Abstract
Long non-coding RNAs (lncRNAs) have been reported to be involved in many important biological processes including proliferation, apoptosis, differentiation, and survival. Recently, nuclear paraspeckle assembly transcript 1 (NEAT1), a novel lncRNA, serves as a crucial regulator in tumors. However, the biological role of NEAT1 in liver fibrosis is largely unknown. In this study, the role of NEAT1 was explored in primary mouse hepatic stellate cells (HSCs) and carbon tetrachloride (CCl4)-induced mouse liver fibrosis models. We found that NEAT1 expression was significantly increased in CCl4-induced mice and activated HSCs. Loss of NEAT1 suppressed liver fibrosis in vivo and in vitro. Conversely, NEAT1 overexpression accelerated HSC activation, including increased cell proliferation and collagen expression. Further studies indicated that the microRNA-122 (miR-122)-Kruppel-like factor 6 (KLF6) axis was involved in the effects of NEAT1 on HSC activation. The effects of NEAT1 on HSC activation were almost blocked down by miR-122 mimics or KLF6 knockdown. Interestingly, both NEAT1 and KLF6 are targets of miR-122. In addition, miR-122 led to a significant reduction in NEAT1 level while NEAT1 overexpression resulted in the suppression of miR-122 expression. Pull-down assay confirmed a direct interaction between miR-122 and NEAT1. NEAT1 contributes to HSC activation via the miR-122-KLF6 axis. In human fibrotic liver samples, increased NEAT1 levels positively correlated with liver fibrosis markers. In conclusion, we disclose a novel NEAT1-miR-122-KLF6 signaling cascade and its implication in liver fibrosis.
Key messages
-
NEAT1 was significantly increased in CCl4-induced mice and activated HSCs.
-
Loss of NEAT1 suppressed liver fibrosis in vivo and in vitro.
-
KLF6 and miR-122 were required for the effects of NEAT1 on HSC activation.
-
NEAT1 contributes to HSC activation via competitively binding miR-122.
-
We disclose a novel NEAT1-miR-122-KLF6 signaling cascade.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Regulatory non-coding RNAs (ncRNAs), such as microRNAs (miRNAs), have been widely studied in recent years [1,2,3]. miRNAs are highly conserved non-coding RNAs of approximately 20–24 nucleotides. miRNAs regulate gene expression at post-transcriptional level by blocking translation at the initiation or post-initiation steps, inducing messenger RNA (mRNA) deadenylation and decay [4]. miRNAs are frequently deregulated in liver fibrosis and involved in a wide range of biological processes, including cell proliferation, apoptosis, and differentiation [1, 5]. For instance, miR-29b suppresses the activation of hepatic stellate cells (HSCs) through the de-methylation of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) [6]. Therefore, miRNAs are involved in HSC activation and act as regulators of the activation of HSCs in liver fibrosis.
Long non-coding RNAs (lncRNAs), commonly defined as a non-protein-coding RNA, are transcribed RNA molecules (> 200 nucleotides in length). Recent studies have shown that lncRNAs regulate vital biological processes, including proliferation, survival, apoptosis, and differentiation [7,8,9]. LncRNAs are found to be implicated in liver fibrosis [10,11,12]. For example, growth arrest-specific transcript 5 (GAS5) inhibits liver fibrosis through a mechanism of competing endogenous RNA (ceRNAs) [11].
Nuclear paraspeckle assembly transcript 1 (NEAT1) is a novel lncRNA, which is required for the formation of paraspeckles [13]. Recently, NEAT1 has been reported to serve as a crucial regulator in cancers and is upregulated in gastric adenocarcinomas and human laryngeal squamous cell cancer [14, 15]. Upregulation of NEAT1 plays a key role in promoting cell proliferation and survival and inhibiting apoptosis, thereby contributing to tumorigenesis [16]. High expression level of NEAT1 is associated with the clinical features of hepatocellular carcinoma [17]. However, the function and role of NEAT1 in liver fibrosis are still unclear. In this study, we aimed to explore the role of NEAT1 in liver fibrosis.
Materials and methods
Isolation and culture of primary HSCs and hepatocytes
Primary HSCs were isolated as described previously [18]. The isolated cells were seeded in tissue culture plates and cultured in DMEM with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. The purity of cultures was confirmed by immunocytochemical staining for α-smooth muscle actin (α-SMA), and the purity reached > 98%. Hepatocytes were isolated using a two-step collagenase perfusion technique [19]. Then, gene expression levels including F4/80, CD32b, and CYP3A11 were measured by quantitative real-time PCR (qRT-PCR). Hepatocyte purity was found to be greater than 95%.
Carbon tetrachloride (CCl4) liver injury model
Eight-week-old male C57BL/6J mice (n = 6) received intraperitoneal injection of 7 μL/g of 10% CCl4 (Sigma-Aldrich, St. Louis, MO, USA) in olive oil two times weekly for 6 weeks. Also, mice (n = 6) treated with olive oil treatment were considered as the control mice. As well as oil treatment and CCl4 treatment, mice additionally received CCl4 in combination with adenoviral vectors expressing the scrambled shRNA (Ad-shCtrl) (n = 6) and CCl4 in combination with adenoviral vectors expressing shRNA against NEAT1 (Ad-shNEAT1) (n = 6). Ad-shNEAT1 (1 × 109 pfu/100 μL) was injected every 2 weeks by way of the tail vein for 6 weeks. All animals were provided by the Experimental Animal Center of Wenzhou Medical University. The animal experimental protocol was approved by the University Animal Care and Use Committee. Mice were sacrificed under anesthesia after CCl4 treatment. The livers from mice were removed for further analysis. The liver tissues were used for Masson staining by fixation with 10% formalin. Quantitative analysis for the Masson-positive area was calculated from five fields for each liver slice.
Statistical analysis
Data from at least three independent experiments were expressed as the mean ± SD. Differences between multiple groups were evaluated using one-way analysis of variance. Differences between two groups were compared using a Student t test. Pearson’s test was used for the correlation analysis between two groups. P < 0.05 was considered significant. All statistical analyses were performed with SPSS software (version 13; SPSS, Chicago, IL). Other methods are described in Supplementary materials and methods.
Results
NEAT1 is upregulated during liver fibrosis
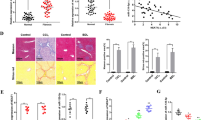
To gain insights into the possible involvement of NEAT1 in liver fibrosis, NEAT1 expression was firstly examined in CCl4-induced mice, a typical model for producing liver fibrosis. Compared with control liver, qRT-PCR analysis indicated increased NEAT1 expression in CCl4-treated liver (Fig. 1a). Next, NEAT1 expression was detected in primary HSCs isolated from CCl4-induced fibrotic liver at different weeks. During CCl4 treatment, there was a significant increase in NEAT1 expression in primary HSCs (Fig. 1b). NEAT1 expression was additionally analyzed in primary HSCs isolated from the livers of healthy mice. Isolated primary HSCs were cultured for up to 8 days. There was also a significant increase in NEAT1 expression during culture days (Fig. 1c). These data suggest that NEAT1 expression is increased in activated HSC and correlates with the progression of liver fibrosis.
NEAT1 was upregulated during liver fibrosis. NEAT1 was detected by qRT-PCR in the livers from CCl4 mice (a), in isolated primary HSCs from the livers of CCl4 mice at different weeks (b), and in primary HSCs during culture days (c). *P < 0.05 compared to the control. Each value is the mean ± SD of three experiments
Silencing NEAT1 alleviates liver fibrosis in vivo
To better understand the role of NEAT1 in the progression of liver fibrosis, Ad-shNEAT1 was injected into CCl4-treated mice via tail vein to inhibit NEAT1. As shown by qRT-PCR analysis, delivery of Ad-shNEAT1 significantly inhibited NEAT1 in vivo (Fig. 2a). Interestingly, silencing of NEAT1 had no effect on ALT value (Fig. 2b). The accumulation of collagen caused by CCl4 was inhibited by NEAT1 knockdown, as illustrated by liver hydroxyproline content and Masson staining (Fig. 2c–e). Moreover, NEAT1 downregulation led to the suppression of CCl4-induced α-SMA and type I collagen (Fig. 2f). Taken together, these results demonstrate that the progression of liver fibrosis can be inhibited by NEAT1 knockdown in vivo.
Silencing NEAT1 inhibited CCl4-induced liver fibrosis in mice. NEAT1 expression (a), ALT value (b), and hydroxyproline (c) were analyzed in CCl4 mice after Ad-shNEAT1 treatment. d, e Accumulation of collagen was assessed by Masson staining. Scale bar, 100 μm. f α-SMA and type I collagen. *P < 0.05 compared to the control and # P < 0.05 compared to the CCl4 group. Each value is the mean ± SD of three experiments
Silencing NEAT1 suppresses the activation of HSCs in vitro
Next, primary HSCs were isolated from the livers of healthy mice and cultured to day 4. Ad-shNEAT1 treatment led to a reduction in NEAT1 while adenoviral vectors expressing NEAT1 (Ad-NEAT1) induced an increase in NEAT1 (Supporting Fig. 1A). EdU assays showed that cell proliferation in the Ad-shNEAT1 group was reduced by 53.8% (Fig. 3a). The mRNA expressions of α-SMA and Col1A1 were decreased by 59 and 81%, respectively, in the Ad-shNEAT1 group relative to the control group (Fig. 3b). Also, the protein levels of α-SMA and type I collagen in HSCs transduced with Ad-shNEAT1 were downregulated by 48 and 74%, respectively (Fig. 3c). Conversely, NEAT1 overexpression promoted HSC proliferation and α-SMA and Col1A1 expression (Supporting Fig. 1B–D). As confirmed by immunocytochemical analysis, NEAT1 overexpression enhanced the levels of red fluorescence and actin fibers in α-SMA protein (Fig. 3d). All the results suggest that NEAT1 accelerates HSC activation.
Effects of NEAT1 on HSC proliferation, HSC transdifferentiation, and collagen expression. Primary 4-day-old HSCs were transduced with Ad-shNEAT1 for 48 h. a HSC proliferation. b The mRNA levels of α-SMA and Col1A1. c Protein levels of α-SMA and Col1A1. d Immunofluorescence staining for α-SMA (red) was evaluated in primary HSCs 48 h after transduction with Ad-NEAT1 by confocal laser microscopy. DAPI-stained nuclei are indicated in blue. Scale bar, 50 μm. *P < 0.05 compared to the control. Each value is the mean ± SD of three experiments
miR-122 is involved in the effects of NEAT1 on HSC activation
miRNAs serve as key regulators in liver fibrosis. Recently, lncRNAs have been reported to function as ceRNAs for miRNAs to regulate the activity of miRNAs [20, 21]. Therefore, whether miRNAs are involved in the role of NEAT1 in regulating liver fibrosis is unclear. Recent studies have shown that miRNAs including miR-19b [22], miR-29b [2], miR-30 [23], miR-101 [3], miR-122 [24], miR-146 [25], miR-150 [26], miR-370 [27], and miR-378 [28], are downregulated in liver fibrosis and act as liver fibrosis suppressors. Due to their inhibitory role in liver fibrosis, we next analyzed whether they are involved in the effects of NEAT1 on liver fibrosis. qRT-PCR analysis showed that miR-122 expression was significantly decreased by NEAT1 overexpression, whereas others are not (Fig. 4a). Reduced miR-122 level caused by NEAT1 overexpression was confirmed by Northern blot (Supporting Fig. 2A). Thus, miR-122 level was firstly examined in liver fibrosis. miR-122 was significantly decreased in primary HSCs isolated from the fibrotic liver during CCl4 treatment (Fig. 4b). With time in culture, there was a significant reduction in miR-122 expression in primary HSCs (Fig. 4c). miR-122 level inversely correlated with NEAT1 expression in liver fibrosis, indicating that miR-122 may be required for the effects of NEAT1 on HSC activation. NEAT1 overexpression induced HSC proliferation, α-SMA and type I collagen were almost suppressed by miR-122 mimics (Fig. 4d–f). As confirmed by immunocytochemical analysis, α-SMA and type I collagen induced by NEAT1 was blocked down by miR-122 (Supporting Fig. 3A). miR-122 mimics alone led to a reduction in cell proliferation, α-SMA, and type I collagen (Supporting Fig. 2B and Supporting Fig. 2C).
miR-122 was involved in the effects of NEAT1 on HSC activation. Primary 4-day-old HSCs were transduced with Ad-NEAT1 for 48 h and treated with miR-122 mimics for additional 48 h. a Expressions of miR-19b, miR-29b, miR-30, miR-101, miR-122, miR-146, miR-150, miR-370, and miR-378 were detected in NEAT1-overexpressing cells. miR-122 was detected by qRT-PCR in isolated primary HSCs from the livers of CCl4 mice at different weeks (b) and in primary HSCs during culture days (c). d NEAT1-induced cell proliferation was inhibited by miR-122. The enhanced mRNA(e) and protein (f) levels of KLF6, α-SMA, and Col1A1 induced by NEAT1 were suppressed by miR-122. *P < 0.05 compared to the Ad-Ctrl and # P < 0.05 compared to the Ad-NEAT1 group. Each value is the mean ± SD of three experiments
NEAT1 accelerates HSC activation through miR-122-mediated KLF6
We demonstrated that miR-122 plays a critical role in liver fibrosis progression, which is consistent with a previous study [29]. Previously, Tsai et al. reported that mice lacking miR-122 will develop liver fibrosis, which can be partially attributed to Kruppel-like factor 6 (KLF6) [29]. Then, whether KLF6 was involved in the effects of NEAT1 on liver fibrosis was examined. KLF6 expression was significantly increased in primary HSCs isolated from the fibrotic liver during CCl4 treatment as well as in primary HSCs during culture days (Fig. 5a, b, Supporting Fig. 2D, and Supporting Fig. 2E). The mRNA and protein expressions of KLF6 were induced by NEAT1 overexpression (Fig. 5c–e). Next, KLF6 small interfering RNA (siRNA) was transfected into HSCs to silence KLF6 expression (Supporting Fig. 1E and Supporting Fig. 1F). Notably, NEAT1-induced HSC proliferation, α-SMA, and type I collagen were inhibited by KLF6 knockdown (Fig. 5c–f). As confirmed by immunocytochemical analysis, α-SMA and type I collagen induced by NEAT1 could be blocked down by KLF6 siRNA (Supporting Fig. 3B). These data suggest that NEAT1 promotes HSC activation, at least in part, via KLF6. Further study was performed to confirm miR-122-KLF6 axis. Bioinformatic analysis (microRNA.org) for miRNA recognition sequences on KLF6 revealed the presence of a putative miR-122 site (Fig. 5g). Then, we cloned the KLF6 target region in the pmirGLO plasmid to generate a KLF6 3′-UTR luciferase reporter containing the miR-122-binding sites (pmirGLO-KLF6-Wt) or mutated sites (pmirGLO-KLF6-Mut). Luciferase activity assays showed that the luciferase activity of pmirGLO-KLF6-Wt was reduced by miR-122, whereas the luciferase activity of pmirGLO-KLF6-Mut was not affected by miR-122 (Fig. 5h), suggesting that KLF6 is a target of miR-122. In addition, miR-122 could inhibit KLF6 in HSCs with or without NEAT1 overexpression (Fig. 4e, f, Supporting Fig. 2C, and Supporting Fig. 3A). Combined with these, NEAT1 contributes to HSC activation via the miR-122-KLF6 axis.
KLF6 was involved in the effects of NEAT1 on HSC activation. Primary 4-day-old HSCs were transduced with Ad-NEAT1 for 48 h and treated with KLF6 siRNA for additional 48 h. KLF6 was detected by qRT-PCR in isolated primary HSCs from the livers of CCl4 mice at different weeks (a) and in primary HSCs during culture days (b). c, d The protein levels of KLF6, α-SMA, and type I collagen. e The mRNA levels of KLF6, α-SMA, and Col1A1. f HSC proliferation. g Schematic diagram of the miR-122 binding site in the 3′-UTR of KLF6 mRNA based on microRNA.org software. h Relative luciferase activities of luciferase reporters bearing wild-type or mutant KLF6 were analyzed 48 h following transfection with the indicated miR-122 mimics or miR-NC in 293T. *P < 0.05 compared to the Ad-Ctrl and # P < 0.05 compared to the Ad-NEAT1 group. Each value is the mean ± SD of three experiments
Interaction between NEAT1 and miR-122
Using bioinformatic analysis (RNA22), it was found that NEAT1 contains one target site for miR-122 (Fig. 6a). Using pmirGLO construct, we generated a NEAT1 luciferase reporter containing the miR-122-binding sites (pmirGLO-NEAT1-Wt) or mutated sites (pmirGLO-NEAT1-Mut) (Fig. 6a). Luciferase activity assays showed that miR-122 mimics induced a reduction in luciferase activity of pmirGLO-NEAT1-Wt without affecting that of pmirGLO-NEAT1-Mut (Fig. 6b). Therefore, NEAT1 is confirmed as a target of miR-122. To further substantiate these results, NEAT1 expression was detected in primary HSCs transfected with miR-122 mimics or inhibitor. Our results showed that miR-122 mimics led to a significant reduction in NEAT1 level (Fig. 6c). Conversely, NEAT1 level was increased by miR-122 inhibitor (Fig. 6c). Similarly, miR-122 was increased by NEAT1 knockdown while miR-122 was reduced by NEAT1 overexpression (Fig. 6d). Interestingly, pri-miR-122 expression was not affected by either Ad-shNEAT1 or Ad-NEAT1, indicating that NEAT1 regulates miR-122 level at the post-transcriptional level (Fig. 6d). In addition, the pull-down assay was applied to detect whether miR-122 could pull down NEAT1. As shown in Fig. 6e, f, Bio-miR-122-Wt pulled down NEAT1 while Bio-miR-122-Mut had no effect on NEAT1, indicating a direct interaction between miR-122 and NEAT1. Likewise, the increased cell proliferation, KLF6, α-SMA, and type I collagen caused by NEAT1 overexpression could not be blocked down by miR-122-Mut (Supporting Fig. 4). Therefore, NEAT1 may regulate KLF6 via competitively binding miR-122 during HSC activation.
Interaction between NEAT1 and miR-122. Primary 4-day-old HSCs were transfected with Ad-NEAT1, Ad-shNEAT1, miR-122 mimics, or miR-122 inhibitor for 48 h. a Schematic diagram of the miR-122-binding site in NEAT1 based on RNA22 software. b Relative luciferase activities of luciferase reporters harboring the wild-type or mutant NEAT1 were analyzed 48 h following transfection with the miR-122 mimics or miR-NC. c NEAT1 expression was analyzed by qRT-PCR in cells transfected with miR-122 mimics or miR-122 inhibitor. d Expressions of miR-122 and pri-miR-122 were analyzed by qRT-PCR in cells transduced with Ad-shNEAT1 or Ad-NEAT1. e Schematic diagram of wild type and the mutated form of miR-122 sequence. f Pull-down assay to validate the direct interaction between NEAT1 and miR-122. Bio-miR-NC is not complementary to NEAT1. *P < 0.05 compared to the control. Each value is the mean ± SD of three experiments
NEAT1-miR-122-KLF6 axis in hepatocytes
Hepatocytes were additionally isolated from the livers of healthy mice, and the NEAT1-miR-122-KLF6 axis was further confirmed in primary hepatocytes. Consistent with our previous results, increased NEAT1 and KLF6 were found in primary hepatocytes isolated from the fibrotic liver during CCl4 treatment, with a reduction in miR-122 (Supporting Fig. 5A-C). In primary hepatocytes, NEAT1 overexpression induced KLF6 and type I collagen was inhibited by miR-122 or KLF6 knockdown (Supporting Fig. 5D and Supporting Fig. 5E). Also, miR-122 alone led to a reduction in KLF6 and type I collagen in primary hepatocytes (Supporting Fig. 2F). Moreover, NEAT1 was inhibited by miR-122 mimics while NEAT1 was enhanced by miR-122 inhibitor (Supporting Fig. 5F). These data confirm the NEAT1-miR-122-KLF6 axis in hepatocytes.
Validation of the aberrant expression of NEAT1, miR-122, and KLF6 in patients with liver cirrhosis
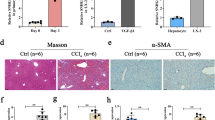
Finally, we sought to determine whether the observations in mice could be verified in patients with liver cirrhosis. Using qRT-PCR, the expressions of NEAT1, miR-122, and KLF6 were detected in human cirrhotic liver tissues. Using H&E and Masson staining, the existence of liver cirrhosis in patients was confirmed (Fig. 7a, b). As observed in rodent fibrotic injury models, the increased expressions of NEAT1 and KLF6, associated with the reduced miR-122 level, were found in patients with liver cirrhosis (Fig. 7c). Notably, NEAT1 expression in cirrhotic liver tissues was positively correlated with the transcriptional level of α-SMA (r = 0.932) (Fig. 7d). A similar positive correlation was observed between NEAT1 expression and the transcriptional level of Col1A1 (r = 0.983) (Fig. 7e). In conclusion, the results indicate that NEAT1 plays a pro-fibrotic role, and may serve as a diagnostic marker for determining the progression of liver fibrosis in humans.
NEAT1 expression is associated with liver fibrosis in humans. a H&E staining for assessing liver fibrosis. Scale bar, 100 μm. b Masson staining for assessing liver fibrosis. Scale bar, 100 μm. c NEAT1 expression was detected by qRT-PCR in healthy controls (n = 15) and cirrhotic patients (n = 15). d Positive correlation between transcriptional level of α-SMA and NEAT1 in cirrhotic human liver tissues. e Positive correlation between transcriptional level of Col1A1 and NEAT1 in cirrhotic human liver tissues. Pearson’s correlation analysis was used for statistical analysis. f The signal pathway was discovered in liver fibrosis. *P < 0.05 compared to the control. Each value is the mean ± SD of three experiments
Discussion
NEAT1 has been reported to act as an oncogene in cancers [14, 15]. In this study, NEAT1 was increased in vivo and in vitro during liver fibrosis, similar with the results in cancers [14, 15]. Inhibition of NEAT1 resulted in the suppression of liver fibrosis in vivo, with a reduction in α-SMA and type I collagen. Consistent with it, there was a significant decrease in α-SMA and type I collagen in vitro after NEAT1 knockdown. By contrast, NEAT1 overexpression accelerated HSC activation, leading to the increase in cell proliferation, α-SMA, and type I collagen. Further studies demonstrated that the effects of NEAT1 on promoting HSC activation were via the miR-122-KLF6 axis. To the best of our knowledge, this is the first study to suggest that NEAT1 plays a pro-fibrotic role in liver fibrosis.
miR-122, which accounts for 72% of total miRNAs in the adult human liver, is the most abundant and liver-specific miRNA in liver [30]. miR-122 plays a vital role in cellular functions such as growth development, differentiation, and metabolism [31,32,33]. Previously, Zeng and colleagues reported that miR-122 suppresses HSC activation and the transforming growth factor-β (TGF-β)-induced expression of fibrosis-related genes, indicating an inhibitory role of miR-122 in liver fibrosis [24]. Herein, miR-122 level was obviously downregulated during liver fibrosis. All these effects of NEAT1 on HSC activation were inhibited by miR-122. Our findings further verified the anti-fibrotic role of miR-122, which are consistent with the previous studies [24, 29]. miRNAs are highly conserved RNAs with high homology between different species, whereas lncRNAs are not. As observed in rodent fibrotic injury models, the reduction in miR-122 and the increase in NEAT1 and KLF6 were also observed in patients with liver cirrhosis, indicating that the functions of NEAT1 may be conserved.
KLF6, as a pro-fibrotic gene in liver fibrosis, contributes to the upregulation of TGF-β receptor I (TβRI) and TβRII in activated HSCs [34, 35]. Increased TβRI and TβRII may lead to the increased activity of TGF-β1, which is characterized as the key cytokine to promote HSC activation through the Smad2/3 pathway [36, 37]. In addition, activated TGF-β signaling accelerates HSC proliferation [25]. Increasing evidence suggests that targeting TβR contributes to the suppression of the TGF-β pathway [38]. In this study, KLF6 was confirmed as a target of miR-122, which is consistent with the previous study [29]. NEAT1-induced HSC activation was significantly inhibited by KLF6 knockdown in primary HSCs. Similarly, NEAT1-induced collagen was suppressed by KLF6 knockdown in primary hepatocytes. We demonstrate that miR-122-mediated KLF6 is required for the effects of NEAT1 on accelerating liver fibrosis.
LncRNAs are found to modulate gene expression through diverse molecular mechanisms such as chromatin modification, transcriptional regulation, and post-transcriptional regulation [39,40,41,42]. Recently, lncRNAs have been demonstrated to act as ceRNAs to sponge miRNAs in many diseases, consequently modulating the de-repression of miRNA targets [43, 44]. Sun et al. found that NEAT1 promotes non-small cell lung cancer progression through regulation of the miR-377-3p-E2F3 pathway [45]. Consistent with the previous study, our results suggest that NEAT1 may function as a ceRNA for miR-122. As shown by the results of luciferase reporter assay and pull-down assay, there is a direct interaction between NEAT1 and miR-122. Moreover, NEAT1 could regulate miR-122 level at the post-transcriptional level. Our data indicate that NEAT1 may modulate KLF6 expression in liver fibrosis through competitively binding miR-122. In our study, NEAT1/miR-122/KLF6 plays a critical role in HSC activation. Interestingly, increased NEAT1 and KLF6 were additionally found in primary hepatocytes from CCl4 mice, associated with reduced miR-122 expression. NEAT1-indcued collagen was inhibited by miR-122 overexpression or KLF6 knockdown in both HSCs and hepatocytes. Therefore, the NEAT1/miR-122/KLF6 axis may participate in the progression of liver fibrosis (Fig. 7f).
In conclusion, our results disclose a novel NEAT1/miR-122/KLF6 signaling cascade in liver fibrosis and further identify the importance of NEAT1 in promoting liver fibrosis.
References
Schickel R, Boyerinas B, Park SM, Peter ME (2008) MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27:5959–5974
Wang J, Chu ES, Chen HY, Man K, Go MY, Huang XR, Lan HY, Sung JJ, Yu J (2015) microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget 6:7325–7338
Tu X, Zhang H, Zhang J, Zhao S, Zheng X, Zhang Z, Zhu J, Chen J, Dong L, Zang Y (2014) MicroRNA-101 suppresses liver fibrosis by targeting the TGFbeta signalling pathway. J Pathol 234:46–59
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655
Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M et al (2011) Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 53:209–218
Zheng J, Wu C, Lin Z, Guo Y, Shi L, Dong P, Lu Z, Gao S, Liao Y, Chen B et al (2014) Curcumin up-regulates phosphatase and tensin homologue deleted on chromosome 10 through microRNA-mediated control of DNA methylation—a novel mechanism suppressing liver fibrosis. FEBS J 281:88–103
Fatica A, Bozzoni I (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15:7–21
Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, Wu D, Liu L (2015) The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology 148(415–426):e418
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, Yang N, Zhou WP, Li WL, Li W et al (2014) Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 60:1278–1290
He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu BM, Xu FY, Zhang L, Lv XW, Li J (2014) Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim Biophys Acta 1842:2204–2215
Yu F, Zheng J, Mao Y, Dong P, Lu Z, Li G, Guo C, Liu Z, Fan X (2015) Long non-coding RNA growth arrest-specific transcript 5 (GAS5) inhibits liver fibrogenesis through a mechanism of competing endogenous RNA. J Biol Chem 290:28286–28298
Zheng J, Dong P, Mao Y, Chen S, Wu X, Li G, Lu Z, Yu F (2015) lincRNA-p21 inhibits hepatic stellate cell activation and liver fibrogenesis via p21. FEBS J 282:4810–4821
Naganuma T, Hirose T (2013) Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol 10:456–461
Ma Y, Liu L, Yan F, Wei W, Deng J, Sun J (2016) Enhanced expression of long non-coding RNA NEAT1 is associated with the progression of gastric adenocarcinomas. World J Surg Oncol 14:41
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H, Xuan L, Wang X, Tian L, Sun Y et al (2016) Long noncoding RNA NEAT1 promotes laryngeal squamous cell cancer through regulating miR-107/CDK6 pathway. J Exp Clin Cancer Res 35:22
Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, Schodel J, Green CM, Camps C, Buffa F et al (2015) Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34:4482–4490
Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G (2015) Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol 8:5395–5402
Chang W, Yang M, Song L, Shen K, Wang H, Gao X, Li M, Niu W, Qin X (2014) Isolation and culture of hepatic stellate cells from mouse liver. Acta Biochim Biophys Sin Shanghai 46:291–298
Bertolino P, Trescol-Biemont MC, Rabourdin-Combe C (1998) Hepatocytes induce functional activation of naive CD8+ T lymphocytes but fail to promote survival. Eur J Immunol 28:221–236
Gao Y, Wu F, Zhou J, Yan L, Jurczak MJ, Lee HY, Yang L, Mueller M, Zhou XB, Dandolo L et al (2014) The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res 42:13799–13811
Yu F, Lu Z, Cai J, Huang K, Chen B, Li G, Dong P, Zheng J (2015) MALAT1 functions as a competing endogenous RNA to mediate Rac1 expression by sequestering miR-101b in liver fibrosis. Cell Cycle 14:3885–3896
Lakner AM, Steuerwald NM, Walling TL, Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL, Schrum LW (2012) Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology 56:300–310
Tu X, Zheng X, Li H, Cao Z, Chang H, Luan S, Zhu J, Chen J, Zang Y, Zhang J (2015) MicroRNA-30 protects against carbon tetrachloride-induced liver fibrosis by attenuating transforming growth factor Beta signaling in hepatic stellate cells. Toxicol Sci 146:157–169
Zeng C, Wang YL, Xie C, Sang Y, Li TJ, Zhang M, Wang R, Zhang Q, Zheng L, Zhuang SM (2015) Identification of a novel TGF-beta-miR-122-fibronectin 1/serum response factor signaling cascade and its implication in hepatic fibrogenesis. Oncotarget 6:12224–12233
He Y, Huang C, Sun X, Long XR, Lv XW, Li J (2012) MicroRNA-146a modulates TGF-beta1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell Signal 24:1923–1930
Zheng J, Lin Z, Dong P, Lu Z, Gao S, Chen X, Wu C, Yu F (2013) Activation of hepatic stellate cells is suppressed by microRNA-150. Int J Mol Med 32:17–24
Lu CH, Hou QR, Deng LF, Fei C, Xu WP, Zhang Q, Wu KM, Ning BF, Xie WF, Zhang X (2015) MicroRNA-370 attenuates hepatic fibrogenesis by targeting smoothened. Dig Dis Sci 60:2038–2048
Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, Ha CS, Kim SW, Yun YH, Jung Y (2016) MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun 7:10993
Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF et al (2012) MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 122:2884–2897
Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12:735–739
Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH (2010) Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 52:1431–1442
Kim N, Kim H, Jung I, Kim Y, Kim D, Han YM (2011) Expression profiles of miRNAs in human embryonic stem cells during hepatocyte differentiation. Hepatol Res 41:170–183
Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R et al (2006) miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3:87–98
Kim Y, Ratziu V, Choi SG, Lalazar A, Theiss G, Dang Q, Kim SJ, Friedman SL (1998) Transcriptional activation of transforming growth factor beta1 and its receptors by the Kruppel-like factor Zf9/core promoter-binding protein and Sp1. Potential mechanisms for autocrine fibrogenesis in response to injury. J Biol Chem 273:33750–33758
Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, Friedman SL (1998) Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci U S A 95:9500–9505
Friedman SL, Yamasaki G, Wong L (1994) Modulation of transforming growth factor beta receptors of rat lipocytes during the hepatic wound healing response. Enhanced binding and reduced gene expression accompany cellular activation in culture and in vivo. J Biol Chem 269:10551–10558
Bataller R, Brenner DA (2005) Liver fibrosis. J Clin Invest 115:209–218
George J, Roulot D, Koteliansky VE, Bissell DM (1999) In vivo inhibition of rat stellate cell activation by soluble transforming growth factor beta type II receptor: a potential new therapy for hepatic fibrosis. Proc Natl Acad Sci U S A 96:12719–12724
Shi X, Sun M, Liu H, Yao Y, Song Y (2013) Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 339:159–166
Huang Y, Nayak S, Jankowitz R, Davidson NE, Oesterreich S (2011) Epigenetics in breast cancer: what’s new? Breast Cancer Res 13:225
Cooper C, Guo J, Yan Y, Chooniedass-Kothari S, Hube F, Hamedani MK, Murphy LC, Myal Y, Leygue E (2009) Increasing the relative expression of endogenous non-coding steroid receptor RNA activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res 37:4518–4531
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA et al (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938
Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY, Bin Z, Wu JB, Tang LY, Gao SM (2015) Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett 589:1981–1987
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369
Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, Li D (2016) Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. https://doi.org/10.18632/oncotarget.10108
Acknowledgements
The project was supported by the Zhejiang Provincial Natural Science Foundation of China (No. LY16H030012 and No. LY13H030006), National Natural Science Foundation of China (No. 81500458/H0317), and Wenzhou Municipal Science and technology Bureau (No. Y20150091).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
All animals were provided by the Experimental Animal Center of Wenzhou Medical University. The animal experimental protocol was approved by the University Animal Care and Use Committee.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 890 kb).
Rights and permissions
About this article
Cite this article
Yu, F., Jiang, Z., Chen, B. et al. NEAT1 accelerates the progression of liver fibrosis via regulation of microRNA-122 and Kruppel-like factor 6. J Mol Med 95, 1191–1202 (2017). https://doi.org/10.1007/s00109-017-1586-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-017-1586-5