Abstract
A series of novel LEE011 derivatives containing pyridine N-oxide were designed, synthesized and evaluated. Systematic study of the structure-activity relationship (SAR) improves the selectivity for CDK4 and led to the identification of compound 9a. The compound showed comparable CDK4 kinase activity with ribociclib and greater selectivity over the closely related CDK6 kinase. The selective CDK4 inhibitor 9a has been demonstrated the antitumor activity via G1 phase cell cycle arrest, as well as dual CDK4/CDK6 inhibitor ribociclib and significantly down-regulated the activity of CDK4-cyclinD-Rb pathway of tumor cells. Taken together, this compound may act as promising lead compound for further development of new CDK4 inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant cancer is now believed to result from perturbations in cell cycle that result in unlimited proliferation(Hanahan and Weinberg 2000; Ortega et al. 2002). The cell cycle describes the various phases of growth, chromosomal replication, and mitosis that are frequent targets of genetic alterations in cancer. The key components which tightly regulate progression through the cell cycle in mammalian cells, are the cyclin dependent kinases (CDKs) (Hunter 1997; Malumbres and Barbacid 2001; Malumbres and Barbacid 2009; Ortega et al. 2002). CDKs belong to the serine/threonine kinase family, involved in important cellular processes, such as cell cycle or transcription regulation, which play key roles in the growth, development, proliferation and apoptosis of cells (Gao et al. 2015; Kamal et al. 2014; Morgan 1997; Ortega et al. 2002; Sherr 1996). CDKs can effectively control the tumor growth through forming complexes with cyclins. Different CDKs operate in distinct phases of the cell cycle. The key regulator of the G1-S transition is CDK4 or the highly homologous enzyme CDK6 (Dyson 1998; Sherr 1996). CDK4-cyclin D complexes phosphorylate retinoblastoma proteins (Rb), which are activated in human cell cycle to initiate G1 phase progression and prepare DNA duplication in S phase(Harbour and Dean 2000; Shapiro 2006). Deregulation of the CDK4-cyclinD-Rb pathway and CDK4 overexpression have been observed in cancer (Peyressatre et al. 2015). Inhibition of CDK4 associated with cell cycle regulation by inhibitors can lead to a G1 arrest and cell cycle progression halt, which provides an effective approach to the control of tumor growth.
Many researchers have focused their efforts on the development of small molecule inhibitors against CDK4. The first generation of CDK4 inhibitors were pan-CDK inhibitors and lead to severe toxic side effects in clinical trials (Fathalla et al. 2013; Geffken et al. 2011). Toxicity caused by poor specificity of these inhibitors has limited their therapeutic efficacy (Lu et al. 2005; Sanchez–Martinez et al. 2015). In an attempt to overcome the toxicity, the third generation CDK4 inhibitors have received significant attentions. More recently, palbociclib and ribociclib have received significant attention for highly specific inhibitors targeting CDK4/6 (Fig. 1) (Asghar et al. 2015; Toogood et al. 2005). Both of them have obtained the “Breakthrough Therapy” designation and the subsequent approval by FDA. While for the treatment of ER+/HER2− metastatic breast cancer, both palbociclib and ribociclib need to combine with aromatase inhibitor, such as letrozole (Hamilton and Infante 2016; Roskoski 2016). The drugs also showed neutropenia as the most frequent side effect in clinical assays. These studies suggest that new inhibitors with highly specificity targeting CDK4 are still urgently needed to design and discover.
As a key component of CDKs family, CDK4 shares the highly conserved ATP binding pockets of the highly homologous enzyme CDK6. In light of this fact, it is not surprising that many of the CDK4 inhibitors also inhibited CDK6. Drug selectivity related to side effects, environmental toxicity, and the emergence of resistance. Therefore, the development of CDK4 inhibitors that are selective over CDK6 may offer an improved therapeutic potential, as well as dual CDK4/CDK6 inhibitors.
Structural analyses of CDK-directed drug binding are conducted to identify molecular interactions (Kelly and Mancera 2006). The X-ray cocrystal structures of CDK6-three third generation drugs showed that each drug interacted with the residue His through hydrogen bonds (Chen et al. 2016). The residues His are only found in CDK4/6 ATP-binding pockets, which differ from other CDKs. The His-95 of CDK4 and His-100 of CDK6 may be responsible for the selectivity of the third generation CDK4/6 drugs to other CDKs. In the complex of CDK6-abemaciclib, an ordered water molecule is observed bridging the imidazole of hinge residue His100 and the ligand’s pyridine nitrogen. There is also have the space for a water molecule In the complex of CDK6-ribociclib at this position. Thus, we propose to introduce oxygen atom to the N position of ribociclib pyridine, to replace the water bridge and form a hydrogen bond with imidazole of hinge residue His100 directly, to study the biological activity and selectivity.
According to the researches, N-oxide moiety plays a key role in anticancer activity. It serving as electron donor or electron acceptor, may involve in the reaction with proteins, consequently affect enzymatic and transcriptional factor activities (Lechner et al. 2005; Li and Wogan 2005). The N-oxide moiety has proved to be responsible for the biological activity of different drug families with antitumor through the production of free radical species and have been described among the compounds classified as bioreductive antitumour agents (BAA) (Gonda et al. 2013). For example, quinoxaline 1, 4-di-N-oxides is a hypoxia-selective agent for anti-tumor therapy (Roskoski 2016) and phenazine-N,N’-dioxide is a pro-drug for anti-tumor therapy (Sarma et al. 2016). On the basis of this knowledge (Roskoski 2016), we reasoned to introduce N-oxide moiety through the structural modifications based on ribociclib, a latest and selective CDK4/6 inhibitor, to study the biological activity and selectivity.
In our study, we replaced the nitrogen atom of pyridine in ribociclib with a N-oxide moiety to form ribociclib analogs. Active and selective compound, 9a, was discovered through in vitro kinase profiling, with excellent CDK4 inhibitory activity (IC50 = 35 nM). The lead compound can induce G0/G1 phase of tumor cell cycle arrest, as well as dual CDK4/6 inhibitor ribociclib. Here, we report the synthesis and biological evaluation of the resulting novel compounds.
Results and discussion
Chemistry
All compounds containing pyridine 1-oxide were synthesized by the following routes outlined in Scheme 1. Displacement of the fluorine atom of commercially available 5-fluoro-2-nitropyridine (1) with 1-Boc-piperazine followed by reduction of nitro group with hydrogen in the presence of 10% Pd/C afforded aminopyridine compound 3.
Preparation of compounds 8, 9a–f, and 10a–n. Reagents and conditions: (a) 1-boc-piperazine, DIEA, DMF, 80 oC, 6 h; (b) H2/Pd, EtOH/EA, rt, 12 h; (c) m-CPBA, THF, rt, 6 h; (d) Na2SO3, rt, 0.5 h; (e) Pd(AcO)2, BINAP, Cs2CO3, 100 oC, 12 h; (f) con HCl, THF; (g) CH3-I or R1-Br (R1 = 9b–f), DIEA, DMF, 80 oC or rt, 6 h; (h) Ac2O, DIEA, rt, 12 h; (i) R2–COOH, EDCI, HOBt, DIEA, rt, 12 h (R2 = 10b–n);
After oxidation reaction of 3 with m-CPBA, compound 4 was treated with sodium sulfite to afford 2-amino-5-(4-(tert-butoxycarbonyl) piperazin-1-yl) pyridine 1-oxide intermidiate 5. Compound 5 react with intermidiate 6 in the presence of Pd(AcO)2, BINAP, Cs2CO3 to generate compound 7. Deprotection of compound 7 with concentrated hydrochloric acid to afford our first title compound 8 (Scheme 1).
Compounds 9a–f were obtained from compound 8 by reacting with iodomethane, 2-bromopropane, 2-bromoethan-1-ol, 1-bromo-2-methoxyethane, methyl 2-bromoacetate and 2-bromoacetonitrile in the presence of DIEA at appropriate temperature. Compound 8 acylated by acetic anhydride to afford 10a. Through condensation reaction of compound 8 with the corresponding carboxylic acids in the presence of condensing agent EDCI and HOBt to afford compounds 10b–n (Scheme 1).
Enzyme inhibitory activity and SAR
It is evident from original compound 8 that the N-oxide substituent of pyrrolo[2,3-d] pyrimidines-2-amine scaffold based on ribociclib showed good inhibition activity targeting CDK4 at 1 μM (Table 1). Then we kept N-oxide substituent constant and focused our efforts on varying the N-4 position of piperazine ring of compound 8 using different substituents. The CDK4 and CDK6 enzyme inhibition of these compounds at 1 μM were shown in Table 1. From Table 1, we can see that compounds 9a and 9b have relative outstanding inhibition activities against CDK4 and that others are obviously weak. The hydrophobic methyl of 9a and isopropyl group of 9b, introduced to the N-4 position on the piperazine ring, keep the inhibitory activity against CDK4. And moderately decreased CDK4 inhibitory activity was found after we prolonged the alkyl chain from acetyl to heptanoyl group (10a–i), indicated that long hydrophobic alkyl chain is not favorable to CDK4 inhibitory activity. Compounds 9c–f and 10j–n were prepared to investigate the influence of substitutions on piperazine N-4 position with hydrophilic group that containing hydrogen bond donors and/or hydrogen bond acceptors in different chain length for CDK4 inhibitory activity. Obviously, the CDK4 inhibitory activity of these compounds were found to be substantially less active than the original compound 8. The half-maximal inhibitory concentrations (IC50) values of compound 8, 9a, 9b against CDK4/6 and 9c, 9d, 10c, 10f–h, 10l, ribociclib against CDK4 were detected (Table 2 and Table S1). The result indicated that compounds 9a showed the similar CDK4 kinase activity with ribociclib. Then we performed all subsequent in vitro biological studies using this compound.
Interestingly, all the prepared compounds had weak CDK6 inhibitory activity, which indicated the N-oxide substituent introduced to the N position on the pyridine ring may improved the selectivity for CDK4 over CDK6. Resulting from high homology, identical substrate specificities and enzymatic activities CDK4 and CDK6 have overlapping functions in development. In light of this fact, many of the third generation CDK4 inhibitors such as abemaciclib, dinaciclib, ribociclib, palbociclib and AG-024322 also inhibited CDK6 (Chen et al. 2016). Development of ATP-competitive CDK4 inhibitors that are selective over CDK6 may offer an improved therapeutic potential, as well as dual CDK4/CDK6 inhibitors.
Molecular docking studies
Molecular docking studies were carried out to investigate the binding modes of compound 9a and ribociclib in CDK4 and CDK6, respectively. The compounds were docked to CDK4 (PDB codae: 2w96) and CDK6 (PDB code: 5L2T) by Discovery Studio (Accelrys, San Diego, CA). The image was created using PyMOL (Delano 2014). For comparison, the binding mode of 9a was superimposed on that of ribociclib as shown in Fig. 2, 9a could tightly bind to the ATP-binding site of CDK4 in a binding mode similar to that of ribociclib. Hydrogen bonds appear to be formed between the aminopyrimidine and carboxamide group with the backbone residue of VAL96, HIS95, and ASP158. Given that these hydrogen bonds may contribute to the activity of compound 9a against CDK4.
In an attempt to find the reasons that the CDK6 inhibitory of compound 9a at 1 μM is only 68%, we docked compound 9a into the CDK6 protein in Fig. 3. Compared with the ribociclib, the hydrogen bonding interaction between pyrimidine group and His100 in 9a was too weak, which may lead to the significant selectivity of compound 9a for CDK4 over CDK6. Namely, compound 9a was more selective than ribociclib. In order to illustrate the SARs more rational, molecular modeling studies are also performed with compound 8 and 9b as the good candidate for CDK4 enzyme inhibition. As shown in Figure S1, compound 8 and 9b showed a similar CDK4 and CDK6 inhibition with 9a. That may be the reason for the similar inhibition activity with 9a. The SARs findings according to the docking poses are coordinating with our SAR results discussed above.
From the above, the N-oxide substituent introduced to the N-1 position on the pyridine ring may improve the selectivity for CDK4 over CDK6. At the meanwhile, the inhibitory activity of compound 9a targeting CDK4 was maintained compared to ribociclib.
Effect on cell cycle progression
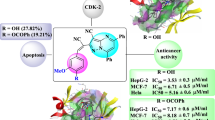
We therefore wondered whether this kinase inhibition effect could partly result in growth arrest induction and cell cycle inhibition. Whether or not the inhibitory activity of the molecules affect the tumor cell proliferation was next explored. As a result, neither the positive control ribociclib nor 9a exhibits good inhibition in tumor cell proliferation. All of the IC50 values in the tumor cell lines are exceed 5 μM (data not shown). All the tested compounds lack cytotoxicity activity. We next investigated the effect of compound 9a treatment on T47D (human breast cancer cell line, Fig. 4) and A549 (human lung cancer cell line, Fig. 5) cell cycle kinetics. Cells were treated with compound 9a with varying concentrations, ribociclib and vehicle (DMSO) for 24 h, which were stained with propidium iodide, and subjected to flow cytometric analysis to determine the distribution of cells in various phases of the cell cycle. Compared to the vehicle-treated cells, cancer cells treated with compound 9a demonstrate an increase in the percentage of cells in G0/G1 and a loss of S-phase cells. The result demonstrated the ability of compound 9a affect the activities of CDK4 in cells and led to support the hypothesis of a blockage during the G1 phase. Ribociclib treatment for 24 h led to a massive accumulation of cells in G0/G1. Compared to ribociclib, our compound 9a showed similar effects on G0/G1-phase inhibition of cancer cells. Taken all the data together, the selective CDK4 inhibitor 9a has been shown antitumor activity via G1 phase cell cycle arrest, as well as dual CDK4/CDK6 inhibitor ribociclib. Thus the selective CDK4 inhibitor 9a showed the antitumor activity via G1 phase cell cycle arrest.
Effect of compound 9a on cell cycle progression of human breast cancer cell line (T47D). a 24 hours cell cycle arrest analysis of 9a was performed on T47D. Cells were seeded in 6-plates at a density of 5 × 105 cells/ml and treated with increasing doses of 9a or rapamycin for 24 h. Then cells were fixed with 70% ethanol overnight at 4 ℃ and analyzed by flow cytometry after propidium iodide staining. b Thepercentage of cell cycle distribution was calculated by GraphPad software
Effect of compound 9a on cell cycle progression of human lung cancer cell line (A549). a 24 hours cell cycle arrest analysis of 9a was performed on A549. Cells were seeded in 6-plates at a density of 5 × 105 cells/ml and treated with increasing doses of 9a or rapamycin for 24 h. Then cells were fixed with 70% ethanol overnight at 4 ℃ and analyzed by flow cytometry after propidium iodide staining. b The percentage of cell cycle distribution was calculated by GraphPad software
Effect on the CDK4-cyclinD-Rb pathway
As compound 9a emerged as one of the most potent inhibitor, being much more potent than the positive control compound of its clear cell cycle suppression in A549, we decided to characterize the cellular effects of compound 9a in more detail. To further confirm the CDK4 inhibitory, we performed western blot analysis to evaluate the CDK4-cyclinD-Rb pathway inhibition (Fig. 6). Compound 9a exhibited stronger inhibition than control. Treated cells exhibited dose-dependent reductions in the level of the protein CDK4 and Cyclin D1, which is consistent with the known mechanisms of action for CDK4 inhibitors (Graf et al. 2010). Compound suppressed CDK4-cyclinD-Rb pathway at 1μM in cell. To evaluate if the antiproliferative activity of compound 9a is caused by inhibition of cellular CDK4, we selected inhibition of Rb phosphorylation at the CDK4-specific sites Ser780. The results of this study show that compound 9a effectively suppressed phosphorylation of Rb Ser780 at 1 μM, confirming that 9a as a specific CDK4 inhibitor.
Conclusion
In summary, through analyzing the co-crystal structures of CDK4/6-inhibitors, we have designed and synthesized a new class of 2-((7-cyclopentyl-6-(dimethyl carbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino) pyridine 1-oxides as novel CDK4 kinase inhibitors. Compound 9a was discovered with potent inhibition on CDK4 and showed good selectivity against CDK6. SAR study demonstrated that the N-oxide substituent introduced to the N-1 position on the pyridine ring improved the selectivity for CDK4 over CDK6 and maintained the inhibitory activity of CDK4. In cell cycle assays, it was evident that compound 9a could block the human breast and lung cancer cell line in G0/G1 phase, which resulted in decreased S-phase populations to exhibit the antitumor activity. Compound showed similar cell cycle suppression to ribociclib. In addition, 9a could significantly down-regulated the activity of CDK4-cyclinD-Rb pathway of tumor cells. All of the studies suggest that this compound could act as promising lead compound for further development of new CDK4 inhibitors and may be useful in drug development.
Experimental
Biology
Materials and methods
Human breast cancer cell line (T47D cells) and lung cancer cell lines (A549 cells) were purchased from ATCC. All the cell lines were recently authenticated by cellular morphology and the short tandem repeat analysis at Microread Inc. (Beijing, China; May 2014) according to the guideline from ATCC.
Kinase inhibition assays
Kinase inhibition profiles were determined using KinaseProfiler services provided by Eurofins, and ATP concentrations used are the Km of corresponding kinases.
Molecular docking
In this investigation, virtual screening based on molecular docking was carried out using the 3D structure of human CDK4/6(PDB entry: 2W96/5L2T). All the calculations were carried out using the platform of Discovery Studio 3.1 (Accelrys Inc. San Diego, CA, USA). Before molecular docking, the receptor protein was prepared by the DS 3.1 software package with standard preparation procedures (protein preparation protocol), which include removing water molecules, adding hydrogen atoms to the protein, and assigning force field (here the CHARMm force field was adopted). Compound libraries used for docking were prepared with the “Prepare Ligands” module in DS 3.1. Parameter values for “Change Ionization, Generate Tautomers, and Generate Isomers” were set to false. Other parameters were set to their default values. The binding site of CDK4/6 was defined as a sphere containing the residues that stay within 10 Å from its original ligand, which is large enough to cover the catalytic site. LipdockScore incorporated into the DS 3.1 program package was employed to evaluate and rank the binding poses.
Cell cycle assay
The different cell lines were plated onto 100 mm2 dishes at a cell density of 1.0 × 106 cells/dish. All cells were treated with increasing concentrations of the indicated compounds 24 h post plating. Cells were harvested 24 h post-treatment, washed in phosphate buffered saline (PBS), and fixed in ice cold 70% ethanol for at least 24 h. The fixed cells were then washed with room temperature PBS and stained with propidium iodide (50 mg/mL) in the presence of RNase A (0.5 mg) for 30 min at 37 °C. The stained cells were then analyzed using a Flow Cytometer. and the resulting data analyzed with cell cycle analysis software (Modfit, BD).
Western blotting
Cell lysates from different cell lines were prepared with RIPA buffer in the presence of protease inhibitor cocktails and Phosphatase Inhibitor Cocktail 2 and 3 (P8340, P5726, and P0044, Sigma-Aldrich, St Louis, MO, USA). Protein (20–50 μg) was separated by 8–15% Tris-acrylamide gels and transferred onto PVDF membrane the membrane was blocked in 5% skim milk, subsequently incubated with primary antibodies at 4 °C over night followed by incubation with peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG and developed with Pierce ECL reagent (cat. #17153, Millipore, Billerica, MA, USA). Antibody information: CDK4(Santa Cruz, H3112), Cyclin D1(Cell Signaling; catalog no.92G2), Rb (Cell Signaling; catalog no. 9309), anti-phosphospecific RbSer780 (Cell Signaling Technology; catalog no. 9307)
Statistical analysis
Values were expressed as means ± s.e.m. Significance was determined by χ2 test, others were determined by Student’s t-test. A value of P < 0.05 was used as the criterion for statistical significance. Asterisk (*) indicates significant difference with P < 0.05, asterisk (**) indicates significant difference with P < 0.01.
Chemistry
General methods
1H NMR (400 MHz) and 13C NMR (101 MHz) spectra were taken on a Bruker AV-400 MHz spectrometer and chemical shifts were reported in ppm downfield from internal Me4Si. The elemental analyses were carried out on a LEEMAN EA3000 CHNSO analyzer. High-resolution mass spectra (HRMS) were recorded on a VG ZAB-HS mass spectrometer under electron spray ionization (ESI). All of the solvents were purified and distilled according to the standard procedure. The commercially obtained materials were used directly without further purification unless otherwise noted. The purity of tested compound was assessed to be >95% by HPLC analysis on a Shimadzu Prominence-i LC-2030C 3D system (column, InertSustain C18, 4.6 mm × 250 mm, 5 μM; mobile phase, gradient elution of methanol/H2O; low rate, 1.0 mL/min; UV wavelength, 190−800 nm; temperature, 40 °C; injection volume, 10 μL).
General procedure for the synthesis of compound 2
To a solution of 5-bromo-2-nitropyridine (40.0 g, 197 mmol) in DMSO (150 mL) were added 1-Boc-piperazine (47.4 g, 252 mmol) and DIPEA (38 mL, 219 mmol) . The reaction mixture was heated at 80 °C for 11 h. The reaction mixture was poured into ice-water and then extracted with EtOAc. The combined extracts were washed with water and brine. The organic layer was dried over Na2SO4, filtered, and concentrated under reduced pressure. Purification by column chromatography (1:9 methanol/ dichloromethane) gave 1-Boc- 4-(6-nitro-pyridin-3-yl)-piperazine (49.9 g, 82%) as yellow solid. 1H NMR (400 MHz, CDCl3): δ 8.11 (d, J = 9.1 Hz, 1H), 8.08 (d, J = 2.7 Hz, 1H), 7.18 (dd, J = 9.1, 2.8 Hz, 1H), 3.65–3.56 (m, 4H), 3.48–3.38 (m, 4H), 1.45 (s, 9H).
General procedure for the synthesis of compound 3
A solution of 1-methyl-4-(6-nitro-pyridin-3-yl)-piperazine (45.3 g, 147 mmol) in ethanol (1.4 L) and ethyl acetate (1.4 L) was hydrogenated in the presence of 10% Pd/C (4.7 g) using an H2 balloon. After 16 h, the reaction mixture was filtered through a pad of Celite and rinsed with methanol (2 × 15 mL). The filtrate was concentrated and purified by column chromatography (1:9 methanol/ dichloromethane) to afford the title compound (37.6 g, 92%) as yellow solid. 1H NMR (400 MHz, CDCl3): δ 7.75 (t, J = 2.7 Hz, 1H), 7.14 (dt, J = 8.6, 3.3 Hz, 1H), 6.47 (dd, J = 8.8, 3.4 Hz, 1H), 4.15 (br s, 2H), 3.54 (t, J = 4.7 Hz Hz, 4H), 2.93 (t, J = 4.7 Hz, 4H), 1.45 (s, 9H).
General procedure for the synthesis of compound 5
A solution of tert-butyl 4-(6-aminopyridin-3-yl)piperazine-1-carboxylate (5.0 g, 18.0 mmol) in tetrahydrofuran (150 mL) was cooled to 0 °C, 3-Chloroperoxybenzoic acid (15.5 g, 54 mmol, 60%) was added carefully in several portions and stirred overnight at room temperature to get compound 4. Then 50 mL saturated sodium sulfite was added and stirred at room temperature. After 1 h, the reaction mixture was concentrated and purified by column chromatography (1:9 methanol/dichloromethane) to afford the title compound 5 (3.14 g, 56%) as brown solid. 1H NMR (400 MHz, Chloroform-d) δ 8.26 (s, 1H), 8.10 (d, J = 9.0 Hz, 1H), 7.90 (t, J = 1.9 Hz, 1H), 7.33–7.27 (m, 1H), 3.58 (t, J = 4.7 Hz, 4H), 3.08 (t, J = 5.1 Hz, 4H), 1.47 (s, 9H).
General procedure for the synthesis of compound 7
50 mL two neck-flask was charged with 2-chloro-7-cyclopentyl-N,N-dimethyl-7H-pyrrolo[2,3-d] pyrimidine-6-carboxamide(835 mg, 2.85 mmol), 2-amino-5-(4-(tert-butoxycarbonyl) piperazin-1-yl) pyridine 1-oxide (840 mg, 2.85 mmol), Palladium diacetate(16 mg, 0.071 mmol), BINAP (89 mg, 0.143 mmol), cesium carbonate (1.39 g, 4.3 mmol) and 1,4-dioxane (20 mL) and stirred at 100 °C under inert atmosphere overnihgt. The reaction mixture was concentrated and purified by column chromatography to afford the title compound 7 (1.1 g, 68%) as tan solid. 1H NMR (400 MHz, Chloroform-d) δ 9.63 (s, 1H), 8.73 (d, J = 1.3 Hz, 1H), 8.65 (d, J = 9.4 Hz, 1H), 7.95 (d, J = 2.3 Hz, 1H), 7.05 (dd, J = 9.5, 2.5 Hz, 1H), 6.46 (d, J = 1.3 Hz, 1H), 4.86–4.72 (m, 1H), 3.59 (t, J = 5.0 Hz, 4H), 3.15 (s, 6H), 3.06 (t, J = 5.0 Hz, 4H), 2.66–2.43 (m, 2H), 2.15–1.96 (m, 4H), 1.85–1.64 (m, 2H), 1.48 (s, 9H). 13C NMR (101 MHz, CDCl3) δ 163.83, 154.53, 153.19, 151.76, 151.55, 141.72, 139.75, 132.98, 126.73, 118.85, 113.68, 113.18, 100.74, 80.24, 58.01, 49.49, 39.42, 35.20, 30.20, 28.41, 24.61.
General procedure for the synthesis of compound 8
5-(4-(tert-butoxycarbonyl)piperazin-1-yl)-2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino)pyridine 1-oxide (700 mg, 1.27 mmol) was dissolved in 10 mL tetrahydrofuran and cooled to 0 °C, 0.5 mL concentrated hydrochloric acid was added. After 15 min the reaction mixture was stirred at room temperature for 2 h. Then saturated sodium carbonate was aadded to adjust pH > 8. The reaction mixture was concentrated and purified by column chromatography to afford the title compound 8 (510.2 mg, 89%) as light brown solid. 1H NMR (400 MHz, Chloroform-d) δ 9.60 (s, 1H), 8.72 (s, 1H), 8.63 (d, J = 9.4 Hz, 1H), 7.99 (d, J = 2.5 Hz, 1H), 7.03 (dd, J = 9.5, 2.4 Hz, 1H), 6.45 (s, 1H), 4.77 (p, J = 8.9 Hz, 1H), 3.29–3.17 (m, 8H), 3.14 (s, 6H), 2.94 (s, 1H), 2.86 (s, 1H), 2.60–2.45 (m, 2H), 2.14–1.91 (m, 4H), 1.83–1.62 (m, 2H).13C NMR (101 MHz, CDCl3) δ 163.86, 153.25, 151.75, 151.65, 141.71, 139.72, 132.92, 126.83, 118.36, 113.65, 113.07, 100.74, 58.00, 49.30, 44.95, 36.49, 30.20, 24.61. ESI-HRMS m/z calcd for C23H31N8O2+ 451.2564, found 451.2560 [M + H]+. Calcd for C23H30N8O2: C, 61.32; H, 6.71; N, 24.87; O, 7.10, found: C, 61.15; H, 6.65; N, 24.89; O, 7.05. HPLC purity 97%.
General procedure for the synthesis of compounds 9a
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino)-5-(piperazin-1-yl)pyridine 1-oxide(200 mg, 0.44 mmol) was dissolved in 10 mL methanol followed by adding formaldehyde(330 µL, 37% in water). After 10 min the sodium cyanoborohydride was added and stirred overnight. The reaction mixture was concentrated and purified by column chromatography to afford the title compound 9a (88.5 mg, 43%) as light brown solid. 1H NMR (400 MHz, Chloroform-d) δ 9.62 (s, 1H), 8.71 (s, 1H), 8.60 (d, J = 9.4 Hz, 1H), 7.93 (d, J = 2.5 Hz, 1H), 7.01 (dd, J = 9.4, 2.5 Hz, 1H), 6.45 (s, 1H), 4.77 (p, J = 9.0 Hz, 1H), 3.23–3.06 (m, 11H), 2.59 (t, J = 5.0 Hz, 4H), 2.56–2.48 (m, 2H), 2.35 (s, 2H), 2.12–1.96 (m, 4H), 1.78–1.62 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 163.88, 153.36, 151.78, 151.63, 141.75, 139.32, 132.80, 126.27, 117.87, 113.55, 113.04, 100.74, 57.98, 54.61, 49.12, 46.01, 39.41, 30.17, 24.59. ESI-HRMS m/z calcd for C24H33N8O2+ 465.2721, found 465.2715 [M + H]+. Calcd for C24H32N8O2: C, 62.05; H, 6.94; N, 24.12; O, 6.89, found: C, 61.98; H, 6.95; N, 24.05; O, 6.85. HPLC purity 96%.
General procedure for the synthesis of compounds 9b–9f
Compound 8 (0.3 mmol), R1 halogen substitutes (0.36 mmol), ethyldiisopropylamine (0.6 mmol) were dissolved in 10 mL DMF. The reaction was heated at 80 oC (for 9b–9d) or room temprature (9e, 9f) over night under the protection of N2. The reaction mixture was evaporated under vacuum to give crude product, which was purified by column chromatography on silica gel to give products 9b–9f.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino)-5-(4-isopropylpiperazin-1-yl)pyridine 1-oxide (9b)
Light brown solid, yield 62%. 1H NMR (400 MHz, Methanol-d4) δ 8.74 (s, 1H), 8.56 (d, J = 9.4 Hz, 1H), 7.97 (s, 1H), 7.39 (d, J = 9.5 Hz, 1H), 6.67 (s, 1H), 4.80–4.68 (m, 1H), 3.54–3.38 (m, 8H), 3.32–3.28 (m, 1H), 3.14 (s, 6H), 2.48–2.35 (m, 2H), 2.12–1.97 (m, 4H), 1.82–1.64 (m, 2H), 1.41 (d, J = 6.6 Hz, 6H). 13C NMR (101 MHz, CD3OD_SPE) δ 164.08, 152.57, 151.87, 151.24, 140.78, 139.01, 133.38, 125.97, 120.58, 113.86, 113.34, 100.99, 58.21, 57.97, 46.38, 38.67, 34.19, 29.93, 24.32, 16.14. ESI-HRMS m/z calcd for C26H37N8O2+ 493.3034, found 493.3033 [M + H]+. Calcd for C26H36N8O2: C, 63.39; H, 7.37; N, 22.75; O, 6.50, found: C, 63.25; H, 7.40; N, 22.69; O, 6.47. HPLC purity 97%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino)-5-(4-(2-hydroxyethyl)piperazin-1-yl)pyridine 1-oxide (9c)
Light brown solid, yield 71%. 1H NMR (400 MHz, Chloroform-d) δ 9.63 (s, 1H), 8.72 (s, 1H), 8.61 (d, J = 9.5 Hz, 1H), 7.93 (d, J = 2.6 Hz, 1H), 7.01 (dd, J = 9.5, 2.6 Hz, 1H), 6.45 (s, 1H), 4.88–4.69 (m, 1H), 3.66 (t, J = 5.3 Hz, 2H), 3.19–3.10 (m, 10H), 2.71–2.65 (m, 4H), 2.63–2.59 (m, 2H), 2.58–2.49 (m, 2H), 2.13–1.98 (m, 4H), 1.76–1.66 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 163.89, 153.37, 151.80, 151.63, 141.72, 139.37, 132.83, 126.23, 117.84, 113.57, 113.07, 100.74, 59.31, 57.88, 52.49, 49.30, 45.90, 35.18, 30.19, 24.60. ESI-HRMS m/z calcd for C25H35N8O3+ 495.2827, found 495.2820 [M + H]+. HPLC purity 96%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino)-5-(4-(2-methoxyethyl)piperazin-1-yl)pyridine 1-oxide (9d)
Light brown solid, yield 70%. 1H NMR (400 MHz, Chloroform-d) δ 9.55 (s, 1H), 8.63 (s, 1H), 8.53 (d, J = 9.5 Hz, 1H), 7.90–7.85 (m, 1H), 6.95 (dd, J = 9.6, 2.4 Hz, 1H), 6.39 (s, 1H), 4.84 – 4.64 (m, 1H), 3.47 (t, J = 5.4 Hz, 2H), 3.29 (s, 3H), 3.14–2.98 (m, 10H), 2.65–2.54 (m, 6H), 2.53–2.39 (m, 2H), 2.03–1.90 (m, 4H), 1.72–1.59 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 163.77, 153.26, 151.68, 151.59, 141.82, 139.05, 132.79, 125.99, 117.87, 113.47, 113.02, 100.70, 69.99, 58.91, 57.79, 53.03, 48.92, 35.13, 30.12, 24.54, 8.70. ESI-HRMS m/z calcd for C26H37N8O3+ 509.2983, found 509.2980 [M + H]+. HPLC purity 98%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino)-5-(4-(2-methoxy-2-oxoethyl)piperazin-1-yl)pyridine 1-oxide (9e)
Light brown solid, yield 79%. 1H NMR (400 MHz, Chloroform-d) δ 9.59 (s, 1H), 8.68 (s, 1H), 8.58 (d, J = 9.5 Hz, 1H), 7.90 (d, J = 2.6 Hz, 1H), 6.98 (dd, J = 9.5, 2.6 Hz, 1H), 6.42 (s, 1H), 4.85–4.65 (m, 1H), 3.70 (s, 3H), 3.26 (s, 2H), 3.18–3.13 (m, 4H), 3.11 (s, 6H), 2.76–2.68 (m, 4H), 2.59–2.42 (m, 2H), 2.08–1.95 (m, 4H), 1.74–1.56 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 170.42, 163.84, 153.35, 151.78, 151.59, 141.71, 139.37, 132.83, 126.29, 117.86, 113.54, 113.03, 100.69, 59.09, 57.94, 52.45, 51.76, 49.09, 35.12, 30.17, 24.57. ESI-HRMS m/z calcd for C26H35N8O4+ 523.2776, found 523.2776 [M + H]+. HPLC purity 96%.
5-(4-(cyanomethyl)piperazin-1-yl)-2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino)pyridine 1-oxide (9f)
Light brown solid, yield 83%. 1H NMR (400 MHz, Chloroform-d) δ 9.64 (s, 1H), 8.73 (s, 1H), 8.64 (d, J = 9.4 Hz, 1H), 7.95 (d, J = 2.6 Hz, 1H), 7.02 (dd, J = 9.5, 2.4 Hz, 1H), 6.46 (s, 1H), 4.79 (t, J = 8.9 Hz, 1H), 3.59 (s, 2H), 3.21–3.10 (m, 10H), 2.77 (t, J = 4.9 Hz, 4H), 2.54 (t, J = 10.2 Hz, 2H), 2.15–1.98 (m, 4H), 1.79–1.65 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 163.88, 153.35, 151.83, 151.60, 141.40, 139.77, 132.94, 126.58, 118.23, 114.32, 113.65, 113.09, 100.70, 58.00, 51.37, 49.08, 45.91, 35.16, 30.21, 24.60. ESI-HRMS m/z calcd for C25H32N9O2+ 490.2673, found 490.2677 [M + H]+. HPLC purity 97%.
General procedure for the synthesis of compounds 10a
Compound 8 (135 mg, 0.3 mmol), acetic anhydride (30.6 µL, 0.33 mmol), ethyldiisopropylamine (93.2 µL, 0.6 mmol) were dissolved in 10 mL DCM. The reaction was stirred at room temperature overnight. The reaction mixture was evaporated under vacuum to give crude product, which was purified by column chromatography on silica gel to give products 10a as yellow solid, yield 83%. 1H NMR (400 MHz, Chloroform-d) δ 9.63 (s, 1H), 8.72 (s, 1H), 8.64 (d, J = 9.5 Hz, 1H), 7.94 (d, J = 2.6 Hz, 1H), 7.03 (dd, J = 9.4, 2.4 Hz, 1H), 6.45 (s, 1H), 4.78 (p, J = 8.9 Hz, 1H), 3.77 (t, J = 5.2 Hz, 2H), 3.62 (t, J = 5.1 Hz, 2H), 3.20–3.03 (m, 10H), 2.61–2.45 (m, 2H), 2.13 (s, 3H), 2.12–1.96 (m, 4H), 1.79–1.64 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 168.99, 163.81, 153.18, 151.75, 151.56, 141.33, 139.98, 133.01, 126.87, 118.61, 113.70, 113.13, 100.72, 57.99, 49.56, 45.85, 35.13, 30.20, 24.60, 21.32. ESI-HRMS m/z calcd for C25H33N8O3+ 493.2670, found 493.2666 [M + H]+. HPLC purity 99%.
General procedure for the synthesis of compounds 10b–10n
To a solution of compound 8 (0.3 mmol) in 5 mL DMF, R2–COOH (0.36 mmol), EDCI (0.45 mmol), HOBt (0.45 mmol), DIEA (0.6 mmol) was added and stirred overnight. The reaction mixture was evaporated under vacuum to give crude product, which was purified by column chromatography on silica gel to give title compounds 10b–10n.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo [2,3-d] pyrimidin-2-yl) amino)-5-(4-propionylpiperazin-1-yl) pyridine 1-oxide (10b)
Light brown solid, yield 56%. 1H NMR (400 MHz, Chloroform-d) δ 9.66 (s, 1H), 8.74 (s, 1H), 8.65 (d, J = 9.5 Hz, 1H), 7.96 (s, 1H), 7.03 (dd, J = 9.5, 2.4 Hz, 1H), 6.46 (s, 1H), 4.79 (p, J = 8.8 Hz, 1H), 3.79 (t, J = 5.1 Hz, 2H), 3.63 (t, J = 5.0 Hz, 2H), 3.15 (s, 6H), 3.09 (q, J = 4.8 Hz, 4H), 2.61–2.47 (m, 2H), 2.39 (q, J = 7.4 Hz, 2H), 2.14–1.97 (m, 4H), 1.78–1.63 (m, 2H), 1.17 (t, J = 7.5 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 172.32, 163.83, 153.22, 151.77, 151.58, 141.39, 139.97, 133.00, 126.88, 118.54, 113.70, 113.14, 100.74, 58.00, 49.62, 44.96, 35.17, 30.21, 26.45, 24.61, 9.42. ESI-HRMS m/z calcd for C26H35N8O3+ 507.2827, found 507.2826 [M + H]+. HPLC purity 98%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino)-5-(4-isobutyrylpiperazin-1-yl) pyridine 1-oxide (10c)
Light brown solid, yield 61%. 1H NMR (400 MHz, Chloroform-d) δ 9.63 (s, 1H), 8.72 (s, 1H), 8.64 (d, J = 9.4 Hz, 1H), 7.95 (d, J = 2.6 Hz, 1H), 7.02 (dd, J = 9.5, 2.6 Hz, 1H), 6.45 (s, 1H), 4.90–4.67 (m, 1H), 3.82–3.64 (m, 4H), 3.13 (s, 6H), 3.13–3.04 (m, 4H), 2.87–2.74 (m, 1H), 2.60–2.44 (m, 2H), 2.14–1.94 (m, 4H), 1.81–1.61 (m, 2H), 1.14 (d, J = 6.8 Hz, 6H). 13C NMR (101 MHz, CDCl3) δ 175.48, 163.82, 153.23, 151.75, 151.60, 141.36, 139.92, 132.96, 126.83, 118.44, 113.68, 113.10, 100.72, 57.98, 49.63, 44.98, 35.17, 30.20, 30.07, 24.60, 19.40. ESI-HRMS m/z calcd for C27H37N8O3+ 521.2983, found 521.2983 [M + H]+. HPLC purity 97%.
5-(4-butyrylpiperazin-1-yl)-2-((7-cyclopentyl-6-(dimethylcarbamoyl)- 7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino)pyridine 1-oxide (10d)
Light brown solid, yield 63%. 1H NMR (400 MHz, Chloroform-d) δ 9.61 (s, 1H), 8.70 (s, 1H), 8.62 (d, J = 9.4 Hz, 1H), 7.92 (d, J = 2.5 Hz, 1H), 7.00 (dd, J = 9.5, 2.4 Hz, 1H), 6.44 (s, 1H), 4.84–4.70 (m, 1H), 3.75 (t, J = 5.1 Hz, 2H), 3.61 (t, J = 5.0 Hz, 2H), 3.12 (s, 6H), 3.11–3.02 (m, 4H), 2.61–2.42 (m, 2H), 2.36–2.27 (m, 2H), 2.10–1.91 (m, 4H), 1.80–1.57 (m, 4H), 0.95 (t, J = 7.4 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 171.54, 163.80, 153.22, 151.73, 151.60, 141.36, 139.91, 132.94, 126.81, 118.38, 113.66, 113.07, 100.71, 57.96, 49.61, 45.13, 35.13, 30.18, 24.59, 18.69, 13.99. ESI-HRMS m/z calcd for C27H37N8O3+ 521.2983, found 521.2972 [M + H]+. HPLC purity 98%.
(E)-5-(4-(but-2-enoyl) piperazin-1-yl)-2-((7-cyclopentyl-6- (dimethylcarbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl)amino)pyridine 1-oxide (10e)
Light brown solid, yield 42%. 1H NMR (400 MHz, Chloroform-d) δ 9.61 (s, 1H), 8.69 (s, 1H), 8.61 (d, J = 9.4 Hz, 1H), 7.99–7.88 (m, 1H), 7.00 (dd, J = 9.4, 2.3 Hz, 1H), 6.95–6.81 (m, 1H), 6.43 (s, 1H), 6.26 (dq, J = 14.9, 1.7 Hz, 1H), 4.74 (q, J = 8.9 Hz, 1H), 3.87–3.66 (m, 4H), 3.19–3.01 (m, 10H), 2.62–2.43 (m, 2H), 2.11–1.94 (m, 4H), 1.87 (dd, J = 6.8, 1.6 Hz, 3H), 1.78–1.62 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 165.57, 163.80, 153.20, 151.72, 151.60, 142.46, 141.34, 139.86, 132.94, 126.80, 121.01, 118.42, 113.65, 113.07, 100.72, 57.96, 53.64, 49.61, 35.15, 30.18, 24.58, 18.29. ESI-HRMS m/z calcd for C27H35N8O3+ 519.2827, found 519.2828 [M + H]+. HPLC purity 96%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo [2,3-d] pyrimidin-2-yl) amino)-5-(4-(3-methylbutanoyl) piperazin-1-yl)pyridine 1-oxide (10f)
Light brown solid, yield 77%. 1H NMR (400 MHz, Chloroform-d) δ 9.65 (s, 1H), 8.73 (s, 1H), 8.65 (d, J = 9.4 Hz, 1H), 7.96 (d, J = 2.6 Hz, 1H), 7.03 (dd, J = 9.4, 2.4 Hz, 1H), 6.46 (s, 1H), 4.79 (p, J = 8.8 Hz, 1H), 3.80 (t, J = 5.2 Hz, 2H), 3.65 (t, J = 5.1 Hz, 2H), 3.15 (s, 6H), 3.09 (t, J = 5.0 Hz, 4H), 2.63–2.49 (m, 2H), 2.25 (d, J = 7.0 Hz, 2H), 2.16–1.97 (m, 6H), 1.79–1.59 (m, 2H), 0.99 (d, J = 6.5 Hz, 6H). 13C NMR (101 MHz, CDCl3) δ 171.07, 163.84, 153.24, 151.78, 151.59, 141.36, 139.99, 132.98, 126.89, 118.46, 113.70, 113.11, 100.72, 58.00, 49.72, 45.37, 41.98, 35.38, 30.21, 25.80, 24.61, 22.77. ESI-HRMS m/z calcd for C29H41N8O3+ 549.3296, found 549.3294 [M + H]+. HPLC purity 98%.ESI-HRMS m/z calcd for C28H39N8O3+ 535.3140, found 535.3138 [M + H]+. HPLC purity 99%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo [2,3-d] pyrimidin-2-yl) amino)-5-(4-pentanoylpiperazin-1-yl) pyridine 1-oxide (10g)
Light brown solid, yield 56%. 1H NMR (400 MHz, Chloroform-d) δ 9.60 (s, 1H), 8.68 (s, 1H), 8.60 (d, J = 9.4 Hz, 1H), 7.92 (d, J = 2.6 Hz, 1H), 6.99 (dd, J = 9.5, 2.5 Hz, 1H), 6.42 (s, 1H), 4.74 (p, J = 8.8 Hz, 1H), 3.74 (t, J = 5.2 Hz, 2H), 3.67–3.54 (m, 2H), 3.11 (s, 6H), 3.10–2.99 (m, 4H), 2.59–2.39 (m, 2H), 2.32 (t, J = 8.0 Hz, 2H), 2.09–1.93 (m, 4H), 1.75–1.62 (m, 2H), 1.59 (p, J = 7.8 Hz, 2H), 1.32 (h, J = 7.4 Hz, 2H), 0.89 (t, J = 7.3 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 171.72, 163.78, 153.18, 151.70, 151.60, 141.35, 139.84, 132.94, 126.80, 118.44, 113.64, 113.06, 100.71, 57.95, 49.55, 45.15, 35.14, 32.95, 30.16, 27.37, 24.57, 22.54, 13.88. ESI-HRMS m/z calcd for C28H39N8O3+ 535.3140, found 535.3135 [M + H]+. HPLC purity 98%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino)-5-(4-hexanoylpiperazin-1-yl) pyridine 1-oxide (10h)
Light brown solid, yield 73%. 1H NMR (400 MHz, Chloroform-d) δ 9.62 (s, 1H), 8.73 (s, 1H), 8.66 (d, J = 9.5 Hz, 1H), 7.96 (d, J = 2.6 Hz, 1H), 7.04 (dt, J = 9.5, 2.2 Hz, 1H), 6.47 (d, J = 1.0 Hz, 1H), 4.79 (t, J = 8.8 Hz, 1H), 3.79 (t, J = 5.1 Hz, 2H), 3.64 (t, J = 5.1 Hz, 2H), 3.15 (s, 6H), 3.09 (d, J = 5.3 Hz, 4H), 2.60–2.47 (m, 2 H), 2.41–2.31 (m, 2H), 2.14–1.98 (m, 4H), 1.79–1.60 (m, 4H), 1.45–1.28 (m, 4H), 0.91 (t, J = 6.7 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 171.77, 163.83, 153.19, 151.76, 151.58, 141.37, 139.92, 133.01, 126.79, 118.79, 113.73, 113.19, 100.72, 58.00, 49.64, 45.15, 35.19, 33.26, 31.65, 30.22, 25.00, 24.61, 22.49, 13.98. ESI-HRMS m/z calcd for C29H41N8O3+ 549.3296, found 549.3294 [M + H]+. HPLC purity 98%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo [2,3-d] pyrimidin-2-yl) amino)-5-(4-heptanoylpiperazin-1-yl) pyridine 1-oxide (10i)
Light brown solid, yield 68%. 1H NMR (400 MHz, Chloroform-d) δ 9.57 (s, 1H), 8.67 (s, 1H), 8.60 (d, J = 9.4 Hz, 1H), 7.93 (d, J = 2.6 Hz, 1H), 7.00 (dd, J = 9.4, 2.4 Hz, 1H), 6.41 (s, 1H), 4.74 (p, J = 8.9 Hz, 1H), 3.66 (dt, J = 54.0, 5.0 Hz, 4H), 3.09 (s, 6H), 3.04 (d, J = 5.5 Hz, 4H), 2.57–2.39 (m, 2H), 2.31 (t, J = 7.7 Hz, 2H), 2.08–1.91 (m, 4H), 1.74–1.50 (m, 4H), 1.33–1.15 (m, 6H), 0.81 (t, J = 6.7 Hz, 3H). 13C NMR (101 MHz, CDCl3) δ 171.71, 163.77, 153.18, 151.71, 151.57, 141.35, 139.80, 132.96, 126.78, 118.51, 113.65, 113.08, 100.69, 57.93, 49.53, 45.13, 35.12, 33.21, 31.56, 30.17, 29.08, 25.22, 24.57, 22.48, 14.00. ESI-HRMS m/z calcd for C30H43N8O3+ 563.3453, found 563.3453 [M + H]+. HPLC purity 99%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino)-5-(4-(2-(dimethylamino)-2-oxoacetyl) piperazin-1-yl)pyridine 1-oxide (10j)
Light brown solid, yield 76%. 1H NMR (400 MHz, Chloroform-d) δ 9.56 (s, 1H), 8.66 (s, 1H), 8.59 (d, J = 9.4 Hz, 1H), 7.89 (d, J = 2.5 Hz, 1H), 6.98 (dd, J = 9.4, 2.5 Hz, 1H), 6.40 (s, 1H), 4.79–4.64 (m, 1H), 3.75 (t, J = 5.0 Hz, 2H), 3.51 (t, J = 5.0 Hz, 2H), 3.10–3.06 (m, 10H), 2.97 (s, 3H), 2.95 (s, 3H), 2.57–2.38 (m, 2H), 2.04–1.92 (m, 4H), 1.72–1.60 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 164.22, 163.73, 163.33, 153.11, 151.67, 151.56, 141.12, 140.10, 133.02, 127.19, 118.71, 113.69, 113.04, 100.66, 57.92, 49.48, 45.61, 37.20, 35.12, 30.17, 24.56. ESI-HRMS m/z calcd for C27H36N9O4+ 550.2885, found 550.2884 [M + H]+. HPLC purity 97%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino)-5-(4-(3-methoxy-3-oxopropanoyl) piperazin-1-yl)pyridine 1-oxide (10k)
Light brown solid, yield 58%. 1H NMR (400 MHz, Chloroform-d) δ 9.62 (s, 1H), 8.71 (s, 1H), 8.63 (d, J = 9.4 Hz, 1H), 7.93 (d, J = 2.5 Hz, 1H), 7.01 (dd, J = 9.5, 2.5 Hz, 1H), 6.44 (s, 1H), 4.76 (t, J = 8.8 Hz, 1H), 3.79 (t, J = 5.1 Hz, 2H), 3.73 (s, 3H), 3.61 (t, J = 5.0 Hz, 2H), 3.51 (s, 2H), 3.16–3.05 (m, 10H), 2.58–2.46 (m, 2H), 2.13–2.00 (m, 4H), 1.78–1.61 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 167.83, 164.27, 163.80, 153.20, 151.72, 151.61, 141.18, 140.05, 132.96, 126.98, 118.46, 113.68, 113.06, 100.72, 57.97, 52.61, 49.45, 46.02, 41.51, 35.18, 30.19, 24.60. ESI-HRMS m/z calcd for C27H35N8O5+ 551.2725, found 551.2727 [M + H]+. HPLC purity 98%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino)-5-(4-(4-methoxy-4-oxobutanoyl) piperazin-1-yl)pyridine 1-oxide (10l)
Light brown solid, yield 66%. 1H NMR (400 MHz, Chloroform-d) δ 9.61 (s, 1H), 8.69 (s, 1H), 8.61 (d, J = 9.4 Hz, 1H), 7.91 (d, J = 2.5 Hz, 1H), 6.99 (dd, J = 9.5, 2.5 Hz, 1H), 6.43 (s, 1H), 4.84–4.65 (m, 1H), 3.75 (t, J = 5.1 Hz, 2H), 3.71–3.61 (m, 5H), 3.14–2.99 (m, 10H), 2.65 (s, 4H), 2.56–2.43 (m, 2H), 2.08–1.94 (m, 4H), 1.77–1.59 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 173.46, 169.72, 163.79, 153.22, 151.73, 151.61, 141.31, 139.96, 132.93, 126.85, 118.35, 113.65, 113.04, 100.70, 57.96, 51.81, 49.52, 44.87, 35.15, 30.18, 28.99, 27.83, 24.59. ESI-HRMS m/z calcd for C28H37N8O5+ 565.2881, found 565.2878 [M + H]+. HPLC purity 97%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino)-5-(4-(5-methoxy-5-oxopentanoyl) piperazin-1-yl)pyridine 1-oxide (10m)
Light brown solid, yield 73%. 1H NMR (400 MHz, Chloroform-d) δ 9.59 (s, 1H), 8.68 (s, 1H), 8.60 (d, J = 9.4 Hz, 1H), 7.90 (d, J = 2.5 Hz, 1H), 6.98 (dd, J = 9.4, 2.5 Hz, 1H), 6.42 (s, 1H), 4.74 (t, J = 8.8 Hz, 1H), 3.73 (t, J = 5.1 Hz, 2H), 3.62 (s, 5H), 3.10 (s, 6H), 3.05 (dt, J = 10.0, 5.0 Hz, 4H), 2.56–2.44 (m, 2H), 2.42–2.34 (m, 4H), 2.10–1.87 (m, 6H), 1.74–1.56 (m, 2H). 13C NMR (101 MHz, CDCl3) δ 173.68, 170.67, 163.77, 153.20, 151.71, 151.60, 141.32, 139.90, 132.93, 126.79, 118.33, 113.63, 113.04, 100.70, 57.94, 51.56, 49.51, 45.00, 35.13, 33.04, 32.10, 30.17, 24.57, 20.30. ESI-HRMS m/z calcd for C29H39N8O5+ 579.3038, found 579.3028 [M + H]+. HPLC purity 96%.
2-((7-cyclopentyl-6-(dimethylcarbamoyl)-7H-pyrrolo[2,3-d] pyrimidin-2-yl) amino)-5-(4-(6-methoxy-6-oxohexanoyl) piperazin-1-yl) pyridine 1-oxide (10n)
Light brown solid, yield 69%. 1H NMR (400 MHz, Chloroform-d) δ 9.60 (s, 1H), 8.70 (s, 1H), 8.62 (d, J = 9.4 Hz, 1H), 7.92 (d, J = 2.5 Hz, 1H), 7.00 (dd, J = 9.5, 2.5 Hz, 1H), 6.43 (s, 1H), 4.76 (t, J = 8.8 Hz, 1H), 3.74 (t, J = 5.1 Hz, 2H), 3.64–3.58 (m, 5H), 3.12 (s, 6H), 3.10–3.01 (m, 4H), 2.58–2.43 (m, 2H), 2.41–2.30 (m, 4H), 2.10–1.92 (m, 4H), 1.73–1.61 (m, 6H). 13C NMR (101 MHz, CDCl3) δ 173.78, 171.04, 163.80, 153.23, 151.75, 151.57, 141.33, 139.94, 132.98, 126.84, 118.41, 113.67, 113.08, 100.68, 57.96, 51.50, 49.55, 45.07, 35.14, 33.75, 32.77, 30.20, 29.64, 24.62, 24.58. ESI-HRMS m/z calcd for C30H41N8O5+ 593.3194, found 593.3201 [M + H] +. HPLC purity 98%.
References
Asghar U, Witkiewicz AK, Turner NC, Knudsen ES (2015) The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 14:130–146
Chen P, Lee NV, Hu W, Xu M, Ferre RA, Lam H, Bergqvist S, Solowiej J, Diehl W, He Y-A et al. (2016) Spectrum and degree of CDK drug interactions predicts clinical performance. Mol Cancer Ther 15:2273–2281
Delano WL (2014) The PyMOL molecular graphics system, version 1.7; Schrödinger LLC, New York
Dyson N (1998) The regulation of E2F by pRB-family proteins. Genes Dev 12:2245
Fathalla OAE-FM, Ismail MAH, Anwar MM, Abouzid KAM, Ramadan AAK (2013) Novel 2-thiopyrimidine derivatives as CDK2 inhibitors: molecular modeling, synthesis, and anti-tumor activity evaluation. Med Chem Res 22:659–673
Gao J, Fang C, Xiao Z, Huang L, Chen C-H, Wang L-T, Lee K-H (2015) Discovery of novel 5-fluoro-N2,N4-diphenylpyrimidine-2,4-diamines as potent inhibitors against CDK2 and CDK9. MedChemComm 6:444–454
Geffken D, Soliman R, FSG Soliman, Abdel-Khalek MM, DAE Issa (2011) Synthesis of new series of pyrazolo[4,3-d]pyrimidin-7-ones and pyrido[2,3-d]pyrimidin-4-ones for their bacterial and cyclin-dependent kinases (CDKs) inhibitory activities. Med Chem Res 20:408–420
Gonda M, Nieves M, Nunes E, Lopez de Cerain A, Monge A, Lavaggi ML, Gonzalez M, Cerecetto H (2013) Phenazine N,N[prime or minute]-dioxide scaffold as selective hypoxic cytotoxin pharmacophore. Structural modifications looking for further DNA topoisomerase II-inhibition activity. MedChemComm 4:595–607
Graf F, Mosch B, Koehler L, Bergmann R, Wuest F, Pietzsch J (2010) Cyclin-dependent kinase 4/6 (Cdk4/6) inhibitors: perspectives in cancer therapy and imaging. Mini Rev Med Chem 10:527–539
Hamilton E, Infante JR (2016) Targeting CDK4/6 in patients with cancer. Cancer Treat Rev 45:129–138
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Harbour JW, Dean DC (2000) The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 14:2393
Hunter T (1997) Oncoprotein networks. Cell 88:333–346
Kamal A, Reddy VS, Santosh K, Bharath Kumar G, Shaik AB, Mahesh R, Chourasiya SS, Sayeed IB, Kotamraju S (2014) Synthesis of imidazo[2,1-b][1,3,4]thiadiazole-chalcones as apoptosis inducing anticancer agents. MedChemComm 5:1718–1723
Kelly MD, Mancera RL (2006) Comparative analysis of the surface interaction properties of the binding sites of CDK2, CDK4, and ERK2. ChemMedChem 1:366–375
Lechner M, Lirk P, Rieder J (2005) Inducible nitric oxide synthase (iNOS) in tumor biology: the two sides of the same coin. Semin Cancer Biol 15:277–289
Li C-Q, Wogan GN (2005) Nitric oxide as a modulator of apoptosis. Cancer Lett 226:1–15
Lu H, Chang DJ, Baratte B, Meijer L, Schulze-Gahmen U (2005) Crystal structure of a human cyclin-dependent kinase 6 complex with a flavonol inhibitor, fisetin. J Med Chem 48:737–743
Malumbres M, Barbacid M (2001) Milestones in cell division: to cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer 1:222–231
Malumbres M, Barbacid M (2009) Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9:153
Morgan DO (1997) Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13:261–291
Ortega S, Malumbres M, Barbacid M (2002) Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta 1602:73
Peyressatre M, Prével C, Pellerano M, Morris MC (2015) Targeting cyclin-dependent kinases in human cancers: from small molecules to peptide inhibitors. Cancers 7:179–237
Roskoski Jr R (2016) Cyclin-dependent protein kinase inhibitors including palbociclib as anticancer drugs. Pharmacol Res 107:249–275
Sanchez-Martinez C, Gelbert LM, Lallena MJ, de Dios A (2015) Cyclin dependent kinase (CDK) inhibitors as anticancer drugs. Bioorg Med Chem Lett 25:3420–3435
Sarma B, Saikia B, Khatioda R, Bora P (2016) Pyridine N-oxides as coformers in the development of drug cocrystals CrystEngComm 18:8454–8464
Shapiro GI (2006) Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol 24:1770–1783
Sherr CJ (1996) Cancer cell cycles. Science 274:1672
Toogood PL, Harvey PJ, Repine JT, Sheehan DJ, VanderWel SN, Zhou H, Keller PR, McNamara DJ, Sherry D, Zhu T (2005) Discovery of a potent and selective inhibitor of cyclin-dependent kinase 4/6. J Med Chem 48:2388–2406
Acknowledgements
This work was supported by the Project of Science and Technology Assistance in Developing Countries (KY201501006) and the National Natural Science Foundation of China (81470354) and the Natural Science Foundation of Tianjin (17JCQNJC13500) and National key scientific research project (2013CB967201).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Guo, Q., Li, Y., Zhang, C. et al. Selective and novel cyclin-dependent kinases 4 inhibitor: synthesis and biological evaluation. Med Chem Res 27, 1666–1678 (2018). https://doi.org/10.1007/s00044-018-2180-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-018-2180-2