cell or fresh tissue extracts, (ii) use of a DNA segment amplified by PCR as a UDG substrate, (iii) assembly of an in vitro DNA repair reaction, and (iv) evaluation of UDG activity by qPCR. In particular, UDG substrates can be easily prepared by the PCR amplification of a DNA region containing several dTTPs. To this end, we added dUTPs to the canonical nucleotide mixture used for PCR reaction. At the same time, measurement of UDG activity is readily derived from the qPCR amplicon yield obtained following the incubation of DNA substrates containing dUTP with cell or tissue extracts. The use of qPCR allows the absolute quantification of the undigested amplicons in the samples treated with different protein extracts. Moreover, the use of low-cost reagents and traditional molecular biology techniques makes our assay more convenient than other protocols proposed until now.
Another advantage of the developed assay is that our method does not use radioisotopes for the detection of UDG enzyme activity. Frosina et al. developed a quantitative assay for measuring BER activity based on the use of mammalian cell extracts and the construction of a radioactive plasmid containing a single AP site or DNA lesions repaired via an AP site formation as a repair reaction substrate. Despite the assay reliability, it requires technically demanding and time-consuming procedures, such as the construction of DNA substrates containing lesions, the radiolabeling of DNA substrates, and DNA purification by density gradient centrifugation with cesium chloride. In addition, the protocol needs radioactive substrates for the detection of repair products [20]. On the other hand, Matsumoto described a convenient and safe in vitro BER assay which does not require radiolabelled substrates or nucleotides. Nevertheless, DNA repair reaction products are analyzed by agarose gel electrophoresis and detected by staining with SYBR Green providing a semi-quantitative measurement of BER activity. Also, this protocol requires a time-consuming procedure for the construction of the circular DNA substrate containing the AP site and the DNA sample purification with the cesium chloride method [22].
The Bohr’s research team reported several protocols to measure the repair activity of UDG, oxoguanine DNA glycosylase (OGG1), and endonuclease III homolog (NTH1) in mouse tissues based on the oligonucleotide incision assay using a synthetic duplex oligo. Although such assays take less time than those discussed above and provide a quantitative measure of BER enzymatic activity, they require the radiolabeling of duplex oligo [18, 19, 50, 51].
A few years ago, Leung et al. published an interesting review describing a variety of methods based on luminescent detection of BER enzyme activity. Luminescence technologies guarantee a good combination of high sensitivity, simplicity, safety, and rapid response time and are usually inexpensive [52]. Nevertheless, even in this case, our method results less expensive and more rapid than luminescent oligonucleotide-based protocols. For example, the assay described by Hu and co-workers requires the purchase of G-quadruplex-DNA and the DNA specific fluorescent binder N-methyl mesoporphyrin IX as well as the protocol by Leung et al. needs the use of iridium(III) complex as a G-quadruplex-selective probe [53, 54]. Again, Zhang and co-workers proposed the use of the target-activated autocatalytic DNAzyme amplification and the rolling circle amplification strategies for the sensitive and fluorescent detection of UDG [55]. The fluorescent ultrahigh sensitivity assay developed by Wang’s research group requires the multi-step excision repair-initiated enzyme-assisted bicyclic cascade signal amplification for the ultrasensitive detection of UDG activity [40]. Of interest is also the method described by Zhang et al. for the simultaneous detection of multiple DNA glycosylase activity using exonuclease-assisted recycling signal amplification in combination with intrinsically fluorescent nucleotides [56]. Despite their distinct advantages of high sensitivity, selectivity, and safety, these latest protocols are more time consuming than the assay we developed. Moreover, they require the synthesis of fluorophore-labeled or modified oligonucleotide substrates and/or probes that are more expensive compared to the home-made DNA substrate required by our protocol.
Recently, Esadze and collaborators described an intriguing and efficient fluorescence assay to determine the activity of human UNG2 in cell extracts. Nevertheless, the method requires the use of a fluorescent substrate (FAM-labeled and containing a quencher) that is more expensive compared to our home-made substrate. In addition, our method is based on PCR-based assays that should have a lower limit of detection (LOD) and limit of quantification (LOQ) compared with fluorescent assays. However, it has to be noted that the paper by Esadze et al. has the merit of having described the first measurements that directly assess the nanoscale DNA translocation by UDG in human cells [41].
Despite the advantages offered by the method we developed, we are aware that it does not allow to differentiate which UDG enzyme is removing the uracil bases in the assay. In any case, our method allows to obtain an overall quantitative measure of the activity of UDG—the enzyme responsible for the initial steps of genomic uracil processing—useful for demonstrating differences in the activation of BER mechanism between different cell or tissue samples.
In conclusion, in this paper, we have described a new in vitro assay to evaluate the enzymatic activity of UDGs in both cell and fresh tissue extracts. We also showed the effectiveness of our method in assessing UDG activity in differentiated cell lines with a reduced activity of DNA repair mechanisms. Moreover, we demonstrated that it is suitable to evaluate the repair activity in extracts from different kinds of tissues. The developed assay has the advantage of simplifying the protocols proposed so far, providing a quantitative measurement of UDG activity without using a radioactive-based detection method and producing results rapidly. Finally, we demonstrated that the protocol can be applied to normal and cancer cell lines as well as fresh tissues, thus also allowing the evaluation of UDG activity in clinical samples. The knowledge of DNA repair activity in human samples is of major importance since unrepaired DNA lesions leads to replication errors, and this, in turn, may result in mutagenesis, premature aging, and other diseases, such as neurodegeneration and cancer.
Abbreviations
- BER:
-
Base excision repair
- dUTP:
-
Deoxyuridine 5′-triphosphate nucleotide
- dTTP:
-
Deoxythymidine 5′-triphosphate
- UDG:
-
Uracil-DNA glycosylase
- AP:
-
Apurinic/apyrimidinic
- HhH:
-
Helix-hairpin-helix
- MPG:
-
3-Methyl-purine glycosylase
- NEIL:
-
Endonuclease VIII-like
- MSCs:
-
Mesenchymal stromal cells
- UNG:
-
Uracil N-glycosylase
- SMUG1:
-
Single-strand-selective mono-functional uracil-DNA glycosylase 1
- TDG:
-
Thymine DNA glycosylase
- MBD4:
-
Methyl-CpG-binding domain 4
References
Friedberg EC (2003) DNA damage and repair. Nature 421:436–440
Kim YJ, Wilson DM 3rd (2012) Overview of base excision repair biochemistry. Curr Mol Pharmacol 5:3–13
Dianov GL, Hubscher U (2013) Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res 41:3483–3490
Markkanen E, Fischer R, Ledentcova M, Kessler BM, Dianov GL (2015) Cells deficient in base-excision repair reveal cancer hallmarks originating from adjustments to genetic instability. Nucleic Acids Res 43:3667–3679
Caldecott KW (2008) Single-strand break repair and genetic disease. Nat Rev Genet 9:619–631
Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW (2005) DNA repair, genome stability, and aging. Cell 120:497–512
Robertson AB, Klungland A, Rognes T, Leiros I (2009) DNA repair in mammalian cells: base excision repair: the long and short of it. Cell Mol Life Sci 66:981–993
Dalhus B, Laerdahl JK, Backe PH, Bjoras M (2009) DNA base repair--recognition and initiation of catalysis. FEMS Microbiol Rev 33:1044–1078
Jacobs AL, Schar P (2012) DNA glycosylases: in DNA repair and beyond. Chromosoma 121:1–20
Brooks SC, Adhikary S, Rubinson EH, Eichman BF (2013) Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta 1834:247–271
Sire J, Querat G, Esnault C, Priet S (2008) Uracil within DNA: an actor of antiviral immunity. Retrovirology 5:45
Prorok P, Alili D, Saint-Pierre C, Gasparutto D, Zharkov DO, Ishchenko AA, Tudek B, Saparbaev MK (2013) Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc Natl Acad Sci U S A 110:E3695–E3703
Whitaker AM, Schaich MA, Smith MR, Flynn TS, Freudenthal BD (2017) Base excision repair of oxidative DNA damage: from mechanism to disease. Front Biosci 22:1493–1522
Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE (1999) Post-replicative base excision repair in replication foci. EMBO J 18:3834–3844
Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T (2001) Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J 20:4278–4286
Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Genuardi M, Karbowski M, Yeung AT, Matsumoto Y, Bellacosa A (2000) Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J Cell Physiol 185:473–480
Waters TR, Swann PF (2000) Thymine-DNA glycosylase and G to A transition mutations at CpG sites. Mutat Res 462:137–147
Imam SZ, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA (2006) Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol Aging 27:1129–1136
Souza-Pinto NC, Croteau DL, Hudson EK, Hansford RG, Bohr VA (1999) Age-associated increase in 8-oxo-deoxyguanosine glycosylase/AP lyase activity in rat mitochondria. Nucleic Acids Res 27:1935–1942
Frosina G, Cappelli E, Ropolo M, Fortini P, Pascucci B, Dogliotti E (2006) In vitro base excision repair assay using mammalian cell extracts. Methods Mol Biol 314:377–396
Matsumoto Y (1999) Base excision repair assay using Xenopus laevis oocyte extracts. Methods Mol Biol 113:289–300
Matsumoto Y (2006) Base excision repair in mammalian cells. Methods Mol Biol 314:365–375
Alessio N, Stellavato A, Squillaro T, Del Gaudio S, Di Bernardo G, Peluso G, De Rosa M, Schiraldi C, Galderisi U (2018) Hybrid complexes of high and low molecular weight hyaluronan delay in vitro replicative senescence of mesenchymal stromal cells: a pilot study for future therapeutic application. Aging 10:1575–1585
Galderisi U, Di Bernardo G, Cipollaro M, Peluso G, Cascino A, Cotrufo R, Melone MA (1999) Differentiation and apoptosis of neuroblastoma cells: role of N-myc gene product. J Cell Biochem 73:97–105
Diggle CP, Bentley J, Kiltie AE (2003) Development of a rapid, small-scale DNA repair assay for use on clinical samples. Nucleic Acids Res 31:e83
Melone MA, Giuliano M, Squillaro T, Alessio N, Casale F, Mattioli E, Cipollaro M, Giordano A, Galderisi U (2009) Genes involved in regulation of stem cell properties: studies on their expression in a small cohort of neuroblastoma patients. Cancer Biol Ther 8:1300–1306
Cirillo A, Di Salle A, Petillo O, Melone MA, Grimaldi G, Bellotti A, Torelli G, De’ Santi MS, Cantatore G, Marinelli A et al (2014) High grade glioblastoma is associated with aberrant expression of ZFP57, a protein involved in gene imprinting, and of CPT1A and CPT1C that regulate fatty acid metabolism. Cancer Biol Ther 15:735–741
Galderisi U, Giordano A (2014) The gap between the physiological and therapeutic roles of mesenchymal stem cells. Med Res Rev 34:1100–1126
Alessio N, Pipino C, Mandatori D, Di Tomo P, Ferone A, Marchiso M, Melone MAB, Peluso G, Pandolfi A, Galderisi U (2018) Mesenchymal stromal cells from amniotic fluid are less prone to senescence compared to those obtained from bone marrow: an in vitro study. J Cell Physiol 233:8996–9006
Squillaro T, Peluso G, Galderisi U (2016) Clinical trials with mesenchymal stem cells: an update. Cell Transplant 25:829–848
Alessio N, Squillaro T, Ozcan S, Di Bernardo G, Venditti M, Melone M, Peluso G, Galderisi U (2018) Stress and stem cells: adult Muse cells tolerate extensive genotoxic stimuli better than mesenchymal stromal cells. Oncotarget 9:19328–19341
Alessio N, Capasso S, Di Bernardo G, Cappabianca S, Casale F, Calarco A, Cipollaro M, Peluso G, Galderisi U (2017) Mesenchymal stromal cells having inactivated RB1 survive following low irradiation and accumulate damaged DNA: hints for side effects following radiotherapy. Cell Cycle 16:251–258
Squillaro T, Alessio N, Di Bernardo G, Ozcan S, Peluso G, Galderisi U (2018) Stem cells and DNA repair capacity: muse stem cells are among the best performers. Adv Exp Med Biol 1103:103–113
Abbotts R, Thompson N, Madhusudan S (2014) DNA repair in cancer: emerging targets for personalized therapy. Cancer Manag Res 6:77–92
Forster JI, Koglsberger S, Trefois C, Boyd O, Baumuratov AS, Buck L, Balling R, Antony PM (2016) Characterization of differentiated SH-SY5Y as neuronal screening model reveals increased oxidative vulnerability. J Biomol Screen 21:496–509
Nouspikel T (2007) DNA repair in differentiated cells: some new answers to old questions. Neuroscience 145:1213–1221
Fortini P, Dogliotti E (2010) Mechanisms of dealing with DNA damage in terminally differentiated cells. Mutat Res 685:38–44
Jori FP, Napolitano MA, Melone MA, Cipollaro M, Cascino A, Giordano A, Galderisi U (2004) Role of RB and RB2/P130 genes in marrow stromal stem cells plasticity. J Cell Physiol 200:201–212
Murillo JR, Goto-Silva L, Sanchez A, Nogueira FCS, Domont GB, Junqueira M (2017) Quantitative proteomic analysis identifies proteins and pathways related to neuronal development in differentiated SH-SY5Y neuroblastoma cells. EuPA Open Proteom 16:1–11
Wang LJ, Ren M, Zhang Q, Tang B, Zhang CY (2017) Excision repair-initiated enzyme-assisted bicyclic cascade signal amplification for ultrasensitive detection of uracil-DNA glycosylase. Anal Chem 89:4488–4494
Esadze A, Rodriguez G, Weiser BP, Cole PA, Stivers JT (2017) Measurement of nanoscale DNA translocation by uracil DNA glycosylase in human cells. Nucleic Acids Res 45:12413–12424
Gorniak JP, Cameron KM, Waldron KJ, von Zglinicki T, Mathers JC, Langie SA (2013) Tissue differences in BER-related incision activity and non-specific nuclease activity as measured by the comet assay. Mutagenesis 28:673–681
Langie SA, Cameron KM, Waldron KJ, Fletcher KP, von Zglinicki T, Mathers JC (2011) Measuring DNA repair incision activity of mouse tissue extracts towards singlet oxygen-induced DNA damage: a comet-based in vitro repair assay. Mutagenesis 26:461–471
Tamai M, Adachi E, Tagawa Y (2013) Characterization of a liver organoid tissue composed of hepatocytes and fibroblasts in dense collagen fibrils. Tissue Eng A 19:2527–2535
Ceglia L (2008) Vitamin D and skeletal muscle tissue and function. Mol Asp Med 29:407–414
Frosina G, Fortini P, Rossi O, Carrozzino F, Abbondandolo A, Dogliotti E (1994) Repair of abasic sites by mammalian cell extracts. Biochem J 304(Pt 3):699–705
Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J 15:6662–6670
Klungland A, Lindahl T (1997) Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J 16:3341–3348
Allinson SL, Dianova II, Dianov GL (2001) DNA polymerase beta is the major dRP lyase involved in repair of oxidative base lesions in DNA by mammalian cell extracts. EMBO J 20:6919–6926
de Souza-Pinto NC, Hogue BA, Bohr VA (2001) DNA repair and aging in mouse liver: 8-oxodG glycosylase activity increase in mitochondrial but not in nuclear extracts. Free Radic Biol Med 30:916–923
Stuart JA, Karahalil B, Hogue BA, Souza-Pinto NC, Bohr VA (2004) Mitochondrial and nuclear DNA base excision repair are affected differently by caloric restriction. FASEB J 18:595–597
Leung CH, Zhong HJ, He HZ, Lu L, Chanb DSH, Ma DL (2013) Luminescent oligonucleotide-based detection of enzymes involved with DNA repair. Chem Sci 4:3781
Hu D, Huang Z, Pu F, Ren J, Qu X (2011) A label-free, quadruplex-based functional molecular beacon (LFG4-MB) for fluorescence turn-on detection of DNA and nuclease. Chemistry 17:1635–1641
Leung KH, He HZ, Ma VP, Zhong HJ, Chan DS, Zhou J, Mergny JL, Leung CH, Ma DL (2013) Detection of base excision repair enzyme activity using a luminescent G-quadruplex selective switch-on probe. Chem Commun 49:5630–5632
Zhang L, Zhao J, Jiang J, Yu R (2012) A target-activated autocatalytic DNAzyme amplification strategy for the assay of base excision repair enzyme activity. Chem Commun 48:8820–8822
Zhang Y, Li CC, Tang B, Zhang CY (2017) Homogeneously sensitive detection of multiple DNA glycosylases with intrinsically fluorescent nucleotides. Anal Chem 89:7684–7692
Acknowledgements
M.A.B.M. and T.S. acknowledge PON I&C 2014-2020 “Micro/nanoformulati innovativi per la valorizzazione di molecole bioattive, utili per la salute e il benessere delle popolazione, ottenute da prodotti di scarto della filiera ittica (FOR.TUNA)” project, Grant/Award Number: F/050347/03IX32 – Ministero dello Sviluppo Economico (MiSE).
Author information
Authors and Affiliations
Contributions
TS and MF planned and performed experiments and wrote the paper; SDG performed experiments; NA and GDB analyzed data; MABM and GP contributed reagents and other essential material; UG conceived and supervised the study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Squillaro, T., Finicelli, M., Alessio, N. et al. A rapid, safe, and quantitative in vitro assay for measurement of uracil-DNA glycosylase activity. J Mol Med 97, 991–1001 (2019). https://doi.org/10.1007/s00109-019-01788-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-019-01788-8