Abstract
Background and purpose
The aim of this study was to evaluate an automatic multi-atlas-based segmentation method for generating prostate, peripheral (PZ), and transition zone (TZ) contours on MRIs with and without fat saturation (±FS), and compare MRIs from different vendor MRI systems.
Methods
T2-weighted (T2) and fat-saturated (T2FS) MRIs were acquired on 3T GE (GE, Waukesha, WI, USA) and Siemens (Erlangen, Germany) systems. Manual prostate and PZ contours were used to create atlas libraries. As a test MRI is entered, the procedure for atlas segmentation automatically identifies the atlas subjects that best match the test subject, followed by a normalized intensity-based free-form deformable registration. The contours are transformed to the test subject, and Dice similarity coefficients (DSC) and Hausdorff distances between atlas-generated and manual contours were used to assess performance.
Results
Three atlases were generated based on GE_T2 (n = 30), GE_T2FS (n = 30), and Siem_T2FS (n = 31). When test images matched the contrast and vendor of the atlas, DSCs of 0.81 and 0.83 for T2 ± FS were obtained (baseline performance). Atlases performed with higher accuracy when segmenting (i) T2FS vs. T2 images, likely due to a superior contrast between prostate vs. surrounding tissue; (ii) prostate vs. zonal anatomy; (iii) in the mid-gland vs. base and apex. Atlases performance declined when tested with images with differing contrast and MRI vendor. Conversely, combined atlases showed similar performance to baseline.
Conclusion
The MRI atlas-based segmentation method achieved good results for prostate, PZ, and TZ compared to expert contoured volumes. Combined atlases performed similarly to matching atlas and scan type. The technique is fast, fully automatic, and implemented on commercially available clinical platform.
Zusammenfassung
Hintergrund und Zweck
Ziel der Studie war es, eine automatische multi-atlasbasierende Segmentierungsmethode zur Erzeugung von Prostata-, peripheren und Übergangszonenkonturen (PZ/TZ) auf Magnetresonanztomographie(MRT)-Bildern mit und ohne Fettsättigung (±FS) zu beurteilen und MRT-Systeme verschiedener Hersteller zu vergleichen.
Methoden
T2-gewichtete (T2) und fettgesättigte (T2FS) MRTs wurden auf 3T-Systemen von GE (GE, Waukesha, WI, USA) und Siemens (Erlangen, Deutschland) aufgenommen. Manuelle Prostata- und PZ-Konturen wurden verwendet, um Atlasbibliotheken zu erstellen. Nach dem Einlesen eines MRT-Testdatensatzes identifiziert das Verfahren zur Atlassegmentierung automatisch die Atlasobjekte, die am besten zum Testobjekt passen, gefolgt von einer normalisierten, intensitätsbasierenden, frei deformierbaren Registrierung. Die Konturen werden dem Testobjekt angepasst und die „Dice Similarity Coefficients“ (DSC) und der Hausdorff-Abstand zwischen atlasgenerierten und manuellen Konturen verwendet, um die Übereinstimmung zu beurteilen.
Ergebnisse
Drei Atlanten wurden basierend auf GE_T2 (n = 30), GE_T2FS (n = 30) und Siem_T2FS (n = 31) erstellt. Wenn die Testbilder mit dem gewählten Kontrast und Atlashersteller übereinstimmten, wurden DSC von 0,81 und 0,83 für T2 ± FS erzielt (Ausgangswert). Atlanten erreichten eine höhere Genauigkeit beim Segmentieren von: (i) T2FS-Bildern verglichen mit T2-Bildern, wahrscheinlich aufgrund des besseren Kontrasts zwischen Prostata und umgebendem Gewebe auf T2FS-Bildern; (ii) Prostata verglichen mit zonaler Anatomie; (iii) der Drüsenmitte verglichen mit Basis und Apex. Die Qualität der Atlanten ging zurück, wenn sie mit Bildern mit unterschiedlichem Kontrast und MRT-Gerät getestet wurden. Umgekehrt zeigten kombinierte Atlanten eine ähnliche Übereinstimmung und Qualität wie der Ausgangswert.
Schlussfolgerung
Die atlasbasierende MRT-Segmentierungsmethode erzielte gute Ergebnisse für Prostata, PZ und TZ im Vergleich zu konturierten Volumina. Kombinierte Atlanten erreichten eine ähnliche Übereinstimmung und Genauigkeit wie passender Atlas- und Scan-Typ. Die Technik ist schnell, vollautomatisch und auf einer kommerziell erhältlichen klinischen Plattform integriert.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Accurate prostate segmentation on MRI datasets is required for many clinical and research applications including diagnosis, staging, and treatment planning for prostate cancer. The prostate has two distinct regions observable on imaging: the peripheral zone (PZ), characterized by a high signal on T2-weighted MRI, and the transition zone (TZ), which appears darker than the PZ on T2. T2 contrast in the prostate reflects the different amounts of macromolecular and free water present: the PZ is composed of highly glandular ductal tissues, which appear bright on T2, while the TZ is composed of more stromal than ductal tissues and appears hypointense. The different imaging properties of the prostate zones are well recognized and reflected in the recommendations of the Prostate Imaging, Reporting and Diagnosis System (PIRADS) from the European Society of Urogenital Radiology (ESUR; [1, 2]). Prostate cancer identification and staging on MRI rely on accurate zonal classification.
Automatic segmentation of the prostate, PZ, and TZ on MR images provides an opportunity to broaden the current scope of research by facilitating studies that include large populations of subjects and/or studies that incorporate serial imaging of the prostate to provide a longitudinal picture of disease progression and response. This is of paramount importance for the application of high-throughput approaches for extraction of radiomics features [3]. Manual segmentation is not feasible, as it is time consuming. The prostate and zonal contours are necessary for identification of the dominant lesions on MRI to allow for precise targeting of MRI–ultrasound fusion (MRI-US)-targeted biopsies [4,5,6] and delivery of a targeted radiation boost or other focal treatments to the designated area [7, 8]. Another potential application includes precise contouring of radiation targets for treatment planning, which is necessary for both intensity-modulated radiotherapy (IMRT) and volumetric arc therapy (VMAT) techniques as well as being critical in hypofractionated radiotherapy of the prostate, where large daily radiation doses are utilized; automatic segmentation may aid in these efforts [9]. Additionally, with MRI-guided adaptive radiation treatments—now a possibility with systems like ViewRay (Oakwood Village, OH, USA) and Elekta (Stockholm, Sweden)—automatic segmentation techniques are a key component of an efficient adaptive treatment planning program [10]. Other adaptive treatment planning applications, such as adaptive planning for intensity-modulated particle therapy, could also significantly benefit from automatic prostate segmentation [11].
Due to this increased role of MRI in prostate cancer diagnosis, treatment, and research, prostate MR image segmentation has become an area of intense research [12]. The Prostate MR Image Segmentation (PROMISE12) challenge aimed to standardize evaluation and objectively compare algorithm performance of the segmentation of prostate MRI. Several promising automatic, semi-automatic, and interactive approaches were evaluated [12], including atlas-based segmentation techniques [13,14,15]. Because of the excellent depiction of the prostate and surrounding anatomy, the high signal-to-noise ratio (SNR), and high spatial resolution [12, 16, 17], T2-weighted MRI is the sequence of choice for building a prostate atlas [12, 18,19,20,21,22]. More recently, several studies also provided segmentation of the prostate zonal structures [21, 23,24,25]. The presented approaches vary from model-based [26, 27] to atlas-based segmentation [15, 18,19,20,21, 28].
The goal of this work was to implement a robust procedure for prostate and prostate zone segmentation in a clinical imaging platform MIM (MIM Software Inc, Cleveland, OH, USA). State-of-the-art techniques are streamlined through an efficient implementation of multi-atlas-based segmentation. Most prostate segmentation developments are carried out in custom platforms/software and are inaccessible to clinicians and researchers. With the objective of creating broad access to automatic segmentation, the performance of the atlas approach is evaluated using (i) T2-weighted sequences with and without fat saturation (±FS), and (ii) data from different MRI manufacturers. A universal MIM atlas that is able to segment the prostate and prostate zones regardless of acquisition protocols, magnetic field strength, or type of scanners will allow unprecedented access to clinicians and researchers.
Methods
Study cohort and MRI acquisition
The Institutional Review Board (IRB) approved a protocol entitled “Development of Methods for Analysis and Interpretation of in vivo Imaging of Prostate Cancer” for retrospective review of MRI examinations from patients with biopsy-proven prostate cancer, protocol #20090554. A total of 30 consecutive patients evaluated for radiation treatment from May 2012 through November 2013 scanned on a Discovery MR750 3T MRI (GE, Waukesha, WI, USA) and 31 patients scanned from December 2008 through January 2014 on a Magnetom 3T Trio (Siemens, Erlangen, Germany) were included. Patients’ clinical characteristics are summarized in Supporting Table S1.
Transverse T2-weighted MRI, acquired on GE with (T2FS) and without (T2) fat saturation, were at identical spatial resolution: 0.7 × 0.7 × 2.5 mm3, 72 axial slices, no gap. Only the T2FS sequence was analyzed for Siemens. Imaging of the pelvis was acquired with parameters (Table 1) based on recommended specifications for clinical applications of prostate MRI [29].
Study design
The goal of the study was to determine the performance of atlas segmentation methods for delineation of the prostate and prostate zones (PZ and TZ). The central zone is not treated separately from TZ in this analysis because it is difficult to differentiate from TZ [30]. An analysis scheme is presented in Fig. 1. The three types of data are GE_T2, GE_T2FS, and Siem_T2FS. Correspondingly, three atlases were generated: αGE_T2 (30 subjects), αGE_T2FS (30 subjects), and αSiem_T2FS (31 subjects), based on manually contoured prostate and PZ performed by an expert radiation oncologist with 26 years of experience. In addition, two combined atlases were created: αContrast(combined) = αGE_T2 & αGE_T2FS (60 subjects) and αVendor(combined) = αGE_T2FS & αSiem_T2FS (61 subjects). The five atlases are schematically presented in Fig. 1. The performance of all atlases were evaluated using both the Dice similarity coefficient (DSC; [31]) and Hausdorff distance metrics [32]. The DSC value is a simple and useful summary measure of spatial overlap, which is often applied to measure accuracy and reproducibility of image segmentation [33]. DSC values are calculated using the equation shown below. The Hausdorff distance represents a measure of the spatial distance between two sets of points; in this manuscript, the mean Hausdorff distance is used [34]. To compute the mean Hausdorff distance, the edge of the two contours that are to be compared must be discretized into individual points. The distance from each point on contour A to the closest point on contour B is measured and then all of these distances are averaged.
-
1.
Baseline performance: The baseline performance of each atlas for segmenting patients with the same scan type and vendor as those used in the atlas itself was established (Fig. 1a). Atlas performance was evaluated by calculation of both DSC and Hausdorff distance metrics between manually drawn and automatic contours (via the atlas) for the three volumes of interest (VOIs): prostate, PZ, and TZ.
-
2.
Contrast neutrality: The goal is to compare the performance of atlases, αGE_T2 and αGE_T2FS, based on differing sequences, i.e., GE_T2FS and GE_T2 sequences, respectively (comparisons shown in Fig. 1b).
-
3.
Vendor neutrality: T2FS studies acquired on GE and Siemens were compared (Fig. 1c).
-
4.
Combined atlas: Determining the robustness of the segmentation when using an atlas comprised of subjects from differing sequences or a combined atlas including subjects from both MRI vendors. The combined atlases αContrast(combined) and αVendor(combined) were compared (Fig. 1d).
The performance of five atlases was evaluated. Each atlas was created by combining imaging data from all or a subset of three sets of data: GE_T2, GE_T2FS, and Siem_T2FS. a Baseline performance: Matching contrast and vendor: Three atlases—αGE_T2, αGE_T2FS, and αSiem_T2FS—were generated from each dataset and the auto-segmentation evaluated on the native images for each atlas; b Differing contrast: The auto-segmentation was evaluated on ±FS images; c Differing Vendor: The auto-segmentation is evaluated on the images from different vendor; d Combined atlases: αContrast(combined) and αVendor(combined) are evaluated on each dataset. FS fat saturation, GE GE, Waukesha, WI, USA, Siem Siemens, Erlangen, Germany
Atlas generation
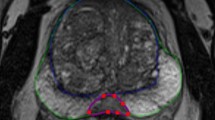
Commercially available software (MIM_Maestro_v6) was used to build the individual vendor/contrast MRI atlases. Prostate and PZ were outlined by an expert radiation oncologist on three sets of images: GE_T2, GE_T2FS, and Siem_T2FS. The TZ volume was created by performing a Boolean operation on the prostate and PZ. For each atlas (αGE_T2, αGE_T2FS, and αSiem_T2FS), a subject free of artifacts and with normal positioning was selected as a template subject to which all other atlas subjects were aligned. The full pelvic MRI for each atlas subject with manually defined prostate, PZ, and TZ contours was automatically rigidly aligned to the template and added to the atlas. The combined atlases, αContrast(combined) and αVendor(combined), were created by union of the individual atlases.
Atlas segmentation
The atlas-based segmentation utilized the deformable image registration function of MIM. A leave-one-out approach was implemented, where the target subject is removed from the atlas prior to segmentation. A schematic demonstrating the different steps of this workflow is shown in Fig. 2. The segmentation begins by aligning the patient scan to the template using a rigid transformation, based on maximizing normalized mutual information. The nine most similar atlas subjects are registered to the test case using a normalized intensity-based free-form deformable algorithm ([35, 36]; Fig. 2, second step). The VOIs are then transformed to the test case utilizing this deformable registration (Fig. 2, third step) and combined using simultaneous truth and performance level estimation (STAPLE [37]) methods ([38, 39]; Fig. 2, fourth step). STAPLE considers the original contours and computes a probabilistic estimate of the true representation of their combination. A measure of the positive effect each contour would have on the result is also estimated. The estimate of the “true representation” is formed by optimally combining the existing contours with weight given to their expected positive effect [37].
Comparative analysis
As specified above, the similarity metrics DSC [31] and Hausdorff distance [32] were used to evaluate the atlas-segmented contours in the comparisons (i–iv) for the VOIs. The DSC/Hausdorff distance between the single expert manual contours on ±FS is confounded by intra-reader variability and image-contrast effects. Thus, the “true” similarity metrics stemming from intra-reader variability are higher than the metrics reported by this analysis. As a measure of inter-reader variability, the volumes were contoured on GE_T2FS by a second expert radiation oncologist with 10 years of experience. The similarity metrics between intra- and inter-reader contours are used as benchmarks for the atlas performance. Analyses were carried out to see whether these metrics differ in three sections of the prostate (base, mid, and apex). These three sections of the prostate were generated by dividing the prostate into three equal parts along the superior/inferior axis. Similarly, the αGE_T2 and αGE_T2FS atlases were tested for variability in these sections.
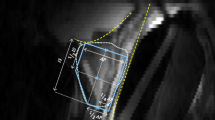
To infer the variability in the atlas performances, the contrast characteristics within and surrounding the prostate were investigated. Two rind contours were created using the manually delineated prostate contour on both the T2 and T2FS scans from both vendors as shown on Supporting Figure S1. This was accomplished by expanding the prostate contour by 3 mm to make an outer rind and shrinking the prostate contour by 3 mm to create the inner rind. Contrast ratios between the two rinds, presented as the ratio of the means from the image intensities in both contours, were estimated for T2 and T2FS scans, see equation below. Additionally, the contrast characteristics separating the TZ and PZ were investigated by utilizing the manually delineated TZ and PZ contours for both the T2 and T2FS scans from both vendors and measuring contrast ratios between these structures to determine whether atlas performance was related to contrast separation (Supporting Figure S1).
Results
Intra- and inter-reader reproducibility of manual contours
The results from the comparisons of the physician-drawn VOIs on T2 and T2FS are shown in Supporting Table S2. The prostate volumes on GE_T2FS were consistently, albeit nominally, larger than the prostate volumes on GE_T2, which resulted in significant differences in the following volumes: prostate, TZ, mid-gland area of the prostate, and apex area of the prostate. No significant differences in volumes were detected in either the PZ or the base region of the prostate. The manual prostate VOI demonstrates excellent reproducibility, with DSC results of 0.94 and average Hausdorff distance <1.0 mm. As expected, the PZ comparisons resulted in higher variability: DSC/Hausdorff distance = 0.78/1.3. Also shown in Supporting Table S2 is the reproducibility of the prostate contour between the two sequences in three sections of the prostate. Again, the volumes on GE_T2FS were larger than on GE_T2. The contours in the base were less reproducible relative to the other two sections. The results from the inter-reader study are summarized in Supporting Table S3. There were no differences in the volumes of the contoured structures. The average DSC of 0.88 between the readers was in good agreement with previously published studies [15]. This DSC serves as a reference point for comparisons of the automatic segmentation results, as described in Fig. 1.
Atlas baseline performance
The results from the performance of the three atlases (αGE_T2, αGE_T2FS, and αSiem_T2FS, Fig. 1a) when considering images native to the particular atlas are summarized in Table 1. Overall, the three atlases performed well for the prostate volume, with DSC results ranging from 0.79 to 0.83. Note that these results are within 15% of the ideal case of comparing manual contours from a single expert or intra-observer reliability (DSC = 0.94, Supporting Table S2) and within 10% of the agreement between two experts or inter-observer reliability (DSC = 0.88, Supporting Table S3). DSCs were lower for PZ and TZ, ranging from 0.54 to 0.57 and 0.70 to 0.75, respectively. While αGE_T2FS outperformed αGE_T2, there was no significance in the metrics. αGE_T2FS also outperformed αSiem_T2FS, with only one of nine measurements reaching significance (Table 2): the GE_T2FS atlas significantly outperformed the Siemens_T2FS atlas using the Hausdorff metric when segmenting the transition zone. These comparisons will serve as a benchmark for the subsequent analysis and be referred to as baseline performance.
The performance metrics were evaluated in three different sectors of the prostate. The mid-gland sector performed the best with DSC results, ranging from 0.87 to 0.90, while the lowest DSC results originated from the base, ranging from 0.68 to 0.76. In all sectors analyzed, the results were superior for αGE_T2FS, although there was no significance between αGE_T2FS and αSiem_T2FS (Table 2). Next, the contrast between the areas immediately inside and outside the prostate and between PZ and TZ was estimated for each patient on GE_T2 and GE_T2FS (Supporting Table S4). GE_T2 scans showed minimal differences between inside and outside the prostate, while FS scans showed, on average, 36% increased contrast between the rinds. FS scans also demonstrated a significant contrast increase between PZ and TZ: 1.27 contrast ratio for GE_T2FS scans as compared to a 1.12 contrast ratio for GE_T2 scans. Total of four contrast comparisons were made and all of them achieved significance.
Contrast neutrality
The performance of an atlas generated by contoured images on T2 scans (Fig. 1b) in segmenting the T2FS sequence and vice versa is summarized in Table 3. The αGE_T2 and αGE_T2FS performance on the differing sequence type was underwhelming, with DSC results ranging from 0.09 to 0.38. The performance of the atlas αContrast(combined) was, however, within the ranges of the baseline measurements in the previous section. Upon further investigation, it was determined that there was a strong trend toward finding matches from the same sequence type in the combined atlas: 264/270 or 98% from T2 images and 254/270, or 90% from the T2_FS images matched images from the same sequence in the combined atlas.
Vendor dependence
The performance of an atlas generated by contoured images on GE_T2FS in segmenting the T2FS sequence on Siemens and vice versa (Fig. 1c) is summarized in Table 4. The αGE_T2FS and αSiemens_T2FS performances on the differing MRI vendor were lower than the native image comparisons, but markedly higher than the sequence neutrality results (DSC ranging from 0.43 to 0.58). Again, the performance of the combined vendor atlas αVendor(combined; Fig. 1d), was higher, reaching DSC/Hausdorff distance measures similar to the baseline measurements. Again, this was due to the fact that 97% of GE scans matched with GE atlas subjects and 100% of Siemens scans matched with Siemens atlas subjects.
Discussion
The implemented method uses multi-atlas-based segmentation and, as shown by Rohlfing et al. [40], the multi-atlas-based segmentation is more successful than using a single or average atlas image. An advantage of the atlas approach is that it can easily be scaled up using the MIM platform, which has access to thousands of contours generated for radiotherapy of prostate cancer. The existing workflow will allow for constant enrichment of the multi-atlas method with new cases. The novel implementation, optimized and streamlined in a commercial imaging platform, resulted in fully automatic and fast (on average less than 90 sec per patient) implementation. The procedure uses a large array of existing robust utilities in MIM for image normalization and deformable fusion. The segmentation results (DSC of 0.83) are comparable to those previously reported in atlas-based approaches: DSC of 0.85 in Klein et al. [15], 0.82 in Chilali et al. [21], 0.87 in Cheng et al. [19], 0.87 in Xie et al. [18], 0.83 in Tian et al. [28], and 0.87–0.88 in Korsager et al. [20]. It is of note that DSC = 0.83 in this work was calculated over the entire prostate, while some of the referenced studies, e.g., Cheng [19], report results in a two-dimensional slice. As demonstrated in this report, DSC varies in the different regions of the prostate. In such cases, the mid-gland DSC of 0.90, as reported here, should be compared to other studies. In addition, Chilali et al. [21] also report similar DSC for TZ (= 0.70) and PZ (= 0.62) segmentations.
As expected, the atlas more accurately segmented the prostate as compared to the PZ, and the atlas performed slightly better in the mid-gland area of the prostate as compared to the base and apex. The atlas demonstrated more accurate results when segmenting the prostate contour on T2FS images as compared to T2 images, likely due to a superior contrast separation between the prostate vs. the surrounding tissue. The investigation of the impact of image contrast on atlas segmentation accuracy is novel and provides important insights. Interestingly, the decreased accuracy in zonal segmentation compared to whole prostate cannot be explained by the differences in contrast, as suggested by Chilaliet al. [21]: the contrast between PZ and TZ was similar to the contrast of the prostate and its surrounding. Similarly, for the FS data, the gains in PZ/TZ contrast did not translate into better atlas performance, indicating that there are other factors at play beyond the image contrast. In part due to the irregular shape and relatively small volume of the PZ, small contouring differences can result in a large decrease in the similarity metrics. On the other hand, the performance of the atlas was affected more by the contrast than by the vendor. Another interesting result is that manual contours of images with lower contrast seem to underestimate the volumes.
The study has several limitations. Atlases were generated using manual segmentation performed by a single experienced operator. The intra- and inter-reader variability in contouring the prostate is a well-recognized challenge [41, 42]. A mitigating factor is that because of the superior contrast of soft tissues on MRI, the inter-reader variability is reduced relative to other imaging modalities [43, 44]. The limited inter-reader study reported here yielded similar results to previously studies [15]. Another limitation of the work is that different subjects were used for the GE and Siemens atlases. While this hinders direct comparison between vendors, the main finding that the target scan is matched almost exclusively by scans from the same vendor still holds.
There are a plethora of factors that affect the process of prostate segmentation: the large anatomic variability between subjects, differences in rectum and bladder filling, as well as variability in imaging data acquired with different sequences, resolution, magnetic field, etc. The findings here show that a large multi-atlas database containing different contrast types from multiple vendors reaches a similar performance by forcing the target scan to match with atlas scans of the same contrast and vendor. Retrieving the relevant information from the Digital Imaging and Communications in Medicine (DICOM) header of the test scan to create a customized subset of atlases matched by contrast, vendor, field strength, etc. will result in efficient and fast segmentation. Future implementation of this functionality into a commercial platform will allow for universal use.
Conclusion
The MRI atlas-based segmentation method achieved good results for both the whole prostate, PZ, and TZ compared to expert-contoured VOIs. The robustness of the proposed segmentation methods is demonstrated by use of combined atlases that perform similarly to matching atlas and scan type. The technique is fast, fully automatic, and implemented on commercially available clinical imaging platforms.
References
Barentsz JO, Richenberg J, Clements R et al (2012) ESUR prostate MR guidelines 2012. Eur Radiol 22:746–757
Weinreb JC, Barentsz JO, Choyke PL et al (2016) PI-RADS prostate imaging—reporting and data system: 2015, version 2. Eur Urol 69:16–40
Stoyanova R, Takhar M, Tschudi Y et al (2016) Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res 5:432–447
Rastinehad AR, Turkbey B, Salami SS et al (2014) Improving detection of clinically significant prostate cancer: magnetic resonance imaging/transrectal ultrasound fusion guided prostate biopsy. J Urol 191:1749–1754
Wysock JS, Rosenkrantz AB, Huang WC et al (2014) A prospective, blinded comparison of magnetic resonance (MR) imaging-ultrasound fusion and visual estimation in the performance of MR-targeted prostate biopsy: the PROFUS trial. Eur Urol 66:343–351
Haffner MC, Mosbruger T, Esopi DM et al (2013) Tracking the clonal origin of lethal prostate cancer. J Clin Invest 123:4918–4922
Lips IM, van der Heide UA, Haustermans K et al (2011) Single blind randomized phase III trial to investigate the benefit of a focal lesion ablative microboost in prostate cancer (FLAME-trial): study protocol for a randomized controlled trial. Trials 12:255
Bauman G, Haider M, Van der Heide UA, Menard C (2013) Boosting imaging defined dominant prostatic tumors: a systematic review. Radiother Oncol 107:274–281
Hocht S, Aebersold DM, Albrecht C et al (2017) Hypofractionated radiotherapy for localized prostate cancer. Strahlenther Onkol 193:1–12
Pathmanathan AU, van As NJ, Kerkmeijer LGW et al (2018) Magnetic resonance imaging-guided adaptive radiation therapy: a “game changer” for prostate treatment? Int J Radiat Oncol Biol Phys 100:361–373
Hild S, Graeff C, Rucinski A et al (2016) Scanned ion beam therapy for prostate carcinoma: comparison of single plan treatment and daily plan-adapted treatment. Strahlenther Onkol 192:118–126
Litjens G, Toth R, van de Ven W et al (2014) Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Med Image Anal 18:359–373
Ou YDJ, Erus G, Davatzikos C (2012) Multi_atlas segmentation of the prostate; a zooming process with robust registration and atlas selection. MICCAI grand challenge: prostate MR image segmentation 2012. https://promise12.grand-challenge.org/. Accessed: 1 Oct 2012
Ou Y, Sotiras A, Paragios N, Davatzikos CDRAMMS (2011) Deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal 15:622–639
Klein S, van der Heide UA, Lips IM, van Vulpen M, Staring M, Pluim JPW (2008) Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys 35:1407–1417
Hegde JV, Mulkern RV, Panych LP et al (2013) Multiparametric MRI of prostate cancer: an update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer. J Magn Reson Imaging 37:1035–1054
Bhavsar A, Verma S (2014) Anatomic imaging of the prostate. Biomed Res Int 2014:728539
Xie Q, Ruan D (2014) Low-complexity atlas-based prostate segmentation by combining global, regional, and local metrics. Med Phys 41:41909
Cheng R, Turkbey B, Gandler W et al (2014) Atlas based AAM and SVM model for fully automatic MRI prostate segmentation. Conf Proc Ieee Eng Med Biol Soc 2014:2881–2885
Korsager AS, Fortunati V, van der Lijn F et al (2015) The use of atlas registration and graph cuts for prostate segmentation in magnetic resonance images. Med Phys 42:1614–1624
Chilali O, Puech P, Lakroum S, Diaf M, Mordon S, Betrouni N (2016) Gland and zonal segmentation of prostate on T2W MR images. J Digit Imaging 29:730–736. https://doi.org/10.1007/s10278-016-9890-0
Wu K, Garnier C, Alirezaie J, Dillenseger JL (2014) Adaptation and evaluation of the multiple organs OSD for T2 MRI prostate segmentation. Conf Proc Ieee Eng Med Biol Soc 2014:4687–4690
Makni N, Iancu A, Colot O, Puech P, Mordon S, Betrouni N (2011) Zonal segmentation of prostate using multispectral magnetic resonance images. Med Phys 38:6093–6105
Litjens G, Debats O, van de Ven W, Karssemeijer N, Huisman H (2012) A pattern recognition approach to zonal segmentation of the prostate on MRI. Med Image Comput Comput Assist Interv 15:413–420
Maan B, van der Heijden F, Futterer JJ (2012) A new prostate segmentation approach using multispectral Magnetic Resonance Imaging and a statistical pattern classifier. Proc SPIE Int Soc Opt Eng 8314. https://doi.org/10.1117/12.911194
Chowdhury N, Toth R, Chappelow J et al (2012) Concurrent segmentation of the prostate on MRI and CT via linked statistical shape models for radiotherapy planning. Med Phys 39:2214–2228
Toth R, Madabhushi A (2012) Multifeature landmark-free active appearance models: application to prostate MRI segmentation. IEEE Trans Med Imaging 31:1638–1650
Tian Z, Liu L, Fei B (2015) A fully automatic multi-atlas based segmentation method for prostate MR images. Proc Spie Int Soc Opt Eng 9413. https://doi.org/10.1117/12.2082229
Rothke M, Blondin D, Schlemmer HP, Franiel T (2013) PI-RADS classification: structured reporting for MRI of the prostate. Fortschr Röntgenstr 185:253–261
Rosenkrantz AB, Taneja SS (2014) Radiologist, be aware: ten pitfalls that confound the interpretation of multiparametric prostate MRI. Ajr Am J Roentgenol 202:109–120
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Hausdorff F (1914) Grundzüge der Mengenlehre. Veit, Leipzig
Zou KH, Warfield SK, Bharatha A et al (2004) Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 11:178–189
Huttenlocher DP, Klanderman GA, Rucklidge WJ (1993) Comparing images using the Hausdorff distance. Ieee T Pattern Anal 15:850–863
Piper J, Nelson A, Harper J (2013) Deformable image registration in MIM Maestro™. Evaluation and description. MIMsoftware, Ohio, p 5
Johnson PB, Padgett KR, Chen KL, Dogan N (2016) Evaluation of the tool “Reg Refine” for user-guided deformable image registration. J Appl Clin Med Phys 17:158–170
Warfield SK, Zou KH, Wells WM (2004) Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23:903–921
Hoeks CMA, Barentsz JO, Hambrock T et al (2011) Prostate cancer: multiparametric MR imaging for detection, localization, and staging. Radiology 261:46–66
McGurk RJ, Bowsher J, Lee JA, Das SK (2013) Combining multiple FDG-PET radiotherapy target segmentation methods to reduce the effect of variable performance of individual segmentation methods. Med Phys 40:42501
Rohlfing T, Brandt R, Menzel R, Maurer CR Jr. (2004) Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage 21:1428–1442
Fiorino C, Reni M, Bolognesi A, Cattaneo GM, Calandrino R (1998) Intra- and inter-observer variability in contouring prostate and seminal vesicles: implications for conformal treatment planning. Radiother Oncol 47:285–292
Choi HJ, Kim YS, Lee SH et al (2011) Inter- and intra-observer variability in contouring of the prostate gland on planning computed tomography and cone beam computed tomography. Acta Oncol 50:539–546
Lutgendorf-Caucig C, Fotina I, Stock M, Potter R, Goldner G, Georg D (2011) Feasibility of CBCT-based target and normal structure delineation in prostate cancer radiotherapy: multi-observer and image multi-modality study. Radiother Oncol 98:154–161
Villeirs GM, Van Vaerenbergh K, Vakaet L et al (2005) Interobserver delineation variation using CT versus combined CT + MRI in intensity-modulated radiotherapy for prostate cancer. Strahlenther Onkol 181:424–430
Acknowledgements
This work was supported by National Cancer Institute (R01CA189295 and R01CA190105 to A.P.). We would also like to thank Randi Steinhagen and Dr. rer. nat. Dipl.-Phys. Ellen Ackerstaff for translating our abstract into German.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
A. Swallen, S. Pirozzi, J. Piper, and A. Nelson were employed by MIM Software Inc. during the duration of this investigation. Additionally, J. Piper and A. Nelson have an ownership interest in MIM Software Inc. K.R. Padgett, F.M. Chinea, M.C. Abramowitz, A. Pollack, and R. Stoyanova declare that they have no competing interests.
Caption Electronic Supplementary Material
Supplementary Table S1
: Patient Clinical Characteristics. Supplementary Table S2: Comparison of manually drown contours on GE_T2 and GE_T2FS sequences. In the second part of the table, the reproducibility of the prostate contour in three sections of the prostate is examined. Supplementary Table S3: Comparison of manually drown contours on GE_T2FS by two expert radiation oncologists (inter-reader comparison). Supplementary Table S4: Summary of contrast comparisons. The ratio of the signal intensity of the inner rind and the outer rind is presented along with the ratio of the signal intensity between the peripheral and transition zones (PZ/TZ). Supplementary Figure S1: (top) The inner and outer rinds that were used for contrast comparisons between T2 and T2FS acquisitions; (bottom) Contrast comparisons between manually drawn TZ and PZ contours for the T2 and T2FS scans from the same patient
Rights and permissions
About this article
Cite this article
Padgett, K.R., Swallen, A., Pirozzi, S. et al. Towards a universal MRI atlas of the prostate and prostate zones. Strahlenther Onkol 195, 121–130 (2019). https://doi.org/10.1007/s00066-018-1348-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00066-018-1348-5