Abstract
Climate change prompts warm-tolerant species upward and poleward to either displace or replace cold-tolerant species. Warm-tolerant species may replace cold-tolerant individuals with upward migration, or cold-tolerant genes if the species hybridize. We examined genetic and morphological differences between low elevation, warm-tolerant (Aphaenogaster rudis) and high elevation cold-tolerant (A. picea) ant species that form an upward-shifting ecotone in the southern Appalachian Mountains (USA). The A. picea/A. rudis ecotone shifted upward ca. 200 m between the decades 1970 and 2010, and characteristic morphological traits appeared muddled where the species met, suggesting hybridization. However, we found no evidence of genetic hybridization, and the trait most associated with species identity, pigmentation, remained so across the environmental gradients. Conversely, femur length did not differentiate well between species identities, and it shifted across the environmental gradients. These results suggest that the cold tolerant A. picea, associated with high-elevation and high-latitude, was replaced by the warm-tolerant, low elevation A. rudis species. As such, these results suggest that less competitive cold-tolerant species may be replaced by more competitive cold-intolerant species with climate warming.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Species generally move upward and poleward with climate warming (Brommer 2004; Parmesan et al. 1999; Pelini et al. 2009; Zuckerberg et al. 2009). If all the species move in the same direction at the same rates, their ranges and interactions would simply shift in space without increasing overlap and interaction pressures. However, species move at different rates in response to climate change (Ibanez et al. 2008; Kelly and Goulden 2008; Miller-Rushing et al. 2008; Pelini et al. 2009; Perry et al. 2005; Schweiger et al. 2008; Zhu et al. 2013) so that less competitive (cold-tolerant) species may be replaced by more competitive (cold-intolerant) species with warming (Connell 1975; Menge and Sutherland 1987; Urban et al. 2012). Alternatively, closely related species may interbreed so that genes, rather than species are replaced, and hybrid zones move upward and poleward (Buggs 2007; Harr and Price 2014; Wolf et al. 2001).
Boreal forest communities blanketed the southern Appalachian Mountains during the most recent glacial maximum ~18,000 years ago (Delcourt and Delcourt 1987, 2000). As the Pleistocene ended, species adapted to cold climates migrated northward, and in montane environments, upward. Generally, species that can better tolerate stressful conditions (e.g., cold) are poor competitors (Chase and Leibold 2003; Tilman and Pascala 1993), and better competitors may replace cold-tolerant species with climate change (Urban et al. 2012). High elevation species not always are replaced, however, and in some cases form hybrid zones with low elevation congeners. For example, ecotones between closely related high- and low-elevation salamander species persist in the Southern Appalachian Mountains (USA), and, though their interactions along elevation gradients generally are competitive, they hybridize where distributions overlap (Bruce 2007; Hairston 1949, 1951; Hairston et al. 1992; Highton and Peabody 2000).
Ant complexes can be very cryptic with plastic morphology that obscures species differentiation across environmental gradients (e.g., darker colors with increased elevation) (MontBlanc et al. 2007; Seifert 2009). For example, in eastern North America, approximately 12 Aphaenogaster species are the most abundant arthropods in mesic deciduous forests (King et al. 2013), but eastern North America Aphaenogaster species are hard to differentiate based on morphology, including A. rudis and A. picea; hereafter ‘rudis complex’ (Lubertazzi 2012; Ness et al. 2009; Umphrey 1996). The eastern North America Aphaenogaster taxonomy remains unresolved, but, genetic, geographic and ecological data suggest discrete species with distinct, albeit overlapping, North America distributions (Creighton 1950; Crozier 1977; De Marco and Cognato 2016; Lubertazzi 2012; Umphrey 1996; Warren et al. 2011b).
Ant species, including Aphaenogaster, typically partition habitat (spatially and temporally) by temperature (Dunn et al. 2007; Parr and Gibb 2009; Sanders et al. 2007). In the southern Appalachian mountain region (USA), two Aphaenogaster species appear to segregate latitude and elevation by minimum temperatures (Warren et al. 2011a, b; Warren and Bradford 2013; Warren and Chick 2013). The northerly, high-elevation A. picea forages at temperatures ca. 6 °C below that of the southerly, low-elevation A. rudis (Warren et al. 2011a), and experimental studies indicate that A. picea’s physiological heat and cold tolerance is ca. 2 °C lower than A. rudis (Warren and Chick 2013). The ecotone between A. rudis and A. picea shifted upward and northward with rising minimum temperatures in the southern Appalachian mountain region 1974–2012 (Warren and Chick 2013). Ant congeners commonly hybridize (Cahan and Keller 2003; Julian et al. 2002; Shoemaker et al. 1996), and in measuring the A. rudis/A. picea ecotone across elevation gradients, Warren and Chick (2013) noted that the species were difficult to differentiate at mid-elevations based on morphological characters alone, and suggested that the two closely related species might hybridize. However, in investigating the same elevation gradients, Crozier (1977) found little chromosomal evidence of hybridization.
We used morphometric analysis of ants collected along an A. rudis/A. picea elevation ecotone in northern Georgia in 1974 (Crozier 1977) and 2014 (Warren and Chick 2013) to investigate whether the shift in species appeared consistent with replacement or hybridization. We also used ants collected ca. 75 km north and 75 km south of the elevation ecotone to capture morphology in non-overlapping A. rudis and A. picea colonies. Moreover, we used ants collected with the 2014 ecotone samples, along with ants collected throughout eastern North America (De Marco and Cognato 2016), for genetic analysis of A. rudis and A. picea. If replacement occurred, we would expect no genetic evidence of hybridization and a discrete ‘step’ in species traits with transition between cold-intolerant, low-elevation A. rudis and cold-tolerant, high-elevation A. picea. If hybridization occurred, we would expect evidence of genetic introgression and a continuous shift in traits across the species boundary. We also investigated links between morphological traits that potentially differentiate the Aphaenogaster species and their thermal tolerances.
Methods
Specimen collections
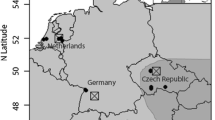
In 2011, A. rudis was collected at Whitehall forest in Athens, GA, and A. picea was collected at Coweeta Hydrologic Laboratory (CWT) in Otto, NC (for methodology and GPS locations see King et al. 2013) (Fig. 1). In 2012, A. rudis (low elevation) and A. picea (high elevation) ants were collected in northern Georgia (NGA) in Chattahoochee National Forest and Black Rock Mountain State Park (for methodology and GPS locations see Warren and Chick 2013). The ant samples were dried and stored frozen at −4 °C until 2013 when they were pinned and mounted for morphometric analysis. A total of 290 ant specimens were used for morphometric analyses: 163 specimens from the 2011 collections and 52 specimens from the 2012 collections. Voucher specimens are stored at the Georgia Museum of Natural History and at SUNY Buffalo State. Location data and morphometric measurements also were taken from 75 voucher specimens collected by Crozier (1977) and stored at the GMNH. An additional 52 specimens (S2) were used for phylogenetic analysis, six of which were collected at the same sites as those used for the morphometric analysis.
Site map of the main study region (not including greater sampling area for genetic analysis) in northern Georgia and western North Carolina (USA). Ant specimens analyzed in this study were collected in 1974 for a genetic gradient study (circle, Crozier 1977), in 2011 to examine social insects (diamond, King et al. 2013), and in 2012 for gradient (triangle, Warren and Chick 2013) and genetic (square, De Marco and Cognato 2016) studies. A small amount of ‘jitter’ was added to the symbols to reduce overplotting. For scale, the distance between Coweeta and Whitehall is 125 km
Morphometrics
We used three morphometric measurements commonly used for ants, and possibly distinctive between Aphaenogaster species (De Marco and Cognato 2016; Umphrey 1996): relative eye length (maximum eye length/maximum head width including outer edges of eyes × 100), maximum femur length (posterior view) and pigmentation. Moreover, these traits correspond with functional abilities as larger relative eye length corresponds with greater night foraging (Greiner et al. 2007), greater femur length corresponds with greater speed/foraging success (Pearce-Duvet et al. 2011) and darker color corresponds with cold tolerance (Kaspari et al. 2015). We digitally measured relative eye metrics and femur lengths using a Leica M125 stereomicroscope with DFC295 digital camera and Leica Application Suite V4. Two traits were measured: Given that A. rudis legs typically are lightly colored and reddish and A. picea legs darker black, and that A. rudis antennal segments are of uniform color whereas the last four segments of A. picea antennae are lighter (Ellison et al. 2012), color and antennae pigmentation indices were generated. Leg pigmentation was coded as 1 = light, 2 = medium, 3 = dark; antennae pigmentation was coded as 1 = segments uniform, 2 = last four segments somewhat lighter than rest, 3 = last four segments clearly lighter than rest. The leg and antennae data were added to create a pigmentation score. Two technicians not involved with Warren and Chick (2013) independently coded specimens.
Phylogenetic analysis
Field-collected Aphaenogaster specimens were sent using an overnight mail carrier to the A. J. Cook Arthropod Research Collection, Michigan State University, East Lansing, Michigan from northern Georgia (A. rudis) and western North Carolina (A. picea). Six of these specimens from NGA and CWT were included in a phylogenetic analysis using DNA data from five genes including the mitochondrial gene cytochrome oxidase subunit 1 (CO1), and nuclear genes: carbamoylphosphate synthase (CAD), elongation factor 1-alpha (EF1αF2), long-wavelength rhodopsin (LWR) and wingless (WG). These loci were included in a broad analysis of North American Aphaenogaster phylogeny (De Marco and Cognato 2016) that also included a larger number of specimens, and are phylogenetically informative for intraspecific and intergeneric relationships (Brady et al. 2006; Ward et al. 2010). All methods for DNA extraction, amplification and PCR (polymerase chain reaction) were followed using methods from De Marco and Cognato (2016). Primers used in PCR came from Brady et al. (2006) and Ward et al. (2010). After PCR, unincorporated deoxyribonucleotide triphosphates (dNTPs) and oligonucleotides were removed from PCR with Exo-SAP (http://www.usbweb.com/category.asp?cat=pcr&id=78200) and directly sequenced on an ABI 3700 automated sequencer using a BigDye (Applied Biosystems, Inc., Foster City, CA) fluorescent chemistry reaction. Both sense and anti-sense strands were sequenced for all individuals. This phylogeny was inferred with Bayesian analysis with Mr. Bayes via the CIPRES Gateway (Huelsenbeck and Ronquist 2001; Miller et al. 2010). Data were partitioned by gene and codon position (Castoe et al. 2004), with models of evolution applied independently to each partition (Nylander et al. 2004), with a best-fit GTR + I + G model, 20 million generations and a burn-in of 5,000,000 generations.
Samples of A. carolinensis and A. miamiana were included in the phylogeny due to an overlap of geographic locations and some morphological characters. Aphaenogaster carolinensis and A. miamiana can be found in North Carolina, along with A. rudis and A. picea. Aphaenogaster miamiana was previously known only from Florida (De Marco and Cognato 2016), but can be distinguished from the others based on heavier sculpturing.
Climate and thermal tolerance
Mean maximum and minimum temperatures for the period June 2011–June 2012 were calculated for each ant collection site (based on GPS coordinates) using PRISM climate data (http://www.prism.oregonstate.edu). For ant thermal tolerance, we used the minimum and maximum physiological temperature tolerances for A. rudis and A. picea ants collected in NGA in 2012 as part of Warren and Chick (2013). In that study, we transferred individuals to 16 mm glass test tubes that were plugged with cotton to reduce thermal refuges and placed the test tubes in an Ac-150-A40 refrigerated water bath (NesLab, ThermoScientific). One vial contained only a copper-constantan Type-T thermocouple (Model HH200A, Omega, Connecticut, USA) to monitor temperature fluctuation inside the test tubes and ensure an accurate temperature reading at which individuals reached their thermal limits. We measured thermal tolerance for 10 individuals (5 for CTmin, 5 for CTmax) from each colony at each site. We estimated thermal tolerance by determining the loss of righting response as the index for thermal tolerance using a ramping rate of 1 °C min−1. We calculated the mean tolerance temperature for each species at each site. See Warren and Chick (2013) for full methodology.
Data analysis
We examined variation among ant morphological traits (pigmentation, femur length and relative eye length), physiological traits (minimum and maximum temperature tolerance), elevation and temperature (minimum and maximum) with principal component analysis (PCA) using the “prcomp” method and “scale” option (standardizes all variables to unit length) in the “R” statistical package (R Development Core Team 2016). There was a large drop in variance explained after the first principal component, which explained 61 % of the variance, but the second principal component explained more variance (16 %) than one variable (12 %), so we examined the first two components, which explained 77 % of the variance. We used the minimum contribution if all variable loadings contributed equally (35 % here) to determine the most important loadings for each principle component.
We used elevation to represent environmental variation as it was the main gradient in the field studies and it correlated with maximum temperature, and ant maximum and minimum temperature tolerances (PCA axis 1). We examined morphological trait values by elevation (1974 and 2012 data) and by species—as determined by karyotype (Crozier 1977) and genotype (De Marco and Cognato 2016) using generalized linear models (GLM) assuming Poisson error distributions. The fitted models were over dispersed, so quasi Poisson error distributions were used. We examined the effect of elevation and species identity on relative eye length, femur length and pigmentation. Given that a shift in species occurred between low-elevation A. rudis and high-elevation A. picea at ca. 750 m elevation (Warren and Chick 2013), we included second-order terms to capture any nonlinear patterning. We considered coefficients with a p value <0.05 ‘significant’ and those with a p-value <0.10 ‘marginally significant’ (sensu Hurlbert and Lombardi 2009).
Results
Phylogenetic analysis
The phylogenetic analysis produced a mostly resolved Bayesian tree with strong support for the outgroups, including Camponotus pennsylvanicus, Formica glacialis, Stenamma diecki, Novomessor cockerelli, Veromessor julianus and Solenopsis invicta (S1). There also was strong support for the A. fulva and A. picea clades, however, A. picea was separated into two smaller clades. The sample from North Carolina was in the smaller clade. Aphaenogaster carolinensis and A. miamiana clades each showed strong support; however, they were within the less supported A. rudis clade. Aphaenogaster carolinensis and A. miamiana were distinguished from A. rudis due to a missing intron in the gene CAD.
Principal component analysis
Principal component analysis on ant morphological and physiological traits, elevation and temperature indicated that most variation occurred across elevation and temperature gradients and correlated well with femur length and temperature tolerance, but not pigmentation or relative eye length (Fig. 2). The most important variables for PC1, which explained most of the variance, were elevation, femur length, minimum and maximum temperature tolerance and maximum temperatures (S3). The most important variables for PC2 were pigmentation and relative eye length, suggesting that elevation and temperature did not explain trait variation well.
Principal component analysis of ant morphological traits [pigment, femur (femur length) and relative eye length], physiological traits (minimum and maximum temperature tolerance), elevation and temperature (minimum and maximum). Arrows pointing in the same direction indicate positive covariation and those pointing in opposite directions indicate negative covariation
Trait variation with elevation and species identity
Elevation correlated strongly with ant maximum (r = 0.92) and minimum (r = 0.97) temperature tolerances, and with maximum ambient temperature (r = 0.97), but poorly with minimum ambient temperature (r = 0.31). The ant pigment was significantly higher in A. picea than A. rudis (Fig. 3a), but did not change with elevation (species: coef. = −0.052, SE = 0.003, t-value = −13.996, p-value <0.001; elevation: coef. = 0.001, SE = 0.007, t-value = 0.515, p-value = 0.607). These results indicated that, although the ants became darker in leg color and last four antennal segments at higher elevations, these patterns were better explained by species identity than by geographic location (Fig. 4a). Relative eye length did not change with species identity or elevation (species: coef. = 0.006, SE = 0.014, t-value = 0.446, p-value = 0.656; elevation: coef. = −0.001, SE = −0.001, t-value = −0.526, p-value = 0.600) (Figs. 3b, 4b). Femur length was significantly longer in A. rudis than A. picea (Fig. 3c), and it decreased significantly with elevation (species: coef. = −0.024, SE = 0.015, t-value = −1.689, p-value = 0.092; elevation: coef. = −0.001, SE = 0.001, t-value = −5.406, p-value <0.001) (Fig. 4b). Variance partitioning indicated that elevation explained 54 % of the variation in femur length whereas species identity only explained 3 %. These results indicated that A. picea had slightly shorter femurs than A. rudis, but more importantly, femur length decreased with elevation.
Changes in Aphaenogaster ant pigmentation (a), the relative eye length (b) and femur length (c) along elevation gradients. The fitted line includes both species, and the legend indicates the residual contribution of A. picea and A. rudis. A small amount of ‘jitter’ was added to the residuals to reduce overplotting
Discussion
Upward movement of an ant ecotone in the southern Appalachian mountain region appears better explained by species replacement than hybridization. Pigmentation best distinguishes between A. picea and A. rudis, and ants get darker with higher elevation, but the change is better explained by species identity than the environmental gradient. Conversely, ant femur lengths get shorter with higher elevation, and the shift is better explained by phenotypic plasticity than species identity.
Crozier (1977) identified an ecotone in the Southern Appalachian Mountains where low elevation Aphaenogaster rudis shift to high elevation A. picea populations—species, he differentiated based on chromosome number and color. Warren and Chick (2013) re-sampled his gradient and found that the A. rudis/A. picea ecotone had shifted ca. 200 m upwards to ca. 750 m elevation due to an increase in minimum temperatures, but they noted that mid-elevation ant morphology appeared muddled between the species, suggesting hybridization. In the current study, we found no evidence of genetic hybridization, nor did Crozier (1977), and the morphological shift in pigmentation at ca. 750 m was consistent with species rather than gene replacement. However, color was highly plastic, relative eye length essentially was invariant, and femur length was more dependent on environment than species. Overall, we found Aphaenogaster spp. morphology highly plastic and somewhat indistinguishable at local scales, but clearly genetically distinct and morphologically differentiated at broader scales.
Ant species in the rudis complex are widespread in North America, hard to morphologically differentiate, and are distributed in a manner that appears linked with post-glacial climates (King et al. 2013; Lubertazzi 2012; Warren and Bradford 2013; Warren and Chick 2013; Warren et al. 2011b). Based on morphology, Creighton (1950) identified six species/subspecies in the rudis complex (A. miamiana, A. picea, A. rudis, A. fulva, A carolinensis and A. texana) and suggested that color was a poor trait for distinguishing between them. Crozier (1977) questioned whether there were more cryptic species hiding in the rudis complex, and used chromosomal and isozyme variation to distinguish light colored coastal plain (A. rudis) and dark colored montane (A. picea) species, but suggested that color differences were too slight to be useful. Umphrey (1996) examined morphometrics in the rudis complex based on species identified with cytogenetic and electrophoretic markers and proposed nine species (four previously undescribed) with geographic location and coloring included as identifying characteristics. Subsequent direct and indirect testing of temperature requirements among the species supported ecological differentiation among the six identified species, particularly A. rudis and A. picea (Warren et al. 2011a, b; Warren and Bradford 2013; Warren and Chick 2013).
Our genetic analyses suggest, as did Umphrey (1996), that additional cryptic species may be hidden within the six identified species. The two A. picea clades that have darker legs and the last four antennal segments lighter in color correspond to the DNA data that separates them from other samples in the A. rudis clade. Morphologically, there is a tremendous amount of variability within the A. rudis clade. We found that pigmentation best distinguished between A. picea and A. rudis as identified by haplo- and genotypes (Crozier 1977; De Marco and Cognato 2016), whereas relative eye length was not useful and femur length appeared more responsive to environment than species. Given that our genetic analysis was based on samples collected at a continental scale and morphological analysis based on samples collected at a regional scale, these results are not conclusive falsification of hybridization between A. picea and A. rudis. A more intensive genetic sampling across the ecotone/environmental gradient is required for greater insight into genetic differentiation at this scale. Moreover, based on the differences in thermal tolerance, traits and distributions, the assumption is that A. picea is a poor competitor that tolerates colder climes better than A. rudis; however, this assumption is not well tested.
The overlap in morphology between species may be explained by the linkage between the color, size and shape traits with physiological temperature tolerance; the strongest link possibly between darker pigmentation and higher elevation. Darker pigmentation (melanization) with higher elevation and latitude is a common inter- and intraspecific difference for many species as an adaptation for cold tolerance (Ellers and Boggs 2004; MontBlanc et al. 2007; Parkash and Munjal 1999; Robinson 2001). The blurring in pigmentation between A. rudis and A. picea at mid elevations may indicate some phenotypic plasticity in melanization, but the species generally remained distinct across the environmental gradient. Moreover, shorter femur length with higher elevation may reflect a plastic response to the higher temperatures at lower elevations so that longer legs are a heat ameliorating advantage.
Both the genetic work of Crozier (1977) and the results presented here show no evidence of hybridizing between sympatric A. rudis and A. picea populations. As such, the movement of A. rudis colonies upward appears to be replacing A. picea colonies. Whether structured by competition or habitat selection, or some combination of both, high-elevation ant species appear replaced by low-elevation congeners.
References
Brady SG, Schultz TR, Fisher BL, Ward PS (2006) Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc Natl Acad Sci USA 273:1643–1649
Brommer JE (2004) The range margins of northern birds shifts polewards. Ann Zool 41:391–397
Bruce RC (2007) Out of the frying pan into the fire: an ecological perspective on evolutionary reversal in life history in plethodontid salamanders (Amphibia: Plethodontidae). Evol Ecol 21:703–726
Buggs RJA (2007) Empirical study of hybrid zone movement. Heredity 99:301–312
Cahan SH, Keller L (2003) Complex hybrid origin of genetic caste determination in harvester ants. Nature 424:306–309
Castoe TA, Doan TM, Parkinson CL (2004) Data partitions and complex models in Bayesian analysis: the phylogeny of gymnophthalmid lizards. Syst Biol 53:448–469
Chase JM, Leibold MA (2003) Ecological niches: linking classical and contemporary approaches. University of Chicago, Chicago
Connell JH (1975) Some mechanisms producing structure in natural communities: a model and evidence from field experiments. In: Cody ML, Diamond JM (eds) Ecology and evolution of communities. Harvard University Press, Cambridge, pp 460–490
Creighton WS (1950) The ants of North America, vol 104. Bull Mus Comp Zool. The Cosmos Press, Inc., Cambridge
Crozier RH (1977) Genetic differentiation between populations of the ant Aphaenogaster ‘rudis’ in the southeastern United States. Genetica 47:17–36
De Marco B, Cognato AI (2016) A multiple gene phylogeny reveals polyphyly among eastern North American Aphaenogaster species (Hymenoptera: Formicidae). Zool Scrip. doi:10.1111/zsc.12168
Delcourt HR, Delcourt PA (1987) Long-term forest dynamics of the temperate zone. Springer, New York
Delcourt HR, Delcourt PA (2000) Eastern deciduous forests. In: Barbour MG, Billings WD (eds) North American terrestrial vegetation. Cambridge University Press, Cambridge
Dunn RR, Parker CR, Sanders NJ (2007) Temporal patterns of diversity: assessing the biotic and abiotic controls on ant assemblages. Biol J Linn Soc 91:191–201
Ellers J, Boggs CL (2004) Functional ecological implications of intraspecific differences in wing melanization in Colias butterflies. Biol J Linn Soc 82:79–87
Ellison AM, Gotelli NJ, Farnsworth EJ, Alpert GD (2012) A field guide to the ants of New England. Yale University Press, New Haven
Greiner B, Narendra A, Reid SF, Dacke M, Ribi WA, Zeil J (2007) Eye structure correlates with distinct foraging-bout timing in primitive ants. Curr Biol 17:R879–R880
Hairston NG (1949) The local distribution and ecology of the Plethodontid salamanders of the Southern Appalachians. Ecol Monogr 19:49–73
Hairston NG (1951) Interspecies competition and its probable influence upon the vertical distribution of Appalachian salamanders of the genus Plethodon. Ecology 32:266–274
Hairston NG, Wiley RH, Smith CK, Kneidel KA (1992) The dynamics of two hybrid zones in Appalachian salamanders of the genus Plethodon. Evolution 46:930–938
Harr B, Price T (2014) Climate change: a hybrid zone moves north. Curr Biol 24:230–232
Highton R, Peabody RB (2000) Geographic protein variation and speciation in salamanders of the Plethodon jordani and Plethodon glutinosus complexes in the southern Appalachian mountains with the description of four new species. In: Bruce RC, Jaeger RG, Houck LD (eds) The biology of Plethodontid Salamanders. Kluwer Academic/Plenum, New York
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Hurlbert SH, Lombardi CM (2009) Final collapse of the Newman-Pearson decision theoretic framework and the rise of the neoFisherian. Ann Zool 46:311–349
Ibanez I, Clark JS, Dietze MC (2008) Evaluating the sources of potential migrant species: implications under climate change. Ecol Appl 18:1664–1678
Julian GE, Fewell JH, Gadau J, Johnson RA, Larrabee D (2002) Genetic determination of the queen caste in an ant hybrid zone. Proc Natl Acad Sci USA 99:8157–8160
Kaspari M, Clay NA, Lucas J, Yanoviak SP, Kay A (2015) Thermal adaptation generates a diversity of thermal limits in a rainforest ant community. Glob Change Biol 21:1092–1102
Kelly AE, Goulden ML (2008) Rapid shifts in plant distribution with recent climate change. Proc Natl Acad Sci USA 105:11823–11826
King JR, Warren RJ II, Bradford MA (2013) Social insects dominate eastern US temperate hardwood forest macroinvertebrate communities in warmer regions. PLoS One 8:e75843
Lubertazzi D (2012) The biology and natural history of Aphaenogaster rudis. Psyche 2012:752815
Menge BA, Sutherland JP (1987) Community regulation—variation in disturbance, competition, and predation in relation to environmental-stress and recruitment. Am Nat 130:730–757
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogetic trees. In: Proceedings of the gateway computing environment workshop (GCE), New Orleans, LA, USA
Miller-Rushing AJ, Lloyd-Evans TL, Primack RB, Satzinger P (2008) Bird migration times, climate change, and changing population sizes. Glob Change Biol 14:1959–1972
MontBlanc EM, Chambers JC, Brussard PF (2007) Variation in ant populations with elevation, tree cover, and fire in a pinyon-juniper-dominated watershed. West N Am Nat 67:469–491
Ness JH, Morin DF, Giladi I (2009) Uncommon specialization in a mutualism between a temperate herbaceous plant guild and an ant: are Aphaenogaster ants keystone mutualists? Oikos 12:1793–1804
Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey J (2004) Bayesian phylogenetic analysis of combined data. Syst Biol 53:47–67
Parkash R, Munjal AK (1999) Phenotypic variability of thoracic pigmentation in Indian populations of Drosophila melanogaster. J Zool Syst Evol Res 37:133–140
Parmesan C et al (1999) Poleward shifts in geographical ranges of butterfly species associated with regional warming. Nature 399:579–583
Parr CL, Gibb H (2009) Competition and the role of dominant ants. In: Lach L, Parr CL, Abbott KL (eds) Ant ecology. Oxford University Press, Oxford, pp 77–96
Pearce-Duvet JMC, Elemans CPH, Feener DH Jr (2011) Walking the line: search behavior and foraging success. Behav Ecol 22:501–509
Pelini SL, Dzurisin JDK, Prior KM, Williams CM, Marsico TD, Sinclair BJ, Hellmann JJ (2009) Translocation experiments with butterflies reveal limits to enhancement of poleward populations under climate change. Proc Natl Acad Sci USA 106:11160–11165
Perry AL, Low PJ, Ellis JR, Reynolds JD (2005) Climate change and distributional shifts in marine fishes. Science 308:1912–1915
R Development Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Robinson CH (2001) Cold adaptation in Arctic and Antarctic fungi. New Phytol 151:341–353
Sanders NJ, Lessard JP, Fitzpatrick MC, Dunn RR (2007) Temperature, but not productivity or geometry, predicts elevational diversity gradients in ants across spatial grains. Glob Ecol Biogeogr 16:640–649
Schweiger O, Settele J, Kudrna O, Klotz S, Kuhn I (2008) Climate change can cause spatial mismatch of trophically interacting species. Ecology 89:3472–3479
Seifert B (2009) Cryptic species in ants (Hymentoptera: Formicidae) revisited: we need a change in the alpha-taxonomic approach. Myrmecol News 12:149–166
Shoemaker DD, Ross KG, Arnold ML (1996) Genetic structure and evolution of a fire ant hybrid zone. Evolution 50:1958–1976
Tilman D, Pascala S (1993) The maintenance of species richness in plant communities. In: Ricklefs RE, Schluter R (eds) Species diversity in ecological communities. University of Chicago Press, Chicago
Umphrey GJ (1996) Morphometric discrimination among sibling species in the fulva-rudis-texana complex of the ant genus Aphaenogaster (Hymenoptera: Formicidae). Can J Zool 74:528–559
Urban MC, Tewksbury JJ, Sheldon KS (2012) On a collision course: competition and dispersal differences create no-analogue communities and cause extinctions during climate change. Proc R Soc B Biol Sci 279:2072–2080
Ward PS, Brady SG, Fisher BL, Schultz TR (2010) Phylogeny and biogeography of dolichoderine ants: effects of data partitioning and relict taxa on historical inference. Syst Biol 59:342–362
Warren RJ II, Bradford MA (2013) Mutualism fails when climate response differs between interacting species. Glob Change Biol 20:466–474
Warren RJ II, Chick L (2013) Upward ant distribution shift corresponds with minimum, not maximum, temperature tolerance. Glob Change Biol 19:2082–2088
Warren RJ II, Bahn V, Bradford MA (2011a) Temperature cues phenological synchrony in ant-mediated seed dispersal. Glob Change Biol 17:2444–2454
Warren RJ II, McAfee P, Bahn V (2011b) Ecological differentiation among key plant mutualists from a cryptic ant guild. Insect Soc 58:505–512
Wolf DE, Takebayashi N, Rieseberg LH (2001) Predicting the risk of extinction through hybridization. Conserv Biol 15:1039–1053
Zhu K, Woodall CW, Ghosh S, Gelfand AE, Clark JS (2013) Dual impacts of climate change: forest migration and turnover through life history. Glob Change Biol 20:251–264
Zuckerberg B, Woods AM, Porter W (2009) Poleward shifts in breeding bird distributions in New York State. Glob Change Biol 15:1866–1883
Acknowledgments
Partial support for this research was provided by the SUNY Buffalo State Office of Undergraduate Research. Additional funding was provided to LDC through National Science Foundation Grant 1136703—Dimensions in Biodiversity: Collaborative Research: The climate cascade: functional and evolutionary consequences of climatic change on species, trait, and genetic diversity in a temperate ant community to PIs: Nate Sanders, Aaron Ellison, Nick Gotelli, Sara Helms Cahan, Bryan Ballif, and Rob Dunn. We also thank Highlands Biological Stations Director Jim Costa and staff for support; Victor Agraz, Deborah Jackson, Mary Schultz, Chris Broecker, Havish Deepnarain, Nicole Dexter, Jesse Helton, Jing Niu, Vidhyaben Patel, Zeph Pendleton, Rachel Power, Paula Reith, Morgan Spinelli for field help. We thank the Georgia Department of Natural Resources Nongame Conservation Section and the US Forest Service Chattahoochee-Oconee National Forest for permission to collect ants. We also thank Georgia Museum of Natural History Curator and Collections Manager Richard Hoebeke for loaning us Aphaenogaster specimens from R. H. Crozier’s collections.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Warren, R.J., Chick, L.D., DeMarco, B. et al. Climate-driven range shift prompts species replacement. Insect. Soc. 63, 593–601 (2016). https://doi.org/10.1007/s00040-016-0504-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00040-016-0504-0