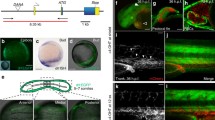
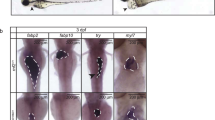
Abstract
Hox genes orchestrate the segmental specification of the muscular circulatory system in invertebrates but it has not proven straightforward to decipher segmental parallels in the vertebrate heart. Recently, patients with HOXB gene cluster deletion were found to exhibit abnormalities including atrioventricular canal defects. Using CRISPR, we established a mutant with the orthologous hoxbb cluster deletion in zebrafish. The mutant exhibited heart failure and atrioventricular regurgitation at 5 days. Analyzing the four genes in the hoxbb cluster, isolated deletion of hoxb1b−/− recapitulated the cardiac abnormalities, supporting hoxb1b as the causal gene. Both in situ and in vitro data indicated that hoxb1b regulates gata5 to inhibit hand2 expression and ultimately is required to pattern the vertebrate atrioventricular boundary. Together, these data reveal a role for segmental specification in vertebrate cardiac development and highlight the utility of CRISPR techniques for efficiently exploring the function of large structural genomic lesions.
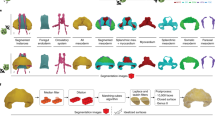
Graphic abstract








Similar content being viewed by others
Data availability
The data underlying this article will be shared on reasonable request to the corresponding authors.
References
Bruneau BG (2003) The developing heart and congenital heart defects: a make or break situation. Clin Genet 63:252–261. https://doi.org/10.1034/j.1399-0004.2003.00066.x
Bruneau BG (2008) The developmental genetics of congenital heart disease. Nature 451:943–948. https://doi.org/10.1038/nature06801
Hoffman JIE, Kaplan S (2002) The incidence of congenital heart disease. J Am Coll Cardiol 39:1890–1900. https://doi.org/10.1016/s0735-1097(02)01886-7
Abdul-Wajid S, Demarest BL, Yost HJ (2018) Loss of embryonic neural crest derived cardiomyocytes causes adult onset hypertrophic cardiomyopathy in zebrafish. Nat Commun 9:4603. https://doi.org/10.1038/s41467-018-07054-8
Cavanaugh AM, Huang J, Chen J-N (2015) Two developmentally distinct populations of neural crest cells contribute to the zebrafish heart. Dev Biol 404:103–112. https://doi.org/10.1016/j.ydbio.2015.06.002
de Pater E, Clijsters L, Marques SR et al (2009) Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 136:1633–1641. https://doi.org/10.1242/dev.030924
Hutson MR, Kirby ML (2003) Neural crest and cardiovascular development: a 20-year perspective. Birth Defects Res C Embryo Today 69:2–13. https://doi.org/10.1002/bdrc.10002
Buckingham M, Meilhac S, Zaffran S (2005) Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 6:826–835. https://doi.org/10.1038/nrg1710
Prummel KD, Nieuwenhuize S, Mosimann C (2020) The lateral plate mesoderm. Development. https://doi.org/10.1242/dev.175059
Tao Y, Schulz RA (2007) Heart development in Drosophila. Semin Cell Dev Biol 18:3–15. https://doi.org/10.1016/j.semcdb.2006.12.001
Ahmad SM (2017) Conserved signaling mechanisms in Drosophila heart development: signaling mechanisms in Drosophila cardiogenesis. Dev Dyn 246:641–656. https://doi.org/10.1002/dvdy.24530
Zaffran S, Kelly RG (2012) New developments in the second heart field. Differentiation 84:17–24. https://doi.org/10.1016/j.diff.2012.03.003
Pajusalu S, Reimand T, Uibo O et al (2015) De novo deletion of HOXB gene cluster in a patient with failure to thrive, developmental delay, gastroesophageal reflux and bronchiectasis. Eur J Med Genet 58:336–340. https://doi.org/10.1016/j.ejmg.2015.04.002
Rooryck C, Burgelin I, Stef M et al (2008) A 580kb microdeletion in 17q21.32 associated with mental retardation, microcephaly, cleft palate, and cardiac malformation. Eur J Med Genet 51:74–80. https://doi.org/10.1016/j.ejmg.2007.09.003
Medina-Martinez O, Bradley A, Ramirez-Solis R (2000) A large targeted deletion of Hoxbl-Hoxb9 produces a series of single-segment anterior homeotic transformations. Develop Biol 13:71–83
Soshnikova N, Dewaele R, Janvier P et al (2013) Duplications of hox gene clusters and the emergence of vertebrates. Dev Biol 378:194–199. https://doi.org/10.1016/j.ydbio.2013.03.004
Burke AC, Nelson CE, Morgan BA, Tabin C (1995) Hox genes and the evolution of vertebrate axial morphology. Development 121:333–346. https://doi.org/10.1242/dev.121.2.333
Krumlauf R (1994) Hox genes in vertebrate development. Cell 78:191–201. https://doi.org/10.1016/0092-8674(94)90290-9
Bertrand N, Roux M, Ryckebüsch L et al (2011) Hox genes define distinct progenitor sub-domains within the second heart field. Dev Biol 353:266–274. https://doi.org/10.1016/j.ydbio.2011.02.029
Diman NYS-G, Remacle S, Bertrand N et al (2011) A retinoic acid responsive Hoxa3 transgene expressed in embryonic pharyngeal endoderm, cardiac neural crest and a subdomain of the second heart field. PLoS One 6:e27624. https://doi.org/10.1371/journal.pone.0027624
Roux M, Laforest B, Capecchi M et al (2015) Hoxb1 regulates proliferation and differentiation of second heart field progenitors in pharyngeal mesoderm and genetically interacts with Hoxa1 during cardiac outflow tract development. Dev Biol 406:247–258. https://doi.org/10.1016/j.ydbio.2015.08.015
Alkan F, Wenzel A, Anthon C et al (2018) CRISPR-Cas9 off-targeting assessment with nucleic acid duplex energy parameters. Genome Biol 19:177. https://doi.org/10.1186/s13059-018-1534-x
Anthon C, Corsi GI, Gorodkin J (2022) CRISPRon/off: CRISPR/Cas9 on- and off-target gRNA design. Bioinformatics 38:5437–5439. https://doi.org/10.1093/bioinformatics/btac697
Pearson JC, Lemons D, McGinnis W (2005) Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6:893–904. https://doi.org/10.1038/nrg1726
McClintock JM, Kheirbek MA, Prince VE (2002) Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development 129:2339–2354. https://doi.org/10.1242/dev.129.10.2339
Moorman AFM, Christoffels VM (2003) Cardiac chamber formation: development, genes, and evolution. Physiol Rev 83:1223–1267. https://doi.org/10.1152/physrev.00006.2003
Perl E, Waxman JS (2019) Reiterative mechanisms of retinoic acid signaling during vertebrate heart development. JDB 7:11. https://doi.org/10.3390/jdb7020011
Holler KL, Hendershot TJ, Troy SE et al (2010) Targeted deletion of Hand2 in cardiac neural crest-derived cells influences cardiac gene expression and outflow tract development. Dev Biol 341:291–304. https://doi.org/10.1016/j.ydbio.2010.02.001
Morikawa Y, Cserjesi P (2008) Cardiac neural crest expression of hand2 regulates outflow and second heart field development. Circ Res 103:1422–1429. https://doi.org/10.1161/CIRCRESAHA.108.180083
Schindler YL, Garske KM, Wang J et al (2014) Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 141:3112–3122. https://doi.org/10.1242/dev.106336
Yelon D, Ticho B, Halpern ME et al (2000) The bHLH transcription factor Hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development 127:2573–2582. https://doi.org/10.1242/dev.127.12.2573
Machon O, Masek J, Machonova O et al (2015) Meis2 is essential for cranial and cardiac neural crest development. BMC Dev Biol 15:40. https://doi.org/10.1186/s12861-015-0093-6
Mosimann C (2015) Chamber identity programs drive early functional partitioning of the heart. Nat Commun 10:8146
Aanhaanen WTJ, Brons JF, Domínguez JN et al (2009) The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res 104:1267–1274. https://doi.org/10.1161/CIRCRESAHA.108.192450
Park JP, Moeschler JB, Berg SZ et al (1992) A unique de novo interstitial deletion del(17) (q21.3q23) in a phenotypically abnormal infant. Clin Genet 41:54–56. https://doi.org/10.1111/j.1399-0004.1992.tb03631.x
Rasmussen M, Vestergaard EM, Graakjaer J et al (2016) 17q12 deletion and duplication syndrome in Denmark-A clinical cohort of 38 patients and review of the literature. Am J Med Genet A 170:2934–2942. https://doi.org/10.1002/ajmg.a.37848
Foo JN, Lee J, Tan LC et al (2016) Large 3-Mb deletions at 22q11.2 locus in Parkinson’s disease and schizophrenia. Mov Disord 31:1924–1925. https://doi.org/10.1002/mds.26822
Jiang T, Zhang X-O, Weng Z, Xue W (2022) Deletion and replacement of long genomic sequences using prime editing. Nat Biotechnol 40:227–234. https://doi.org/10.1038/s41587-021-01026-y
Souidi A, Jagla K (2021) Drosophila heart as a model for cardiac development and diseases. Cells 10:3078. https://doi.org/10.3390/cells10113078
Jia G, Preussner J, Chen X et al (2018) Single cell RNA-seq and ATAC-seq analysis of cardiac progenitor cell transition states and lineage settlement. Nat Commun 9:4877. https://doi.org/10.1038/s41467-018-07307-6
Makki N, Capecchi MR (2010) Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev Biol 341:499–509. https://doi.org/10.1016/j.ydbio.2010.02.014
Holve S, Friedman B, Hoyme HE et al (2003) Athabascan brainstem dysgenesis syndrome. Am J Med Genet 120A:169–173. https://doi.org/10.1002/ajmg.a.20087
Makki N, Capecchi MR (2012) Cardiovascular defects in a mouse model of HOXA1 syndrome. Hum Mol Genet 21:26–31. https://doi.org/10.1093/hmg/ddr434
Tischfield MA, Bosley TM, Salih MAM et al (2005) Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet 37:3
Roux M, Laforest B, Eudes N et al (2017) Hoxa1 and Hoxb1 are required for pharyngeal arch artery development. Mechan Develop 8:1–8
Nemer G, Qureshi ST, Malo D, Nemer M (1999) Functional analysis and chromosomal mapping of Gata5, a gene encoding a zinc finger DNA-binding protein. Mamm Genome 10:993–999. https://doi.org/10.1007/s003359901146
Tremblay M, Sanchez-Ferras O, Bouchard M (2018) GATA transcription factors in development and disease. Development. https://doi.org/10.1242/dev.164384
Laforest B, Nemer M (2011) GATA5 interacts with GATA4 and GATA6 in outflow tract development. Dev Biol 358:368–378. https://doi.org/10.1016/j.ydbio.2011.07.037
Singh MK, Li Y, Li S et al (2010) Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. J Biol Chem 285:1765–1772. https://doi.org/10.1074/jbc.M109.038539
Srivastava D, Thomas T, Lin Q et al (1997) Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 16:154–160. https://doi.org/10.1038/ng0697-154
Zeisberg EM, Ma Q, Juraszek AL et al (2005) Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest 115:1522–1531. https://doi.org/10.1172/JCI23769
McFadden DG, Charité J, Richardson JA et al (2000) A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development 127:5331–5341. https://doi.org/10.1242/dev.127.24.5331
Ladam F, Sagerström CG (2014) Hox regulation of transcription: More complex(es): Hox Transcription Complexes. Dev Dyn 243:4–15. https://doi.org/10.1002/dvdy.23997
Paige SL, Thomas S, Stoick-Cooper CL et al (2012) A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151:221–232. https://doi.org/10.1016/j.cell.2012.08.027
Tong X, Zu Y, Li Z et al (2014) Kctd10 regulates heart morphogenesis by repressing the transcriptional activity of Tbx5a in zebrafish. Nat Commun 5:3153. https://doi.org/10.1038/ncomms4153
Asimaki A, Kapoor S, Plovie E et al (2014) Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci Transl Med 6:240ra74. https://doi.org/10.1126/scitranslmed.3008008
Shin JT, Pomerantsev EV, Mably JD, MacRae CA (2010) High-resolution cardiovascular function confirms functional orthology of myocardial contractility pathways in zebrafish. Physiol Genom 42:300–309. https://doi.org/10.1152/physiolgenomics.00206.2009
Zhang K, Yuan G, Werdich AA, Zhao Y (2020) Ibuprofen and diclofenac impair the cardiovascular development of zebrafish (Danio rerio) at low concentrations. Environ Pollut 258:113613. https://doi.org/10.1016/j.envpol.2019.113613
Han P, Bloomekatz J, Ren J et al (2016) Coordinating cardiomyocyte interactions to direct ventricular chamber morphogenesis. Nature 534:700–704. https://doi.org/10.1038/nature18310
Liu J, Bressan M, Hassel D et al (2010) A dual role for ErbB2 signaling in cardiac trabeculation. Development 137:3867–3875. https://doi.org/10.1242/dev.053736
Panáková D, Werdich AA, MacRae CA (2010) Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca2+ channel. Nature 466:874–878. https://doi.org/10.1038/nature09249
Becker JR, Deo RC, Werdich AA et al (2011) Human cardiomyopathy mutations induce myocyte hyperplasia and activate hypertrophic pathways during cardiogenesis in zebrafish. Dis Model Mech 4:400–410. https://doi.org/10.1242/dmm.006148
Wagner DE, Weinreb C, Collins ZM et al (2018) Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360:981–987. https://doi.org/10.1126/science.aar4362
Zhu Y, Zu Y (2023) Comprehensive bioinformatics analysis reveals PTPN1 (PTP1B) is a promising immunotherapy target associated with t cell function for liver cancer. J Healthcare Eng 2023:1533794. https://doi.org/10.1155/2023/1533794
Acknowledgements
We thank Dr. Bo Zhang (School of Life Sciences, Peking University) for providing the zCas9 plasmid, Dr. Jingwei Xiong (Institute of Molecular Medicine, Peking University) for providing the pUC19-scaffold plasmid for gRNA synthesis, Dr. Yi Zhou(Harvard Medical School) for the data discussion. We thank Jie Wang, Yumeng Rui, Xuebin Ye, Yihao Zhu, Xuhui Han, Wenhao Li, and Liang Jia for data analysis assistance.
Funding
This work was supported by the National Natural Science Foundation of China [No. 32170423 and 31501166 to Y.Z.], Shanghai Sailing Program of Science and Technology Commission of Shanghai Municipality [15YF1405000 to Y.Z.], Chenguang Program of the Shanghai Municipal Education Commission [14CG49 to Y.Z.], SciTech Funding by CSPFTZ Lingang Special Area Marine Biomedical Innovation Platform. Calum A. MacRae was funded by the NIH (OD017870).
Author information
Authors and Affiliations
Contributions
Conceptualization and design, Y.Z.; Investigation, P.H., Y.Z., B.W., D.J., Y.G., H.H., and X.M.; Formal analysis, Y.Z., P.H., B.W., D.J., Y.G., W.Z., D.Y.C. and C.A.M.; Writing, Y.Z., P.H., C.A.M. and B.W.; Resources, Y.Z. W. L. and C.A.M.; Funding acquisition, Y.Z. and C.A.M. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2 (MP4 4285 KB)
Supplementary file3 (MP4 15957 KB)
Supplementary file4 (MP4 15941 KB)
Supplementary file5 (MP4 55574 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, P., Wang, B., Jin, D. et al. Modeling of large-scale hoxbb cluster deletions in zebrafish uncovers a role for segmentation pathways in atrioventricular boundary specification. Cell. Mol. Life Sci. 80, 317 (2023). https://doi.org/10.1007/s00018-023-04933-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04933-2