Abstract
The study investigated the anti-urease and cytotoxic activities of two major essential oil compounds; 1-Nitro-2-phenylehtane (NPE) and Nerolidol, isolated from the acetone extract of Dennettia tripetala (Pepper Fruit) seeds. Structural characterization of these compounds was performed by NMR, E1MS, and FT-IR spectroscopic techniques. Anti-urease activity of the compounds on Jack bean Urease was accessed by the indophenol method; molecular docking analysis using MOE 2015.010 program was performed to explain the possible mechanism of interaction between the compounds and urease enzyme. Cytotoxic activities of the compounds were also assayed on brine shrimp (Artemia salina L.) nauplii and 3T3 (mouse fibroblast cells). NPE and Nerolidol displayed significant inhibitory activities on Jack Bean urease with percentage inhibition of 82.4% and 78.6% and IC50 ± SD of 27.4 ± 1.80 and 34.9 ± 3.57 µM respectively. Molecular docking analysis of the interactions between these compounds and active site of the PDB of the enzyme displayed favorable conformational binding with docking scores of −5.3716 and −7.3547 and ligand efficiencies of −0.4883 and −0.4597 for NPE and Nerolidol, respectively. Both interactions were characterized by hydrogen bonding; NPE also displayed π-stacking and hydrophobic interactions. Cytotoxic activity evaluation of both compounds on brine shrimp and mouse fibroblasts revealed no toxicity for both compounds. Conclusively, NPE and Nerolidol could be employed in the treatment of infections engendered by ureolytic activity and could serve as templates in the rational design of functional derivatives that possess higher potencies than the isolated compounds with minimal toxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ureases (urea amidohydrolase E.C 3.5.1.5) are ubiquitous metalloenzymes produced by plants, fungi, and bacteria but not by animals (Kappaun et al. 2018). They rapidly catalyze the hydrolysis of urea to form ammonia and carbamate; with the urea decomposing into another ammonia molecule and carbon dioxide (Callahan et al. 2005). The enzyme plays a crucial role in the pathogenicity of bacteria by providing nitrogen in the form of ammonia for their growth (Hamif et al. 2012; Konieczna et al. 2012). The ureolytic activity of Helicobacter pylori plays an important role in the pathogenicity of gastrointestinal diseases comprising gastritis, duodenal, peptic ulcer, and gastric cancer (Mobley et al. 1995; Dunn and Phadnis 1998). Human and animal pathogenicity of hepatic encephalopathy, hepatic coma urolithiasis, pyelonephritis, and urinary catheter encrustation are caused by ammonia produced by ureases (Konieczna et al. 2012). High ureolytic activity is also responsible for negative effects in agriculture by causing the release of large amounts of ammonia into the soil. This induces plant damage primarily by depriving plants of their essential nutrients and secondarily via ammonia toxicity, which increases the pH level of the soil (Amtul et al. 2002; Khan et al. 2014).
It is against this backdrop that urease inhibitors have attracted huge attention as potentials for the treatment of infections triggered and fostered by ureolytic activity. Many urease inhibitors have been described in the past including; fluorofamide, hydroxyureas, and hydroxamic acids (Uesato et al. 2002). However, the in vivo use of some of these inhibitors has been ban due to their toxicity or instability (Xiao et al. 2007; Shabana et al. 2010). Plants are well known to contain active metabolites which are beneficial in treating various infectious diseases. Much attention has been focused on exploring novel biological properties of phytochemicals isolated from edible plants; so as to cater for toxicity concerns from the onset. Several naturally occurring medicinal plants, herbs, extracts, and isolated compounds have been shown to possess anti-urease activities (Shabana et al. 2010; Amin et al. 2013; Golbabaei et al. 2013).
Dennettia tripetala G. Baker (Annonaceae) also known as Pepper fruit tree is an indigenous spicy medicinal plant in Nigeria (Iseghohi 2015). The leaves and roots are commonly used by the local herbalist in folk medicine in combination with other medicinal plants to treat various ailments including fever, infantile convulsion, typhoid, worm infestation, vomiting, and stomach upset (Oyemitan et al. 2013; Iseghohi 2015). They are also utilized in the treatment of cough, sore throat, toothache, diabetes, and nausea during pregnancy (Ejechi and Akpomedaye 2005; Iseghohi 2015). Phytochemical evaluations of the seeds, whole fruits, leaves, bark, and root of the plant revealed the presence of saponins, tannins, flavonoids, cardiac glycosides, essential oils, phenolic acids as well as alkaloids (Egharevba and Idah 2015; Akabueze et al. 2016; Omage et al. 2019). The plant has been shown to possess antimicrobial, insecticidal, analgesic, anti-inflammatory, antihyperglycemic, antioxidant, and antiulcer properties (Ukeh et al. 2012; Oyemitan et al. 2013; Anosike et al. 2016; Omage et al. 2019).
In view of the antiulcer property of D. tripetala, the present study investigated the anti-urease activity of two major compounds isolated from the seeds of the plant; the compounds were subjected to molecular docking studies for better recognition of their interaction with the urease enzyme. Their cytotoxic activities were also evaluated to investigate their possible applications in antiulcer therapy as well as in the treatment of other infections induced by ureolytic activity.
Materials and methods
Experimental
Chemicals and reagents
Hexane, dichloromethane, ethylacetate, methanol (Analytical grade; BDH, England); hexane, ethylacetate (HPLC; Samchun Chemicals, Republic of Korea); silica gel 60 (70–230 Mesh); Jack Bean Urease, urea, phenol, sodium nitroprusside, sodium hydroxide, sodium hypochlorite, acetohydroxamic acid (AHA), 3T3 (mouse fibroblast) cells, Dulbecco’s Modified Eagle Medium, penicillin, streptomycin, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), sulfuric acid, DMSO (Sigma Aldrich, USA).
Equipment
TLC (Silica gel 60 F254, 20 × 20 cm Merck, Germany); rotary evaporator (Buchi R200); freeze dryer (LGJ-10, Searchtech Instruments, China); recycling HPLC (LC-908W, Japan Analytical Industry); microplate spectrophotometer (SpectraMax340, Molecular Device, USA). NMR (Bruker Avance Neo 500 MHz Cyroprobe and 300 MHz Spectrometers Biospin USA); EIMS (JEOL, JMS-600H, Japan); FT-IR (TENSOR II Bruker Optics, Germany), computer (Linux OS) Incubator (Fisher Scientific, USA), haemocytometer (Fisher Scientific, USA).
Experimental biological material
Fresh fruits of D. tripetala, were purchased from a local market in Benin-city, Edo State, Nigeria. The plant was identified and authenticated by Dr. H. A. Akinnibosun; and a voucher specimen (UBH–D484), was deposited at the Herbarium in the Department of Plant Biology and Biotechnology, University of Benin, Nigeria.
Extraction, isolation, and structural characterization
Extraction was by maceration over a 48-h period, 10 kg of pulverized seeds were extracted with 50 l of absolute acetone and subsequently filtered using a muslin cloth. The extract was concentrated using a rotary evaporator at reduced pressure and at a temperature of 50 °C; complete evaporation of the solvent was carried out with a freeze-dryer to obtain the crude extract (170 g). The crude extract was subjected to solvent–solvent partitioning according to a modified Kupchan method (Kupchan and Tsou 1973) in a separating funnel with an elutropic series of solvents (hexane, dichloromethane, and ethylacetate) to yield hexane (72.50% of the crude extract), dichloromethane (5.63%), ethylacetate (0.38%), and aqueous (1.25%) fractions (Fig. 1).
Diagrammatic description of the partitioning of the acetone extract of D. tripetala seeds. The crude extract was initially dissolved in methanol–water (9:1) mixture to yield an aqueous–methanol mixture. The mixture was successively mixed with hexane, dichloromethane, and ethylacetate to partition the extract into respective fractions as well as a last aqueous fraction
The hexane fraction was subjected to Silica gel column chromatographic (SG-CC) separation eluting with an increasing gradient from hexane to ethylacetate (100:0–0:100) yielding 54 fractions of 200 ml each. These were pooled into eight main fractions on the basis of their TLC similarity in Hex–EtAOc–MeOH (95:5:0, 90:10:0, 70:25:5). Sub-fraction 1 (SF-1); was rechromatographed with Hex–EtAOc (98:2) to yield three main sub-fractions (SF-1A–C). Compound 1; a pale-yellow liquid (Rf 0.38; TLC, Hex–EtAOc (98:2)) was purified from SF-IB by normal phase (NP)-HPLC eluting with Hex–EtAOc (98:2) on a Silica column (250 (l) × 20 (i.d.) mm) with a flow rate of 4 ml/min with UV detection. The compound was also purified directly from Sub-fraction 2 (SF-2) by NP-HPLC as described above. Sub-fraction 3 (SF-3) was subjected to SG-CC eluting with Hex–DCM in increasing polarity. Fractions obtained were bulked into four main sub-fractions (SF-3A–D). SF-3B; was subjected to NP-HPLC with Hex–EtAOc (90:10) as mobile phase at a flow rate 3 ml/min with UV detection to obtain compound 2 (Rf 0.39; TLC, Hex–EtAOc (90:10)); a pale-yellow oily compound. SG-CC of SF3C yielded five main sub-fractions (SF-3CI -V). Compound 2 was obtained again from SF-3CIII via NP-HPLC, Hex–EtAOc (95:5).
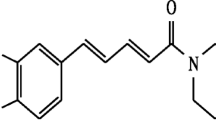
The structural characterization of these compounds was performed by the analyses of data obtained from one-dimensional nuclear magnetic resonance (1D-NMR) including Proton (1H) and Carbon-13 (13C) NMR; and two-dimensional (2D) NMR including correlation spectroscopy (COSY), heteronuclear multiple-bond COSY (HMBC), and heteronuclear single-quantum COSY. Electron ionization mass spectrometry (EIMS) and Fourier transform infrared (FT-IR) data were also obtained for both compounds; spectra data were compared with literature data. The chemical structures of compounds 1 and 2 are presented in Figs 2 and 3, respectively.
1-Nitro-2-phenylethane (Compound 1) was obtained as a pale-yellow oily liquid (157 mg yield). FT-IR: 3030, 2922, 1547, 1496, 1377, 1224, 1184, 1083, 1031, 910, 861, 748, 698, 606, 563 cm−1; 1H NMR (C3D60, 500 MHz): δ = 7.27 (5H, m, H-2, H-3, H-4, H-5, H-6), 4.79 (2H, t, J = 7 Hz, H-2′). 3.31 (2H, t, J = 7 Hz, H-1′); 13C NMR (C3D60, 125 MHz): δ = 137.57 (C, C-1), 129.49 (CH, C-3, C-5), 129.46 (CH, C-2, C-6) 127.81 (CH, C-4), 76.95 (CH2, C-2′), 33.6 (CH2, C-1′). EIMS m/z 149.1 [M]+, 104 (100), HNO2 loss, molecular formula: C8H9NO2.
Nerolidol (Compound 2) was obtained as a yellow oily liquid (108 mg yield). FT-IR: 3386, 2970, 2920, 2855, 2114, 1642, 1446, 1375, 1156, 1108, 994, 918, 836, 741, 689, 549 cm−1; 1H NMR (C3D60, 500 MHz): δ = 5.91 (1H, ddd, J = 17.5; 11; 0.5 Hz, H-2), 5.19 (1H, dd, J = 17.5; 2 Hz, H-1a); 5.14 (1H, tq, J = 7.5; 7.5; 1.0 Hz, H-10); 5.08 (1H, tq, J = 7; 7; 1.5 Hz, H-6), 4.95 (1H, dd, J = 10.5; 2 Hz, H-1b), 2.04 (4H, m, H-5, H-9), 1.95 (2H, brdd, J = 7; 7 Hz, H-8), 1.64 (3H, brd, J = 1 Hz, H-14), 1.57 (6H, brS, H-12, H-13), 1.49 (2H, ddd, J = 7; 7.3 Hz, H-4), 1.21 (3H, S, H-15); 13C NMR (C3D60, 125 MHz): δ = 146.9 (CH, C-2), 135.1 (C, C-7), 131.5 (C, C-11), 125.1 (CH, C-6), 111.2 (CH2, C-1), 72.8 (C, C-3), 40.4 (CH2, C-8), 28.2 (CH3, C-15), 27.3 (CH2, C-9), 25.7 (CH3, C-14), 25.6 (CH, C-10), 23.3 (CH2, C-5), 17.69 (CH3, C-12), 15.9 (CH3, C-13). EIMS m/z 204.2 (M+), 69 (100). H2O loss, molecular formula: C15H26O.
Anti-urease activity
The anti-urease activity of isolated compounds was determined by the indophenol method as described by Weatherburn (1967); which measures the concentration of ammonia produced. AHA served as the standard urease inhibitor. The percentage inhibition was calculated using the formula:
The concentration which induced 50% inhibition (IC50) represented the anti-urease activity.
Molecular docking study of anti-urease activity
Owing to the unavailability of PDBs of Jack Bean Urease in complex with appropriate ligands of interest, the PDB of Bacillus pasteurii urease, a bacterial urease (PDB ID: 4UBP) in complex with the standard inhibitor acetohydroxamate (Fig. 4), was utilized in this study (Benini et al. 2000). Besides, the Jack Bean urease differs from Bacterial Ureases only in the number of repeating subunits which do not influence the ureolytic activity of the enzyme (Follmer et al. 2004). Acetohydroxamate binds at the active site of the enzyme and competitively inhibits substrate binding (Benini et al. 2000); therefore compounds 1 and 2 were docked on this site. The target was validated with redocking experiments; the evaluation of the root mean square deviation between cartesian coordinates of the simulated and crystal pose was found to be less than 1 Å. The builder module in MOE 2015.10 was used to draw the compounds. The compounds were energy minimized, followed by addition of partial charges as per Merck Molecular Force Field (MMFF94) (Halgren 1996). The compounds were docked using MOE 2015.10 after initial protein preparation. The default rigid docking protocol in MOE Suite was utilized for docking. The resulting poses of the compounds were visually inspected to comprehend protein ligand interactions. The interactions were analyzed with the help of the Protein Ligand Interaction Profiler web server (Salentin et al. 2015). All visuals were recorded using Chimera 1.13.1 (Pettersen et al. 2004).
Structure of the Bacillus pasteurii urease inhibited with acetohydroxamic acid at 1.55 Å resolution. The Ni ions in the active site are separated by a distance of 3.53 Å. The inhibitor bridges the two Ni ions in the active site through the hydroxamate oxygen and chelating one Ni ion through the carbony oxygen. The flexible flap flanking the active site cavity is in the open conformation (Source: Benini et al. 2000; RCSB PDB: 4UBP)
Brine shrimp lethality assay
Cytotoxic activities of the compounds were accessed on brine shrimp (Artemia salina L.) larvae based on the method of Mayer et al. (1982). The cytotoxic drug; etoposide served as the positive control. Ten (10) Brime shrimp nauplii were subjected to 10, 100, and 1000 µg/ml sea water (with samples initially dissolved in DMSO) in triplicates; incubated at 25–28 °C under illumination for 24 h. Surviving shrimps were counted and recorded; percentage shrimp mortality was then calculated using the formula:
The concentration which resulted in 50% lethality (LC50) represented the cytotoxicity for Brine shrimp nauplii.
MTT (3T3) colorimetric assay
Cytotoxic activities of both compounds were also evaluated using the standard MTT (3-[4, 5-dimethylthiazole-2-yl]-2, 5-diphenyl-tetrazolium bromide) colorimetric assay utilizing 3T3 (mouse fibroblast cells) (Mosmann 1983). The degree of MTT reduction to formazan was measured, the percentage inhibition was calculated using the formula:
The concentration which induced 50% inhibition in growth (IC50) represented the cytotoxicity for 3T3 cells.
Statistical analysis
All bioassays were performed in triplicates and were expressed as mean ± standard deviation (SD). IC50 values for the anti-urease activity and MTT (3T3) cytotoxicity were calculated using the Soft-Max Pro software (Molecular Device, USA) at 95% confidence interval. Using the GraphPad Prism 8.2.0, data obtained for the anti-urease activity was subjected to one-way analysis of variance (ANOVA) and Turkey’s Post Hoc Test (P < 0.05). The latter software was also used in calculating the LC50 values of the brine shrimp lethality by probit analysis at 95% confidence interval.
Results and discussion
Compound 1 had a molecular weight of 151.1 g/mol, which is consistent with the molecular formula C8H9NO2 and was determined by combined analyses of EIMS, NMR, and FT-IR data. FT-IR spectra revealed absorption frequencies characteristic of the nitro group (1547 cm−1; N=O stretch and 1377 cm−1; N=O bend), mono-substituted aromatic ring (698 and 748 cm−1), aromatic C=C stretch (1496 cm−1) and aromatic C–H stretch (3030 cm−1). 1H NMR (500 MHz, C3D6O) signal at δ 7.27 (multiplet) with five (5) hydrogens was assigned to the aromatic ring mono-substituted at C-1 (δ137.57); and two signals at δ = 3.33 (triplet) and δ = 4.79 (triplet) correlating with 13C NMR (125 MHz, C3D6O) signals at δ 33.74 and δ 76.95, respectively, were assigned to the two methylene (–CH2–) groups with the former at α-position to the aromatic ring and the latter at α-position to the nitro group. Therefore, the structure of compound 1 was assigned as 1-Nitro-2-phenylethane, which agrees with data obtained by Brito et al. (2013) and Oyemitan et al. (2013).
Compound 2 possessed a molecular weight of 222.4 g/mol consistent with the molecular formula: C15H26O and was determined by combined analyses of EIMS, NMR, and FT-IR data. FT-IR spectra revealed a broad peak at 3386 cm−1 which is characteristic of the hydroxyl (O–H) stretch, 1108 cm−1 represents the C–O stretch of a secondary alcohol, 994 and 918 cm−1 are characteristic of C–H bends attached to olefinic (double) bonds, 2920 and 2855 cm−1 are representatives of methylene C–H asymmetric and symmetric stretches respectively, 2970 and 1375 cm−1 signify methyl C–H asymmetric stretch and the presence of three (3) methyl (trimethyl) groups, respectively. 1H NMR signals appearing at δ 1.57 (broad singlet), bearing six (6) protons, δ 1.64 (broad doublet), habouring three (3) protons, and a singlet at δ 1.21, bearing three (3) protons were assigned to four (4) methyl groups at C-12, C-13, C-14, and C-15. It also revealed the presence of five (5) olefinic protons at δ 5.19 (doublet of doublet) and δ 4.95 (doublet of doublet), both attached to C-1; δ 5.91 (doublet of doublet of doublet) attached to C-2, δ 5.08 (triplet of quartet) attached to C-6 and δ 5.14 (triplet of quartet) attached to C-10. 13C Broadband and HMBC NMR revealed the presence of three (3) quaternary carbons at δ 72.8, δ 135.17, and δ 131.59 assigned to C-3, C-7, and C-11, respectively. The signal at δ 72.8 indicates the presence of a hydroxyl functional group at C-3. 1H NMR also revealed the presence of four (4) methylene groups at 1H δ 1.49 (doublet of doublet of doublet) attached to C-4, δ 2.04 (multiplet) bearing four (4) hydrogens attached to C-5 and C-9, and δ 1. 95 (broad doublet of doublet) attached to C-8. Therefore, the structure of compound 2 was designated 3,7,11-trimethyl-1,6,10-dodecatrien-3-ol (Nerolidol) in consistency with NMR data contained in the Biological Magnetic Resonance Data Bank (Ulrich et al. 2008).
The result of the inhibitory activities of compound 1, compound 2, and AHA (Standard) on Jack bean Urease is shown in Table 1. Significant inhibitory activities were recorded for compounds 1 and 2 at 0.5 mM concentrations, although statistically significantly lower than that of the standard (p = 0.0234, p = 0.0005 respectively). Moreover, the inhibitory activity of compound 1 was statistically significantly higher than that of compound 2 (p = 0.0185).
The anti-urease activity of NPE and Nerolidol are reported for the first time in this study. NPE and Nerolidol are major components of the essential oil of D. tripetala leaves, fruits, and seeds (Ekundayo et al. 1992; Egharevba and Idah 2015). Essential oils have been shown to produce promising results as alternative therapies in the treatment of gastric ulcers with less side-effect than conventionally used drugs (Olivera et al. 2014; Chan et al. 2016). Anosike et al. (2016) reported the antiulcer effect of methanol extract of D. tripetala seeds on aspirin-induced ulcer in albino wistar rats. The antiulcer property of Nerolidol using different experimental models such as ethanol-, indomethacin-, and stress-induced ulceration in rat has been reported (Klopell et al. 2007). The urease inhibitory activity of NPE and Nerolidol highlights a crucial mechanism involved in the antiulcer property reported for D. tripetela.
The results of the molecular docking analysis of the inhibition of the Jack Bean urease utilizing the PDB of B. pasteurii urease (PDB ID: 4UBP) by compounds 1 and 2 as well as the standard AHA are presented in Fig. 5 and Table 2. The three compounds were found to dock in favorable conformations with the PDB of the enzyme. The interaction of NPE is characterized by two hydrogen bonds, with His222 (2.95) and Arg339 (2.83). The ligand also mediates hydrophobic interactions with Ala366 and with Met367 which stabilizes the interactions. It also forms π-cation interactions with His323 (Fig. 5a). The interaction of Nerolidol is characterized by a hydrogen bond with Asp363 (1.95) and a hydrophobic interaction with Lys169 (Fig. 5b). The top ranked docked pose of AHA forms three hydrogen bonds with His222, Arg339, and Asp363. It also forms a hydrophobic interaction with Met367 (Fig. 5c). Docking calculations reveal the free energy of binding (∆G°) represented by the docking score (S) of NPE (–5.3716), higher than that of the standard AHA (−4.7963) but less than that of Nerolidol (−7.3547) thus a higher binding affinity for Nerolidol than NPE, with the standard possessing the least affinity. However, we observe an inverse correlation in the binding affinities of these compounds with their anti-urease activities (AHA > NPE > Nerolidol). This is reflected in their ligand efficiencies (S/N), AHA (−0.9591) > NPE (−0.4883) > Nerolidol (−0.4597) which normalizes the potency of these compounds by their molecular sizes (Reynolds 2015).
Molecular docking study of urease inhibitory activity. Hydrogen bonds in black dashes, coral, cyan, and sky blue sticks show the ligands in a, b, and c, respectively; urease residues in pink sticks in all the figures. The top ranked simulated pose of (a) Compound 1 (NPE); hydrogen bonds with His222 (2.95) and Arg339 (2.83); hydrophobic interactions with Ala366 and Met367, π-cation interactions with His323. b Compound 2 (Nerolidol); hydrogen bond with Asp363 (1.95) and a hydrophobic interaction with Lys169. c AHA; hydrogen bonds with His222, Arg339, and Asp363, hydrophobic interaction with Met367
Cytotoxic activities of compound 1, compound 2, and etoposide (standard) on brine shrimp (Artemia salina L.) after 24 h are shown in Table 3. No toxicities were observed for compounds 1 and 2 i.e., LC50 > 1000 µg/ml. This assay is considered a reliable general assay which parallels other cytotoxicity assays (Hamidi et al. 2014). The cytotoxic activities of compounds 1 and 2 and as well as cycloheximide (standard) on mouse fibroblasts (3T3) cells are shown in Table 4. Less than 50% inhibition were observed for both compounds; thus indicating no toxicity for both compounds at 30 µM.
Cytotoxicity results for compound 2 (Nerolidol) are corroborated by Togashi et al. (2010) and Marques et al. (2011) who reported nontoxic effects of the essential oil of Piper clausenianum which contains trans-nerolidol as its major component on fibroblasts and macrophages, and L929 fibroblasts, respectively. Mendanha et al. (2013) reported toxic effects of Nerolidol at higher concentrations on fibroblast cells (IC50 = 0.06 ± 0.01 mM) and hemolytic effect (2.3 ± 0.8 mM) on erythrocyte membrane fluidity. These results are also corroborated by oral LD50 of >5000 and 9976 mg/kg body weight values reported by Lapczynski et al. (2008) for Nerolidol in rats and mice, respectively. Therefore, at normal doses required to elicit beneficial responses, Nerolidol is nontoxic. Besides Nerolidol has been approved by the US Food and Drug Administration as a safe food flavoring agent, thus favouring its inclusion in the diets of individuals with diseases engendered by ureolytic activity specifically gastric ulcers at pharmacologically tolerable concentrations. The toxicity of NPE has been sparingly investigated; Oyemitan et al. (2013) reported a moderate toxicity with an intraperitoneal LD50 of 470 mg/kg rat. However like Nerolidol, NPE is required in relatively low concentrations to elicit targeted biochemical changes as revealed by de Siqueira et al. (2010) and Oyemitan et al. (2013) in their respective studies. Although slightly less potent than the standard AHA, the nontoxicity of these compounds at normal physiological concentrations could bolster their implementation in the treatment of infections engendered by urease producing bacteria.
Conclusion
1-Nitro-2-phenylethane and Nerolidol isolated from Dennettia tripetala seeds are good inhibitors of the urease enzyme. The inhibition of urease by these compounds is mediated by hydrophobic and hydrogen bond interactions between the compounds and amino acid residues of the enzyme’s active site. These compounds present no toxicities at normal physiological concentrations and thus can be employed in antiulcer therapy as well as in the treatment of other ailments afforded by ureolytic activity; Structural characterization and the study of the interaction of these compounds with urease could serve as a guide for the synthesis of functional derivatives which possess higher potencies than isolated compounds with minimal toxicity.
Abbreviations
- NPE:
-
1-Nitro-2-phenylethane
- AHA:
-
Acetohydroxamic acid
- Hex:
-
Hexane
- DCM:
-
Dichloromethane
- EtAOc:
-
Ethyl acetate
- MeOH:
-
Methanol
- SF:
-
Sub-fraction
- SG-CC:
-
Silica gel column chromatography
- PDB:
-
Protein Data Bank
- MOE:
-
Molecular operating environment
References
Akabueze KO, Idu M, Erhabor JO, Timothy O (2016) Antimicrobial and phytochemical attributes of Dennettia Tripetala F. Baker root and bark extracts. J Microbiol Biotech Food Sci 5:297–300
Amin M, Anwar F, Naz F, Mehmood T, Saari N (2013) Anti-Helicobacter pylori and urease inhibition activities of some traditional medicinal plants. Molecules 18:2135–2149
Amtul Z, Atta-ur-Rahman, Siddiqui RA, Choudhary MI (2002) Chemistry and Metabolism of Urease Inhibition. Curr Med Chem 9:1323–1348
Anosike CA, Okagu IU, Uchenna OK (2016) Phytoconstituents, acute toxicity study and protective effect of ethanol extract of Dennettia tripetala seed against aspirin-induced ulcer in rats. Int J Adv Sci Res 1:1–6
Benini S, Rypniewski WR, Wilson KS, Miletti S, Ciurli S, Mangani S (2000) The complex of Bacillus pasteuri urease with acetohydroxamate anion form X-ray data at 1.55 Å resolution. J Bio Inorg Chem 5:110–118
Brito TS, Lima FJ, Aragão KS, de Siqueira RJ, Sousa JC, Maia GS, Filho JD, Lahlou S, Magalhães JC (2013) The vasorelaxant effects of 1-nitro-2-phenylethane involve stimulation of the soluble guanylate cyclase-cGMP pathway. Biochem Pharm 85:780–788
Callahan BP, Yuan Y, Wolfenden R (2005) The burden borne by urease. J Am Chem Soc 127:10828–10829
Chan W, Tan LT, Chan K, Lee L, Goh B (2016) Nerolidol: a sesquiterpene alcohol with multi-faceted pharmacological and biological activities. Molecules 21:529–568
de Siqueira RJ, Macedo FI, Intraminense LdeF, Duarte GP, Magalhaes PJ, Brito TS, da Siva JK, Maia JG, Sousa PJ, Leal-Cardoso JH, Lahlou S (2010) 1-Nitro-2-phenylethane, the main constituent of the essential oil of Aniba canelilla, elicits a vago vagal bradycardiac and depressor reflex in normotensive rats. Eur J Pharm 638:90–98
Dunn BE, Phadnis SH (1998) Structure, function and localization of Helicobacter pylori urease. Yale J Biol Med 71:63–73
Egharevba HO, Idah EA (2015) Major compounds from the essential oil of the fruit and comparative phytochemical studies of the fruits and leaves of Dennettia tripetala barker F. Found in North Central Nigeria. Int J Pharm Phytochem Res 7:1262–1266
Ejechi BO, Akpomedaye DE (2005) Activity of essential oil and phenolic acid extracts of pepperfruit (Dennetia tripetala G. Barker; Anonaceae) against some food-borne microorganisms. Afr J Biotechnol 4:258–261
Ekundayo O, Ajaiyeoba E, Aiyelaagbe O, Stahl-Biskup E (1992) Volatile oil constituents of Dennettia tripetala. Planta Med 58:386–387
Follmer C, Real‐Guerra R, Wasserman GE, Olivera‐Severo D, Carlini CR (2004) Jackbean, soybean and Bacillus pasteurii ureases: biological effects unrelated to ureolytic activity. Eur J Biochem 271:1357–1363
Golbabaei S, Bazl R, Golestanian S, Nabati F, Omrany ZB, Yousefi B, Hajiaghaee R, Rezazadeh S, Amanlou M (2013) Urease inhibitory activities of β-boswellic acid derivatives. DARU J Pharm Sci 21:1–6
Halgren TA (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519
Hamidi MR, Jovanova B, Panovska TK (2014) Toxicological evaluation of the plant products using Brine Shrimp (Artemia salina L.) model. Macedonian Pharm Bull 60:9–18
Hamif M, Shoaib K, Saleem M, Rama NH, Zaib S, Iqbal J (2012) Synthesis, urease inhibition and molecular docking studies of 1,3,4-oxadiazole derivatives. ISRN Pharmacol 928901:1–9
Iseghohi SO (2015) A review of the uses and medicinal properties of Dennettia tripetala (Pepperfruit). Med Sci 3:104–111
Kappaun K, Piovesan AR, Carlini CR, Ligabue-Braun R (2018) Ureases: historical aspects, catalytic and non-catalytic properties—a review. J Adv Res 13:3–17
Khan MA, Khan H, Tariq SA, Pervez S (2014) Urease inihibitory activity of aerial parts of Artemisia scoparia: exploration in an In vitro study. Ulcers 184726:1–5
Klopell FC, Lemos M, Sousa JPB, Comunello E, Maistro EL, Bastos JK, Andrade SFD (2007) Nerolidol, antiulcer constituent Essent oil Baccharis dracunculifolia DC (Asteraceae) Z Naturforsch C 62:537–542
Konieczna I, Zarnowiec P, Kwinkowski M, Kolesinska B, Fraczyk J, Kaminski Z, Kaca W (2012) Bacterial urease and its role in long-lasting human diseases. Curr Prot Pep Sci 13:789–806
Kupchan SM, Tsou G (1973) Tumor inhibitors: a new antileukemic simaroubolide from Brucea antidysenterica. J Org Chem 38:178–179
Lapczynski A, Bhatia SP, Letizia CS, Api AM (2008) Fragrance material review on nerolidol (isomer unspecified). Food Chem Toxicol 46:S247–S250
Marques AM, Barreto ALS, Curvelo JadR, Romanos MTV, Soares RmdA, Kaplan MAC (2011) Antileishmanial activity of nerolidol-rich essential oil from Piper claussenianum. Rev Bras Farmacogn 21:908–914
Mayer BN, Ferrigni NR, Putnam JE, Jacobsen LB, Nicholas PE, McLaughin JL (1982) Brine Shrimp: a convenient general bioassay for active plant constituents. Planta Med 45:31–34
Mendanha SA, Moura SS, Anjos JLV, Valadares MC, Alonso A (2013) Toxicity of terpenes on fibroblast cells compared to their hemolytic potential and increase in erythrocyte membrane fluidity. Toxicol Vitr 27:323–329
Mobley HLT, Island MD, Hausinger RP (1995) Molecular biology of microbial ureases. Microbiol Rev 59:451–480
Mosmann T (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65:55–63
Olivera FdA, Andrade LN, de Sousa EBV, de Sousa DP (2014) Anti-ulcer activity of essential oil constituents. Molecules 19:5717–5747
Omage SO, Orhue NE, Omage K (2019) Evaluation of the phytochemical content, in vitro antioxidant capacity, biochemical and histological effects of Dennettia tripetala fruits in healthy rats. Food Sci Nutr 7:65–75
Oyemitan IA, Elusiyan CA, Akanmu MA, Olugbade TA (2013) Hypnotic, anticonvulsant and anxiolytic effects of 1-nitro-2-phenylethane isolated from the essential oil of Dennettia tripetala in mice. Phytomedicine 20:1315–1322
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Reynolds CH (2015) Ligand efficiency metrics: why all the fuss? Future Med Chem 7:1363–1365
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M (2015) PLIP: fully automated protein-ligand interaction profiler. Nucl Acids Res 43:W443–W447
Shabana S, Kawai A, Kai K, Akiyama K, Hayashi H (2010) Inhibitory activity against urease of quercetin glycosides isolated from Allium cepa and Psidium guajava. Biosci Biotechnol Biochem 74:878–880
Togashi N, Hamashima H, Shiraishi A, Inoue Y, Takano A (2010) Antibacterial activities against Staphylococcus aureus of terpene alcohols with aliphatic carbon chains. J Ess Oil Res 22:263–269
Uesato S, Hashimoto Y, Nishino M, Nagaoka Y, Kuwajima H (2002) N-substituted hydroxyureas as urease inhibitors. Chem Pharm Bull 50:1280–1282
Ukeh DA, Oku EE, Udo IA, Nta AI, Ukeh JA (2012) Insecticidal effect of fruit extracts from Xylopia aethiopica and Dennettia tripetala (Annonaceae) against Sitophilus oryzae (Coleoptera: Curculionidae). Chilean J Agric Res 72:195–200
Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao H, Markley JL (2008) Biological magnetic resonance data bank. Nucl Acids Res 36:D402–D408
Weatherburn MW (1967) Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974
Xiao ZP, Shi DH, Li HQ, Zhang LN, Xu C, Zhu HL (2007) Polyphenols based on isoflavones as inhibitors of Helicobacter pylori urease. Bioorg Med Chem 15:3703–3710
Acknowledgements
The authors are grateful to The World Academy of Sciences (TWAS) and International Centre for Chemical and Biological Sciences (ICCBS), University of Karachi, Karachi, Pakistan for a Postgraduate Fellowship Award to Ugheighele Samuel Edosewele. The interest and support of Professor (Mrs.) E.S. Omoregie of the Department of Biochemistry, University of Benin, is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ugheighele, S.E., Imafidon, K.E., Choudhary, M.I. et al. Anti-urease and cytotoxic activity of 1-Nitro-2-phenylethane and Nerolidol; two major compounds isolated from the seeds of Dennettia tripetala. Med Chem Res 29, 1874–1881 (2020). https://doi.org/10.1007/s00044-020-02607-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-020-02607-3