Abstract
Background
Obesity affects the cargo packaging of the adipocyte-derived exosomes. Furthermore, adipocytes in different adipose tissues have different genetic makeup, the cargo contents of the exosomes derived from different adipose tissues under obesity conditions should be different, and hence their impacts on the pathophysiological conditions.
Methods and results
iTRAQ-based quantitative proteomics show that obesity has more prominent effects on the protein profiles of the exosomes derived from subcutaneous adipose tissue (SAT-Exos) in the high fat diet-induced obesity (DIO) mice than those derived from epididymal adipose tissue (EAT-Exos) and visceral adipose tissue (VAT-Exos). The differentially expressed proteins (DEPs) in SAT-Exos and VAT-Exos are mainly involved in metabolism. Subsequent untargeted metabolomic and lipidomics analyses reveal that injection of these SAT-Exos into the B6/J-Rab27a-Cas9-KO mice significantly affects the mouse metabolism such as fatty acid metabolism. Some of the DEPs in SAT-Exos are correlated with fatty acid metabolism including ADP-ribosylation factor and mitogen-activated protein kinase kinase kinase-3. Pathway analysis also shows that SAT-Exos affect adipocyte lipolysis and glycerophospholipid metabolism, which is in parallel with the enhanced plasma levels of fatty acids, diglycerides, monoglycerides and the changes in glycerophospholipid levels in DIO mice.
Conclusion
Our data provide scientific evidence to suggest SAT-Exos contribute to the changes in plasma lipid profiles under obesity conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Exosomes are nanometer-size, membrane-bound extracellular vesicles that mediate cell-to-cell communications locally and systemically by delivering different functional cargos such as proteins, lipids, long noncoding RNAs and microRNAs to the recipient cells and affect the cellular biological process [1, 2].
The contents of the exosomes that are derived from pre-adipocytes and mature adipocytes are different. Studies show that exosomes derived from preadipocyte have higher level of signaling fatty acids and adipogenesis markers when compared to those derived from mature adipocytes [3]. Besides, the cargo contents of the exosomes derived from mature adipocytes are also different between non-obese and obese subjects. Under obesity conditions, adipocytes have hypertrophic and hyperplastic growth that led to adipocyte dysfunction that dysregulates the assembly and sorting of the biological contents in the exosomes [4,5,6,7]. Hence, these exosomal contents may underlie the pathophysiological changes of the recipient targets under obesity conditions. Many studies have demonstrated that under obesity conditions, the exosomes derived from mature adipocytes have crucial roles in mediating obesity associated comorbidities such as type 2 diabetes, vascular diseases, liver steatosis, inflammation and cancers [8,9,10,11,12]. For example, in the ob/ob mice, the adipocyte-derived exosomes induce insulin resistance and increase pro-inflammatory cytokine secretion [6]. Besides, exosomes secreted from the adipocytes also increase the hypertrophic growth of the adipocytes in an autocrine manner by enhancing lipogenesis [13].
The formation of exosomes involves ESCRT-dependent and ESCRT-independent mechanisms [14]. Besides, tetraspanins and CD81 are involved in sorting target proteins into the exosomes [15, 16]. However, it is not clear whether the same mechanism is used to selectively load the exosomal cargo in different cell types. Nevertheless, it is generally believed that exosomal content resembles that of the donor cells. Indeed, exosomes have been suggested to be cancer biomarkers because the exosomal contents represent the tumor state [17]. The signature of the tumor-derived exosomes (TEXscore) has also been established to predict the overall survival of 12 cancer types [18]. We expect the exosomal content of the adipocyte-derived exosomes should have a high resemblance to the adipocyte. However, adipocytes in different adipose depots have different genetic backgrounds [19,20,21]. That is, the genes are of fat depot-specific expression patterns [21] and are differentially expressed in different adipocytes such as subcutaneous and visceral adipocytes. A study shows that the adipocytes in the visceral adipose tissues (VAT), subcutaneous adipose tissues (SAT) and epididymal adipose tissues (EAT) share only 1907 genes in common, with the genetic profile in SAT is distinct from those in VAT and EAT [20]. Many of these differentially expressed genes are correlated with various obesity-related traits [21]. Since the genetic makeup of the adipocytes in different adipose tissues are different, the exosomal contents in the exosomes that are derived from these adipose tissues and, hence, their biological and pathological impacts should also be different. Nevertheless, less studies have differentiated the biological and pathological effects of these exosomes under obesity conditions.
Exosomes carry a broad array of transmembrane proteins and soluble proteins [1]. The exosomal proteins constitute an essential part of the human adipose tissue secretome [7, 22]. A proteome analysis has identified 897 adipokines in the adipocyte-derived exosomes, which have strong association to human metabolic diseases [22]. Besides, many of these exosomal proteins are implicated in various signaling pathways, integrin signaling and membrane mediated processes [22]. Indeed, the majority of the proteins implicated in metabolism can be found in the adipocyte-derived exosomes [23].
In this study, we aimed to compare the impacts of obesity on the protein profiles of the exosomes that are derived from different adipose tissues. Then, we separately injected these exosomes into constitutive Rab27a knockout mice (B6/J-Rab27a-Cas9-KO) and compared the metabolic profiles of these mice with high fat diet-induced obesity mouse models. The data would suggest the differential contributions of these exosomes to the metabolic profiles and the pathophysiology of obesity. Besides, correlations of the differential exosomal proteins to the metabolic changes under obesity conditions would suggest potential novel targets for exosome-based therapy for obesity.
Methods and materials
Establishing diet-induced obesity (DIO) mouse models
Male C57BL/6 mice (4–5 weeks old) were purchased from the Laboratory Animal Services Centre, Chinese University of Hong Kong. Mice were housed in ventilated cages in the animal room at the Hong Kong Baptist University, with a 12 h light–dark cycle. The animal studies were approved by the Research Ethics Committee at the Hong Kong Baptist University and the Department of Health in the Hong Kong Special Administration Region. All the animal experiments were conducted according to the ethical standards and national guidelines. The mice were randomly selected to have either control diet (D12450J Research Diets), or high fat diet (D12762 Research Diets) for 8 weeks to induce obesity. Both diet and water were supplied ad libitum. Body weight of the mouse was recorded every week.
Hematoxylin and Eosin (H&E) staining of the adipose tissues
Adipose tissues were cut into pieces and then fixed and embedded in paraffin. The embedded tissues were sectioned into 5 μm thick sections using microtome. Before preforming H&E staining (ThermoFisher Scientific), de-paraffinization and re-hydration of the sections were done. The stained sections were imaged using bright-field microscopy [24].
Extraction of adipose tissue derived exosomes from DIO mouse models
After dietary intervention, DIO mice were scarified, and adipose tissues were dissected. Visceral adipose tissues (VAT) were the perirenal fat dissected from the fat pad surrounding the kidneys. Subcutaneous adipose tissues (SAT) were the bilateral superficial subcutaneous white adipose deposits between the skin and muscle fascia just anterior to the lower segment of the hind limbs. Epididymal adipose tissues (EAT) were dissected from the fat pad over the epididymis. These adipose tissues were separately cultured. The culture medium was Dulbecco's modified Eagles medium (ThermoFisher Scientific) supplemented with 50 μg/ml gentamicin and 10% FBS with exosome-depleted fetal bovine serum (ThermoFisher Scientific). The culture was kept in a 37 °C incubator in an atmosphere of 5% CO2 and 95% air [6]. The adipose tissue derived exosomes were then purified form the conditioned medium with differential ultracentrifugation method [6, 25] in which the conditioned medium was first centrifuged at 300g for 10 min; then, the supernatant was centrifuged at 2,000 g for 10 min, followed by another centrifugation at 10,000g for 30 min. The clear supernatant was then centrifugated at 100,000g for 70 min. The exosomes were washed by PBS before another centrifugation at 100,000g for 70 min. Transmission electron microscopy (TEM), dynamic light scattering (DLS) and expressions of exosomal markers were done to confirm the authenticity of the purified exosomes.
Transmission electron microscopy (TEM)
The exosomes were prepared for the TEM [26]. Briefly, exosomes in the conditioned medium were isolated as described above and were mixed with 2.5% glutaraldehyde (Sigma-Aldrich) in 0.1 M sodium cacodylate solution (pH 7.0) (Sigma-Aldrich) for 1 h at 4 °C. Sodium cacodylate (0.1 M) was prepared and was adjusted to pH 7.4. The exosomes were rinsed with the sodium cacodylate (Sigma-Aldrich) and was post-fixed with osmium tetroxide (1%) (Sigma-Aldrich) for 1 h at 4 °C before incubating with graded acetone on shaker. Then, the exosomes were subsequently incubated with solutions of different ratios of acetone and low viscosity embedding mixture. Exosomes were then double stained with uranyl acetate (2%) (Sigma-Aldrich) and lead citrate (2.5%) (Sigma-Aldrich) before being observed under TEM (Philips CM100 Transmission Electron Microscope).
Dynamic light scattering (DLS)
The extracted exosomes were diluted to 0.1 μg/μl using Dulbecco's phosphate-buffered saline and then transferred into a cuvette. The measurement was started after the laser and temperature equilibrium of the Photon Correlation Spectroscopy (PHOTOCOR) was stabilized. Temperature control, refractive index, scattering angle and laser wavelength were standardized. Exosomes were identified by their diameters.
Western blot analysis
Proteins were extracted in lysis buffer containing 150 mM sodium chloride, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 8.0), protease and phosphatase inhibitors (Bio-Rad), incubated on ice for 30 min before centrifugation. Quantification of protein was performed using the Pierce™ BCA Protocol Assay kit (ThermoFisher Scientific). Equal amount of the protein samples (10 µg) was denatured before being separated on 10% sodium dodecyl sulfate–polyacrylamide (SDS-PAGE) gel and transferred onto polyvinylidene difluoride (PVDF) membranes. The transferred proteins were incubated overnight at 4 °C with the respective primary antibody (Cell Signaling and Santa Cruz). Immunodetection was accomplished using horseradish peroxidase-conjugated secondary antibody (Bio-Rad), followed by enhanced chemiluminescence (ECL) detection system (Amersham).
iTRAQ-based quantitative proteomics
Proteins were extracted from the exosomes, and proteolysis and peptide labeling were then performed. The protein samples were dissolved in 0.5 M TEAB and added to the corresponding iTRAQ labeling reagent. For each sample, 100 µg of protein was taken for the iTRAQ peptide labeling, and 20 µg of the labeled peptides will be sent for fractionation for the proteomics analysis. Different sample peptides had different iTRAQ labeling. For peptide fractionation, Shimadzu LC-20AB liquid-phase system was used [27]. A 5 µm 4.6 × 250 mm Gemini C18 column for liquid-phase separation of the samples was used. The samples were reconstituted with mobile phase A (5% ACN pH 9.8), eluted at 1 mL/min flow rate with mobile phase B (95% ACN, pH 9.8). The peptides separated by liquid-phase chromatography were passed to a tandem mass spectrometer Q Exactive HF-X MS (ThermoFisher Scientific) for data dependent acquisition (DDA) mode detection. The protein databases NCBInr compiled by the National Center for Biotechnology Information (NCBI) were used for Blast searches. Protein quantification was done with software IQuant integrated with Mascot Percolator to quantitatively analyze the labeled peptides with isobaric tags and provide significance measures. The peptidespectrum matches (PSMs) were pre-filtered at PSM-level false discovery rate (FDR) of 1%. The pathway enrichment analysis was performed with differentially expressed proteins with p value less than 0.05.
LC–MS/MS-based untargeted metabolomics
Mouse serum metabolites were extracted by methanol and acetonitrile (2:1, v/v), and internal standards were used. The metabolites were resuspended in 50% methanol. After centrifugation, supernatants were subjected to LC–MS analysis with Waters 2D UPLC coupled to Q-Exactive mass spectrometer with a heated electrospray ionization source and controlled by the Xcalibur 2.3 software program (ThermoFisher Scientific). Chromatographic separation was performed on a Waters ACQUITY UPLC BEH C18 column. In the positive mode, mobile phase A was 0.1% formic acid and mobile phase B was acetonitrile. In negative mode, mobile phase A was 10 mM ammonium formate and mobile phase B was acetonitrile. The data quality was evaluated by the repeatability of QC sample detection. The data were processed by Compound Discoverer 3.1 (ThermoFisher Scientific) and R package metaX software. Compounds with coefficient of variation (CV) of the relative peak area greater than 30% were deleted. For univariate analysis, data were subjected for fold change analysis and T test and p value was corrected for the FDR to obtain q value. Probabilistic quotient normalization (PQN) was used for sample normalization. Local polynomial regression fitting signal correction (QC-RLSC) was used as an effective method for data correction in metabolomics area data analysis.
LC–MS/MS-based untargeted lipidomics
The nontargeted LC–MS/MS lipidomics analysis was performed using high-resolution mass spectrometer Q Exactive (ThermoFisher Scientific) for data acquisition in both positive ion and negative ion mode. SPLASH Internal standards (SPLASHTM Lipidomix Mass Spec Standard, Avanti Polar Lipids) were used. To each sample, isopropanol and internal standard were added and subjected to centrifugation. Equal volume of each sample was mixed into a QC sample for the evaluation of the repeatability and stability of the LC–MS analysis. CSH C18 column (1.7 μm 2.1 × 100 mm, Waters) was used. The mobile phase consisted of solvent A (60% acetonitrile aqueous solution, 0.1% formic acid, 10 mM ammonium formate) and solvent B (10% acetonitrile aqueous solution, 90% Isopropanol, 0.1% formic acid, 10 mM ammonium formate) under positive ion mode, and solvent A (60% acetonitrile aqueous solution, 10 mM ammonium formate) and solvent B (10% acetonitrile aqueous solution, 90% Isopropanol, 10 mM ammonium formate) under negative ion mode. Q Exactive mass spectrometer (ThermoFisher Scientific) was used to obtain MS1 and MS2 data. The MS scan method was in the range of m/z 200–2000. The parameters of ESI were sheath gas of 40 L/min, aux gas of 10 L/min, spray voltage 3.80 in positive ion mode and 3.20 in negative ion mode, capillary temperature of 320 °C and aux gas heater temperature of 350 °C. Lipid identification was performed by Software Thermo Scientific LipidSearch v4.1, and data were transferred to metaX for data preprocessing and subsequent analysis.
Principal component analysis (PCA), partial least squares method-discriminant analysis (PLS-DA), and variable importance in projection (VIP) values were employed in the data analysis. For the univariate analysis, fold change analysis, T test and false discovery rate (FDR) correction were performed to obtain the fold change, p value and q value, respectively. The results were screened for differential lipid molecules with criteria set at VIP of the first two principal components of the PLS-DA model equaled or above 1, fold change equaled or above 1.2 or less than 0.83 and the p value less than 0.05.
Joint-pathway analysis for proteomics and metabolic data
To examine the correlation between the differentially expressed proteins (DEPs) and the alternations in metabolites, we performed joint pathway analysis using MetaboAnalyst 3.0, which enabled the visualization of significant genes and metabolites that were enriched in a particular pathway. Functional annotation databases were utilized based on the gene ontology (GO) networks.
Establish constitutive Rab27a knockout mice (B6/J-Rab27a-Cas9-KO)
Constitutive Rab27a knockout mice (B6/J-Rab27a-Cas9-KO) were obtained by CRISPR/Cas9 technology (GemPharmatech). CRISPR-Cas9 system was used to perform the genome editing in the mice by delivering guide RNA to the designated genomic site and knockout Rab27a expression [28, 29]. Knockout of Rab27a in the mouse was confirmed by genotyping. The primers sequences for genotyping PCR study were JS05042-Rab27a-5wt-t, forward (F1) 5′-AGGCTACACGTACTGTTTCAAGGG-3′ and reverse (R1) 5′-AAACCAACAGTGTCAGCCATGTGTC-3′, and JS15042-Rab27a-wt-t, forward (F2) 5′-AGCTTTGTGGATGCTAAGCAAGAC-3′ and reverse (R2) 5′-TCATCTTACCAGGTGCAGATGAG-3′. Western blot analysis was also used to validate the knockout of Rab27a protein in various adipose tissues and organs.
Injection of exosomes to B6/J-Rab27a-Cas9-KO mice
The concentration levels of the adipose derived exosomes, namely those exosomes released from VAT (VAT-Exos), SAT (SAT-Exos) and EAT (EAT-Exos) in healthy and obese mouse models, have not been reported. With reference to the other published studies that investigate the roles of adipocyte-derived exosomes in mediating obesity comorbidities [6, 30], we injected 80 µg of the VAT-Exos, SAT-Exos or EAT-Exos dissected from the DIO mice into the B6/J-Rab27a-Cas9-KO mice via tail vein twice a week for 2 weeks. The mice were fed with control diet during the experiment. Equal volume of PBS served as vehicle control.
Statistical analysis
Results were shown as mean ± SD. Data were analyzed by one-way ANOVA followed by the Dunnett's multiple comparisons. Statistical analyses were carried out using GraphPad Prism (GraphPad Software) version 7.0. *p < 0.05 was considered statistically significant.
Results
Obesity has differential impacts on the protein profiles of the EAT-Exos, VAT-Exos and SAT-Exos
We used C57BL/6J mice to set up HFD-induced obesity (DIO) mouse model that had a significant difference in body weight when compared to control diet (CD) mice (supplementary Fig. 1a). The mass of subcutaneous adipose tissue (SAT), visceral adipose tissue (VAT), epididymal adipose tissue (EAT) (supplementary Fig. 1b), and the size of these adipocytes in the DIO mice were significantly larger than those in CD mice (supplementary Fig. 1c), suggesting that the adipocytes in DIO mice are under hypertrophic growth.
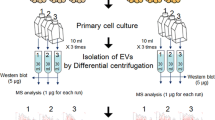
Exosomes derived from epididymal adipose tissue (EAT-Exos), visceral adipose tissue (VAT-Exos) and subcutaneous adipose tissue (SAT-Exos) were purified (Fig. 1a). Transmission electron microscopy (TEM) indicated the exosomes were intact and were morphologically “cup-shape” (Fig. 1b) [31]. TEM (Fig. 1b) and dynamic light scattering (DLS) (Fig. 1c) showed that the diameters of the purified exosomes were consistent with the proposed sizes of exosomes (30–150 nm) [1]. Authenticity of the purified exosomes was also examined by the expressions of exosomal markers CD63 and CD81 (Fig. 1d). The International Society for Extracellular Vesicles has recommended some negative controls for exosomes including calnexin, histone, cytochrome C, Grp94, GM130, and argonaut [32]. We also tried to detect histone and cytochrome c in the purified exosomes. As shown in Fig. 1e, histone and cytochrome c were detected in the adipose tissues but not in the purified exosomes, suggesting that only exosomes are purified in the preparation. To ascertain the exosomes were derived from adipocytes, we examined the purified exosomes with the markers that are specific for adipocyte-derived exosomes, namely adiponectin and fatty acid binding protein 4 (FABP-4) [33] as shown in Fig. 1f. Adipocyte-derived exosomes also have inflammatory adipokines such as macrophage migration inhibitory factor (MIF), monocyte chemoattractant protein-1 (MCP-1) and interleukin 6 (IL-6) [25]. Therefore, we also examined these markers in our purified exosomes (Fig. 1f). The presence of adiponectin, FABP-4, MIF, MCP-1 and IL-6 suggest that the purified exosomes are derived from adipocytes. Besides, we examined whether the adipocyte-derived exosomes would enter the blood circulation. We purified the circulating exosomes from the mice and tested for the adipocyte-specific exosomal markers. As shown in supplementary Fig. 1d, adiponectin and FABP-4 positive exosomes were detected in the circulation, suggesting that the adipocyte-derived exosomes could enter the blood circulation.
Obesity has differential impacts on the protein profiles of the EAT-Exos, VAT-Exos and SAT-Exos. a A flow diagram illustrates the exosome extraction procedures. b TEM and c DLS analysis of the purified exosomes. d Expressions of CD63 and CD81, f adiponectin, FABP4, MIF, MCP-1, IL6 and e absence of histone and cytochrome c expression in the purified exosomes. Volcano plots showing the upregulated and downregulated proteins in the g EAT-Exos, h VAT-Exos and i SAT-Exos under obesity conditions; and the j pathway analysis of the detected exosomal proteins. FABP4 fatty acid binding protein 4, MCP-1 monocyte chemoattractant protein-1, MIF macrophage migration inhibitory factor, EAT-Exos exosomes derived from epididymal adipose tissues, VAT-Exos exosomes derived from visceral adipose tissues, SAT-Exos exosomes derived from subcutaneous adipose tissues of the high fat diet-induced (DIO) mice. n = 3–4 mice in each group
iTRAQ-based quantitative proteomics were used to examine the protein profiles of the EAT-Exos, VAT-Exos and SAT-Exos under obesity conditions. Data showed that a total of 304 proteins were upregulated and 186 were downregulated in EAT-Exos (Fig. 1g), while 306 exosomal proteins were upregulated, and 273 were downregulated in VAT-Exos (Fig. 1h). Interestingly, HFD feeding had a great impact on the protein profiles in the SAT-Exos as 578 proteins were upregulated, and 310 proteins were downregulated (Fig. 1i). Besides, we also compared the exosomal protein profiles in SAT-Exos, VAT-Exos and EAT-Exos under CD and HFD conditions as shown in the supplementary Fig. 2a–f. With eukaryotic orthologous groups (KOG) annotation, we found that the detected proteins in these exosomes were mainly involved in cellular process and participate in binding activity (supplementary Fig. 2g). Among different cellular processes, 13.46% of the exosomal proteins was involved in metabolic process (Table 1) including carbohydrate metabolism, amino acid metabolism and lipid metabolism (Fig. 1j).
Obesity affects the metabolic-associated proteins in SAT-Exos and VAT-Exos
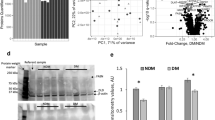
We next compared the impacts of obesity on the protein profiles of EAT-Exos, VAT-Exos and SAT-Exos. Proteins with 1.2-fold change and q value less than 0.05 were determined as differentially expressed proteins (DEPs). Most of the DEPs in the EAT-Exos, VAT-Exos and SAT-Exos were cytosolic proteins (supplementary Fig. 3a to 3c). Although the pathway enrichment analysis showed that DEPs in EAT-Exos, VAT-Exos and SAT-Exos were mainly involved in metabolism (Fig. 2a–c), only 239 and 375 metabolic-associated DEPs were identified in EAT-Exos (supplementary Fig. 3d) and VAT-Exos (supplementary Fig. 3e), respectively, while 544 metabolic-associated DEPs were identified in SAT-Exos (supplementary Fig. 3f). Scatter plot of the top 20 of KEGG enrichment RichFactor also revealed that DEPs in EAT-Exos were associated to not only metabolism but also other biological processes. As shown in Fig. 2d and supplementary Fig. 3 g, DEPs in EAT-Exos were involved in carbon metabolism, endocytosis, bacterial invasion and cortisol synthesis and secretion, endocrine and other factor-regulated calcium reabsorption, peroxisome and fatty acid metabolism. DEPs in VAT-Exos were mainly involved in complement and coagulation cascades, pathogenic bacterial infection and metabolism (Fig. 2e and supplementary Fig. 3h). Interestingly, DEPs in SAT-Exos were mostly involved in metabolism with a few DEPs involved in proteasome (Fig. 2f and supplementary Fig. 3i).
Obesity affects the metabolic-associated proteins in SAT-Exos and VAT-Exos. Pathway enrichment analysis of the differentially expressed proteins (DEPs) in the a EAT-Exos, b VAT-Exos and c SAT-Exos under obesity conditions. Statistics of pathway enrichment of DEPs in d EAT-Exos, e VAT-Exos and f SAT-Exos under obesity conditions. g Representative western blot showing the expressions of ARF1 and MAP3K proteins in the SAT-Exos, VAT-Exos and EAT-Exos, and the relative protein quantifications. ARF-1 ADP-ribosylation factor 1, MAP3K mitogen-activated protein kinase kinase kinase-3, EAT-Exos exosomes derived from epididymal adipose tissues, VAT-Exos exosomes derived from visceral adipose tissues, SAT-Exos exosomes derived from subcutaneous adipose tissues. CD control diet mice, DIO high fat diet-induced obesity mice. CS, CD mice SAT-Exos; HS, DIO mice SAT-Exos; CV, CD mice VAT-Exos; HV, DIO mice VAT-Exos; CE, CD mice EAT-Exos; HE, DIO mice EAT-Exos. n = 3–4 mice in each group
Furthermore, we performed STRING to predict the protein–protein interaction (PPI) of the DEPs, which includes both physical and functional associations. Interestingly, PPI of the DEPs in SAT-Exos predicted that the upregulated DEPs mainly interacted with other DEPs that were upregulated, and downregulated DEPs with other DEPs that were downregulated, the protein–protein interaction was different from those in EAT-Exos and VAT-Exos (supplementary Fig. 3j–l). In this specific case, this observed phenomenon is likely to be a kind of synergism between proteins up or down regulated that may lead to a synergistic effect, which, in turn, may lead to exacerbation of pathological effects under obesity conditions. Supplementary Table 1a–c separately highlighted the DEPs in EAT-Exos, VAT-Exos and SAT-Exos that were increased by at least twofold or reduced by at least 0.3-fold. The results clearly showed that obesity had differential effects on the exosomal proteins in these exosomes. Some highlighted DEPs were also validated. For example, ADP-ribosylation factor 1 (ARF1) was significantly increased in the SAT-Exos and VAT-Exos, while mitogen-activated protein kinase kinase kinase 3 (MAP3K, also known as MEKK3 or MAPKKK3) expression was significantly increased in the SAT-Exos, VAT-Exos and EAT-Exos (Fig. 2g), suggesting that ARF1 and MAP3K may play a role in mediating the effects of the exosomes under obesity conditions.
Our proteomic data strongly suggest that obesity has a more prominent effect on the proteins in the SAT-Exos than those in EAT-Exos and VAT-Exos. The DEPs in the EAT-Exos are involved in different biological functions, while those in SAT-Exos and VAT-Exos are mainly involved in metabolism.
Untargeted metabolomics analysis reveals a significant impact of SAT-Exos on the serum metabolite profile in constitutive Rab27a knockout mouse model
Since the DEPs in SAT-Exos and VAT-Exos under obesity conditions are mainly related to metabolism, we next examined whether these exosomes would affect the metabolic profiles in the mice. To eliminate the effects of endogenous exosomes on the mouse metabolism, we used constitutive Rab27a knockout mouse (B6/J-Rab27a-Cas9-KO) as model. Rab27a is critical for exosome secretion [28]. More importantly, Rab27a-KO mice are shown to have reduced exosome secretion [34]. We first preformed genotyping to validate the knockout of Rab27a in the B6/J-Rab27a-Cas9-KO mice. As shown in Fig. 3a, only homozygous knockout mice were selected for the study. Knockout of Rab27a protein in adipose tissues, heart, liver, spleen, lung, kidney and brain was confirmed by Western blot analysis (Fig. 3b). These data clearly demonstrated that Rab27a, the critical protein for exosome secretion, was successfully knockout in the B6/J-Rab27a-Cas9-KO mice.
Untargeted metabolomics analysis reveals the impact of SAT-Exos on the serum metabolite profile in constitutive Rab27a knockout mouse model. a Genotyping of the male Rab27a knockout mice (B6/J-Rab27a-Cas9-KO) and b expressions of Rab27a protein in the heart, liver, spleen, lung, kidney, brain, subcutaneous adipose tissue (SAT) and epididymal adipose tissue (EAT) in the homozygous knockout mice. Volcano map of the differential metabolites in c EAT-Exos, d VAT-Exos and e SAT-Exos (upper panel, positive ion mode; lower panel, negative ion mode) under obesity conditions. f Correlation analysis of the differential metabolites in the B6/J-Rab27a-Cas9-KO mice after SAT-Exos injection and the differentially expressed proteins in the SAT-Exos derived from high fat diet-induced (DIO) mice. EAT-Exos, exosomes derived from epididymal adipose tissues; VAT-Exos, exosomes derived from visceral adipose tissues; SAT-Exos, exosomes derived from subcutaneous adipose tissues. n = 3–4 mice in each group
Then, we separately injected equal amount of EAT-Exos, VAT-Exos and SAT-Exos that were purified from DIO mice into the B6/J-Rab27a-Cas9-KO mouse models. PBS served as vehicle control. The injections were done twice a week for 2 weeks. Then, we employed untargeted metabolomics analysis to examine the changes of the metabolite profiles in these mice. The quality of the samples was tested by chromatogram overlapping (supplementary Fig. 4a, b) and the coefficient of variation (CV) of the relative peak area in the samples (supplementary Fig. 4c). In the metabolomic study, a total of 1858 compounds were detected in the positive mode, and 739 of them with known identification. In the negative mode, only 561 compounds were detected and 315 of them with known identification. Most of the metabolites detected in the positive mode are amino acids and organic acids in the negative mode. KEGG database was used to annotate the identified metabolites to understand their biological functions. As shown in the supplementary Fig. 4d, e, most of the metabolites were involved in amino acid metabolism, followed by lipid metabolism, carbohydrate metabolism and metabolism of cofactors and vitamins.
To examine the differential effects of the EAT-Exos, VAT-Exos and SAT-Exos on metabolism, we employed PLS-DA to examine the distribution and separation trend of the metabolite samples. PLS-DA provides high rates of sensitivity and specificity as it can substantially reduce the number of discriminatory variables by creating significant VIP (variable importance in projection) scores. As revealed by the models, injections of SAT-Exos, EAT-Exos or VAT-Exos that were purified from DIO mice had significant impacts on the plasma metabolites in the B6/J-Rab27a-Cas9-KO mice as illustrated by the sample clustering in the PLS-DA (Supplementary Fig. 4f–h).
We next screened for the differential metabolites in different grouping pairs of the mice. In the screening process, VIP of the first two PCs of the PLS-DA model above 1 was set as one of the criteria; and only metabolites with fold changes ≥ 1.2 or ≤ 0.83 that reached statistical significance (p value < 0.05) were shortlisted. As shown in Supplementary Table 2, EAT-Exos injection significantly affected a total of 47 metabolites, VAT-Exos injection affected a total of 62 metabolites, and SAT-Exos injection affected a total of 153 metabolites in both positive and negative modes. The metabolites detected in these mice were listed in supplementary Table 3a to 3f. The visual displays of the differential metabolites were shown in the respective volcano plots (Fig. 3c–e) and heat maps (supplementary Fig. 4i–k).
In the pathway enrichment analysis, only the metabolites that showed statistically significant differences between groups were shortlisted for the analysis. The number of affected pathways after EAT-Exos injection was less than those after VAT-Exos and SAT-Exos injections (Table 2). Only 5 signaling pathways were affected by EAT-Exos, while 20 signaling pathways were affected by VAT-Exos or SAT-Exos. For the SAT-Exos, the signaling pathway that was the most significantly affected was metabolic pathway, with a total of 1706 metabolites annotated. The other pathways that were also affected by SAT-Exos included tyrosine metabolism, phenylalanine metabolism, lysine degradation, aminoacyl-tRNA biosynthesis, fatty acid biosynthesis and so on (Table 2).
We next correlated these differential metabolites after SAT-Exos injection with the DEPs in the SAT-Exos that were increased by at least twofold or reduced by 0.3-fold as listed in supplementary Table 1c. We found that these DEPs were correlated to 14 different metabolisms including amino acid metabolisms and lipid metabolisms such as fatty acid degradation and steroid hormone biosynthesis (Fig. 3f).
Taken together, the metabolomic data suggest that the effects of EAT-Exos, VAT-Exos and SAT-Exos on the mouse metabolism are different. Injection of SAT-Exos has more prominent effects on the metabolite profiles and metabolic pathways in the B6/J-Rab27a-Cas9-KO mice when compared to EAT-Exos and VAT-Exos injections. Correlation analysis suggests that DEPs in the SAT-Exos contribute to the metabolic changes including amino acid metabolism and fatty acid metabolism under obesity conditions.
Untargeted lipidomics analysis reveals the impacts of SAT-Exos on the plasma lipid profile in B6/J-Rab27a-Cas9-KO mouse model, which mimic the plasma lipid profile in DIO mice
Since our data suggest that SAT-Exos that are derived from DIO mice have significant effects on fatty acid metabolism, we next performed global lipidomics to compare the plasma lipid profiles between the DIO mice and the B6/J-Rab27a-Cas9-KO mice after SAT-Exos injection.
After injecting SAT-Exos purified from DIO mice into the B6/J-Rab27a-Cas9-KO mice twice a week for two weeks, we performed untargeted lipidomics with the plasma lipid samples. The base peak chromatograms (BPC) of the lipid samples in positive and negative modes were shown in supplementary Fig. 5a, b. The CV distribution of lipid molecules was shown in supplementary Fig. 5c. Multivariate statistical analysis and univariate analysis were used to screen different lipids between groups. PLS-DA models showed the separation trends of the different plasma lipid samples, suggesting that SAT-Exos affects the plasma lipid profiles (supplementary Fig. 5d). Interestingly, SAT-Exos injection reduced the triglyceride (TG) levels; it also changed the levels of glycerophospholipids in the plasma in these mice (supplementary Table 4). Subsequent pathway enrichment analysis also suggests that SAT-Exos affects metabolism such as arachidonic acid metabolism, linoleic acid metabolism, biosynthesis of unsaturated fatty acid, regulation of lipolysis in adipocytes and glycerophospholipid metabolism in the mice (Table 3).
Next, we examined the lipid profiles in the DIO mice, with CD mice served as control. The BPC of the lipid samples in the DIO and CD mice were shown in supplementary Fig. 6a–d. In the lipidomic study, a total of 667 lipid molecules were detected. The lipid profiles for DIO and CD mice were different as indicated by the lipid sample clustering in the PCA (supplementary Fig. 6e). The differences in the lipid molecules between these mice were visualized in volcano plots (Fig. 4a) and the heat maps (supplementary Fig. 6f), respectively.
Untargeted lipidomics analysis of the plasma lipid profiles in DIO mice. a The volcano map of differential metabolites and the b detected changes in lipid sub-classes in the high fat diet-induced (DIO) mice. n = 3–4 mice in each group, *p < 0.05. c A diagram summarizes the main findings of this study
Interestingly, our data (supplementary Table 4) showed that injection of SAT-Exos purified from DIO mice into the B6/J-Rab27a-Cas9-KO mice reduced TG levels and changed glycerophospholipid levels. DIO mice also exhibited similar changes when compared to CD mice. As shown in Fig. 4b, in the DIO mice, the levels of fatty acid (FA), diglyceride (DG), monoglyceride and diglyceride (MGDG) were increased, while TG level was reduced, which suggest a change in the lipolytic activity. Besides, SAT-Exos also changed the plasma levels of glycerophospholipids including dimethylphosphatidylethanolamine (dMePH), phosphatidylserine (PS), lyso-phosphatidylglycerol (LPG), phosphatidylglycerol (PG), lyso-phosphatidylcholine (LPC), phosphatidylethanolamine (PE), lyso-phosphatidylethanolamine (LPE), phosphatidylglycerol (PI), lysodimethylphosphatidylethanolamine (LdMePE) and phosphatidylcholine (PC).
Discussion
This is the first report comparing the protein profiles in EAT-Exos, VAT-Exos and SAT-Exos under obesity conditions. Our data show that obesity has a more prominent effect on the protein profiles in SAT-Exos than those in EAT-Exos and VAT-Exos. Interestingly, DEPs in SAT-Exos and VAT-Exos are mainly involved in metabolism. Subsequent metabolomics and lipidomics suggest that SAT-Exos contribute to the changes in lipid metabolism under obesity conditions such as fatty acid metabolism and the changes in glycerophospholipid levels (Fig. 4c).
Dysregulation of fatty acid disposition and ectopic lipid accumulation in organs and tissues is a major contributing factor to metabolic syndrome under obesity conditions [35]. Clinical and epidemiological findings show that obese patients more likely to develop metabolic complications [36], which is in part, due to the abnormalities in lipid metabolism [37]. The elevated circulating fatty acid level is a key pathophysiological feature in the obese patients [38]. These elevated fatty acids affect the secretion of adipokines and inflammatory cytokines [37]. In obese patients, the circulating palmitic acid has a positive correlation with the levels of C-reactive protein (r = 0.2965; p = 0.035) [37] that is an acute marker of inflammation [39]. Many studies have explored the mechanism underlying the elevated circulating fatty acid levels in the obese patients. It has been suggested that under obesity conditions, adipocytes are having hypertrophic and hyperplastic growth. The hypertrophic adipocytes develop insulin resistance, hence increases lipolysis and fatty acid flux from the adipose tissues to the circulation [40]. Besides, pro-inflammatory signal such as tumor necrosis factor-alpha or some adipokines [36] also induce insulin resistance and enhance lipolysis in the adipocytes.
Our data have identified the DEPs in the SAT-Exos under obesity conditions; among these, 16 DEPs are upregulated by more than 2 folds, and 19 DEPs are reduced by more than 0.3 folds. By correlating these DEPs and the metabolic profiles of B6/J-Rab27a-Cas9-KO mice that have received SAT-Exos injections, we suggest that the DEPs in SAT-Exos may contribute to the metabolic changes under obesity conditions. Some of these metabolic changes are amino acid metabolism while others are lipid metabolism including fatty acid degradation and steroid hormone biosynthesis. With focus on the lipid metabolism, we then performed lipidomics with DIO mice, and we indeed observe the changes in the levels of the plasma fatty acids and glycerophospholipids in these mice. Nevertheless, the changes in these metabolic pathways after SAT-Exos injection may not solely dependent on the primary changes of the SAT-Exos because metabolic pathways work in a complex intertwined network in our body. The highlighted metabolic changes that are induced by SAT-Exos worth investigating, which may suggest novel exosomal-based therapeutic design for treating obesity.
Among the DEPs in the SAT-Exos, ADP-ribosylation factor (Arf) is elevated by 2.1 folds in the SAT-Exos under obesity conditions. Other study has shown that Arf protein levels are markedly increased in the white adipose tissues in ob/ob mice, whereas they are decreased in obesity-resistant mice [41]. More importantly, depletion of Arf inhibits isoproterenol-stimulated lipolysis in adipocytes, which is independent of insulin signaling [41]. Besides, Arf also mediates the endothelin-1 induced lipolysis in the adipocytes [42]. A study shows that endothelin-1 induces lipolysis by activating Arf and extracellular signal-regulated kinases (ERK) in adipocytes, which can be inhibited by the inhibitors of Arf and ERK [42]. Given the lipolytic role of Arf, the elevated level of Arf in the SAT-Exos may underlie the enhanced lipolysis in the adipocytes under obesity conditions.
Besides, mitogen-activated protein kinase kinase kinase 3 (MAP3K) is upregulated by 4.49 folds in the SAT-Exos under obesity conditions. MAP3K is a serine/threonine specific protein kinase; it activates MAP2K that in turns activates MAPK. Other protein kinases involved in the MAPK signaling cascade are ERK1/2, the c-JUN N-terminal kinase 1, 2 and 3 (JNK1/2/3), and the p38 MAPK. Study shows that MAP3K mix-lineage kinase 3 (MLK3) knockout mice are resistant to high fat diet-induced obesity, which is due to the inhibition of obesity-induced JNK activation, reduction in macrophage infiltration into adipose tissue and pro-inflammatory cytokine expressions [43]. Therefore, MAP3K activity contributes to the obesity pathophysiology. However, whether the elevation of MAP3K in the SAT-Exos under obesity conditions contributes to the changes in the plasma levels of fatty acids and glycerophospholipids is yet unknown. Other studies also show that MAPK signaling pathways are associated with the development of liver steatosis [44, 45] and hepatocellular carcinoma [46], further studies can be done to investigate the involvement of the elevated level of MAP3K in SAT-Exos in the disease development.
Our data also show that, unlike the DEPs in EAT-Exos, those in the SAT-Exos and VAT-Exos are mainly involved in metabolism under obesity conditions. In general, it is believed that excessive VAT is a risk factor for metabolic diseases [47]. VAT is directly associated with systemic inflammation by secreting various cytokines [48, 49], which leads to type 2 diabetes, atherosclerosis, cardiovascular disease, and some cancers [50,51,52]. However, study also suggests that the altered fatty acid metabolism under obesity conditions is not a direct consequence of VAT activity, but a dysfunction of SAT in which the abdominal SAT fails to adapt the expand through hyperplasia to store the excess circulating fatty acids [53]. Our study may reveal a novel role of SAT and SAT-Exos in changing the fatty acid metabolism under obesity conditions, which may suggest therapeutic targets for the treatment of obesity and its associated comorbidities.
Besides, based on our data, the role of EAT-Exos in contributing to the metabolic changes under obesity conditions is less prominent. DEPs in EAT-Exos under obesity conditions are involved in various signaling pathways including melanogenesis, Fc gamma R-mediated phagocytosis, calcium signaling pathway, regulation of actin cytoskeleton, gastric acid secretion. Although some of the DEPs in EAT-Exos are involved in metabolism such as carbon metabolism and fatty acid metabolism, injection of EAT-Exos into the B6/J-Rab27a-Cas9-KO mice does not profoundly affect the mouse metabolism. Indeed, the association of EAT and obesity or its comorbid conditions is less reported.
In conclusion, our data strongly suggest that obesity has more prominent effects on the protein profile of the SAT-Exos. DEPs in SAT-Exos may contribute to the altered fatty acid metabolism under obesity conditions. Our study has paved the path for the future development of novel exosomal-based therapeutics for the treatment of obesity.
Fundings
This work was partially supported by Shenzhen Basic Research Program for Shenzhen Virtual University Park (2021Szvup131), FNRA-IG (RC-FNRA-IG/20-21/SCM/01), ITC (PRP/015/19FX), GDNSF (2021A1515010655) and HMRF (08193596) to HYK; and Characteristic Innovation Projects of Universities in Guangdong Province (2020KTSCX030), Administration of Traditional Chinese Medicine of Guangdong Province (20222042), Young Elite Scientists Sponsorship Program by CAST (2021-QNRC2-B15), Guangzhou Basic and Applied Basic Research Project (202201011503) to ST.
Availability of data and material
Online version contains supplementary materials. All the data are available from the corresponding authors upon reasonable request.
Abbreviations
- ARF-1:
-
ADP-ribosylation factor 1
- BPC:
-
Base peak chromatograms
- CV:
-
Coefficient of variation
- DEPs:
-
Differentially expressed proteins
- DG:
-
Diglyceride
- DLS:
-
Dynamic light scattering
- dMePH:
-
Dimethylphosphatidylethanolamine
- EAT-Exos:
-
Exosomes derived from epididymal adipose tissues
- ERK:
-
Extracellular signal-regulated kinases
- FA:
-
Fatty acid
- FABP4:
-
Fatty acid binding protein 4
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- IL6:
-
Interleukin 6
- JNK1/2/3:
-
C-JUN N-terminal kinase 1, 2 and 3
- KOG:
-
Eukaryotic orthologous groups
- LdMePE:
-
Lysodimethylphosphatidylethanolamine
- LPC:
-
Lyso-phosphatidylcholine
- LPE:
-
Lyso-phosphatidylethanolamine
- LPG:
-
Lyso-phosphatidylglycerol
- MAP3K:
-
Mitogen-activated protein kinase kinase kinase-3
- MCP-1:
-
Monocyte chemoattractant protein-1
- MGDG:
-
Monoglyceride and diglyceride
- MIF:
-
Macrophage migration inhibitory factor
- PC:
-
Phosphatidylcholine
- PCA:
-
Principal component analysis
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- PI:
-
Phosphatidylglycerol
- PLS-DA:
-
Partial least squares method-discriminant analysis
- PPI:
-
Protein–protein interaction
- PS:
-
Phosphatidylserine
- PSMs:
-
Peptidespectrum matches
- SAT-Exos:
-
Exosomes derived from subcutaneous adipose tissues
- TEM:
-
Transmission electron microscopy
- TG:
-
Triglyceride
- VAT-Exos:
-
Exosomes derived from visceral adipose tissues
- VIP:
-
Variable importance in projection
References
Pegtel DM, Gould SJ (2019) Exosomes. Annu Rev Biochem 88:487–514
Kalluri R, LeBleu VS (2020) The biology, function, and biomedical applications of exosomes. Science 367:eaau6977
Connolly KD et al (2015) Characterisation of adipocyte-derived extracellular vesicles released pre- and post-adipogenesis. J Extracell Vesicles 4:29159
Wen Z et al (2020) Hypertrophic adipocyte-derived exosomal miR-802-5p contributes to insulin resistance in cardiac myocytes through targeting HSP60. Obesity 28:1932–1940
Sano S et al (2014) Lipid synthesis is promoted by hypoxic adipocyte-derived exosomes in 3T3-L1 cells. Biochem Biophys Res Commun 445:327–333
Deng ZB et al (2009) Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes 58:2498–2505
Kwan HY et al (2021) The impact of obesity on adipocyte-derived extracellular vesicles. Cell Mol Life Sci 78(23):7275–7288
Samuelson I, Vidal-Puig AJ (2018) Fed-EXosome: extracellular vesicles and cell–cell communication in metabolic regulation. Essays Biochem 62:165–175
Gao X, Salomon C, Freeman DJ (2017) Extracellular vesicles from adipose tissue-A prominential role in obesity and type 2 diabetes? Front Endocrinol 8:202
Akbar N, Azzimato V, Choudhury RP, Aouadi M (2019) Extracellular vesicles in metabolic disease. Diabetologia 62:2179–2187
Li CJ, Fang QH, Liu ML, Lin JN (2020) Current understanding of the role of Adipose-derived Extracellular Vesicles in Metabolic Homeostasis and Diseases: communication from the distance between cells/tissues. Theranostics 10:7422–7435
Kita S, Maeda N, Shimomura I (2019) Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J Clin Invest 129:4041–4049
Müller G et al (2011) Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositolanchored proteins transfer RNA stimulating lipid synthesis. Cell Signal 23(7):1207–1223
Wei H et al (2021) Regulation of exosome production and cargo sorting. Int J Biol Sci 17(1):163–177
Perez-Hernandez D et al (2013) The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem 288(17):11649–11661
van den Boorn JG, Dassler J, Coch C, Schlee M, Hartmann G (2013) Exosomes as nucleic acid nanocarriers. Adv Drug Deliv Rev 65(3):331–335
Kobayashi M, Rice GE, Tapia J, Mitchell MD, Salomon C (2015) Exosomes are fingerprints of originating cells: potential biomarkers for ovarian cancer. Res Rep Biochem 5:101–109
Wu J et al (2021) Single-cell analysis of a tumor-derived exosome signature correlates with prognosis and immunotherapy response. J Transl Med 19(1):381
Kwok KHM, Lam KSL, Xu A (2016) Heterogeneity of white adipose tissue: molecular basis and clinical implications. Exp Mol Med 48:e215
Gerhard GS et al (2014) Gene expression profiling in subcutaneous, visceral and epigastric adipose tissues of patients with extreme obesity. Int J Obesity 38:371–378
Schleinitz D, Bottcher Y, Bluher M, Dovacs P (2014) The genetics of fat distribution. Diabetologia 57:1276–1286
Hartwig S et al (2019) Exosomal proteins constitute an essential part of the human adipose tissue secretome. Biochim Biophys Acta Proteins Proteom 1867(12):140172
Lazar I et al (2016) Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: a novel mechanism linking obesity and cancer. Cancer Res 76(14):4051–4057
Berry R, Church C, Gericke MT, Jeffery E, Colman L, Rodeheffer MS (2014) Methods in enzymology (MIE): methods of adipose tissue biology—chapter 7: imaging of adipose tissue. Methods Enzymol. 537:47–73
Kranendonk ME, Visseren FL, van Balkom BW, Nolte-t Noen EV, van Herwaarden JA, de Jager W (2014) Human adipocytes extracellular vesicles in reciprocal signaling between adipocyte and macrophage. Obesity (Silver Spring) 22:1296–1275
Jung MK, Mun JY (2018) Sample preparation and imaging of exosomes by transmission electron microscopy. J Vis Exp 131:56482
Qian YJ, Pi WX, Zhu GF, Wei W, Lu TL, Mao CQ (2022) Quality evaluation of raw and processed Corni Fructus by UHPLC-QTOF-MS and HPLC coupled with color determination. J Pharm Biomed Anal 218:114842
Ostrowski M et al (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12(1):19–30
Zhao S et al (2021) Exosomal miR-21 from tubular cells contributes to renal fibrosis by activating fibroblasts via targeting PTEN in obstructed kidneys. Theranostics 11(18):8660–8673
Wei M et al (2020) Visceral adipose tissue derived exosomes exacerbate colitis severity via pro-inflammatory miRNAs in high fat diet fed mice. ACS Nano 14:5099–5110
Rikkert LG, Nieuwland R, Terstappen LWMM, Coumans FAW (2019) Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J Extracell Vesicles 8:1
Editorial, (2014) Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 3:26913
Gao X, Salomon C, Freeman DJ (2017) Extracellualr vesicles from adipose tissue—a potential role in obesity and type 2 diabetes? Front Endocrinol 8:202
Hu M et al (2018) The harsh microenvironment in infarcted heart accelerates transplanted bone marrow mesenchymal stem cells injury: the role of injured cardiomyocytes-derived exosomes. Cell Death Dis 9:357
Verna EC, Berk PD (2008) Role of fatty acids in the pathogenesis of obesity and fatty liver: impact of bariatric surgery. Semin Liver Dis 28(4):407–426
Roche HM (2005) Fatty acids and the metabolic syndrome. Proc Nutr Soc 64(1):23–29
Lottenberg AM et al (2012) The role of dietary fatty acids in the pathology of metabolic syndrome. J Nutr Biochem 23:1027–1040
Guerendiain M et al (2018) Changes in plasma fatty acid composition are associated with improvements in obesity and related metabolic disorders: a therapeutic approach to overweight adolescents. Clin Nutr 37(1):149–156
Du Clos TW (2000) Function of C-reactive protein. Ann Med 32(4):274–248
Wrzosek M et al (2022) Impact of fatty acids on obesity-associated diseases and radical weight reduction. Obes Surg 32:428–440
Liu Y et al (2010) ADP-ribosylation factor 6 modulates adrenergic stimulated lipolysis in adipocytes. Am J Physiol Cell Physiol 298(4):C921–C928
Davies JCB et al (2014) ADP-ribosylation factor 6 regulates endothelin-1-induced lipolysis in adipocytes. Biochem Pharmacol 90(4):406–413
Gadang V et al (2013) MLK3 promotes metabolic dysfunction induced by saturated fatty acid-enriched diet. Am J Physiol Endocrinol Metab 305(4):E549–E556
Cicuendez B, Ruiz-Garrido R, Mora A, Sabio G (2021) Stress kinases in the development of liver steatosis and hepatocellular carcinoma. Mol Metab 50:101190
Donohoe F et al (2020) Mitogen-activated protein kinase (MAPK) and obesity-related cancer. Int J Mol Sci 21(4):1241
Wang L et al (2016) Mitogen-activated protein kinase kinase 3 induces cell cycle arrest via p38 activation mediated Bmi-1 downregulation in hepatocellular carcinoma. Mol Med Rep 13:243–248
Fox CS et al (2007) Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the framingham heart study. Circulation 116:39–48
Fontana L et al (2007) Visceral fat adipokine secretion is associated with systemic inflammation in obese humans. Diabetes 56:1010–1013
Choi CHJ, Cohen P (2017) Adipose crosstalk with other cell types in health and disease. Exp Cell Res 360:6–11
de Jong PE, Verhave JC, Pinto-Sietsma SJ, Hillege HL, PREVEND study group (2002) Obesity and target organ damage: the kidney. Int J Obes Relat Metab Disord 26(Suppl 4):S21–S24
Ji C, Guo X (2019) The clinical potential of circulating microRNAs in obesity. Nat Rev Endocrinol 15:731–743
Liu Y et al (2021) Multifaceted roles of adipose tissue-derived exosomes in physiological and pathological conditions. Front Physiol 12:669429
Marcadenti A et al (2015) Different adipose tissue depots: metabolic implications and effects of surgical removal. Endocrinol Nutr 62(9):458–464
Acknowledgements
We thank the Centralized Research Facilities, The Science and Engineering Core Service at the Hong Kong University for providing the TEM facilities and Mr. Lee WS for his professional support. We thank the BGI Genomics for providing the facilities and service for the omics studies.
Author information
Authors and Affiliations
Contributions
MC, FZ, BC and KX carried out the experiments and preformed the data analysis in this study. CH performed the DLS in this study. CL, TT, QH, TS and HYK designed the experiments and study. TS and HYK wrote the manuscript, CL, TT, TS, and HYK revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
The animal studies were approved by the Research Ethics Committee at the Hong Kong Baptist University and the Department of Health in the Hong Kong Special Administration Region.
Consent for publication
Have informed consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Table 1
The highlighted differentially expressed proteins in (a) EAT-Exos, (b) VAT-Exos and (c) SAT-Exos that were increased by at least 2-fold or reduced by at least 0.3-fold under the obesity conditions. The arrows indicate either up- or downregulations (XLSX 16 KB)
Supplementary Figure 1
We used C57BL/6J mice to set up HFD-induced obesity (DIO) mouse model. (a) The body weight of the DIO mice and control diet (CD) mice. (b) Mass of the SAT, VAT and EAT in the mice. (c) Hematoxylin and eosin (H&E) staining of the SAT, VAT and EAT in the mice. n = 3-4 mice in each group. The p values are indicated in graph, *p<0.05; **p<0.01. (d) Representative Western blot showing the FABP-4 positive and adiponectin-positive circulating exosomes in the mice. SAT, subcutaneous adipose tissues; VAT, visceral adipose tissues; EAT, epididymal adipose tissues; FABP-4, fatty acid binding protein 4. n = 3-4 mice in each group. Supplementary Figure 2 Volcano plot of the upregulated and downregulated proteins when compared between (a) SAT-Exos and VAT-Exos under CD; (b) SAT-Exos and EAT-Exos under CD; (c) EAT-Exos and VAT-Exos under CD; (d) SAT-Exos and VAT-Exos under HFD; (e) SAT-Exos and EAT-Exos under HFD; (f) EAT-Exos and VAT-Exos under HFD. (g) Eukaryotic orthologous groups (KOG) annotation of the detected proteins in these exosomes. CD, control diet; HFD, high fat diet. EAT-Exos, exosomes derived from epididymal adipose tissues; VAT-Exos, exosomes derived from visceral adipose tissues; SAT-Exos, exosomes derived from subcutaneous adipose tissues of the high fat diet-induced obesity (DIO) mice. n = 3-4 mice in each group. Supplementary Figure 3 The categories of the proteins detected in (a) EAT-Exos, (b) VAT-Exos and (c) SAT-Exos. Pathway analysis of the differentially expressed proteins in (d) EAT-Exos, (e) VAT-Exos and (f) SAT-Exos under obesity conditions. Network analysis of the pathway enrichment of DEPs in (g) EAT-Exos, (h) VAT-Exos and (i) SAT-Exos. Network analysis of the DEPs in (j) EAT-Exos, (k) VAT-Exos and (l) SAT-Exos under obesity conditions with accessions shown. EAT-Exos, exosomes derived from epididymal adipose tissues; VAT-Exos, exosomes derived from visceral adipose tissues; SAT-Exos, exosomes derived from subcutaneous adipose tissues of the high fat diet-induced obesity (DIO) mice. n = 3-4 mice in each group. Supplementary Figure 4 We separately injected equal amount of EAT-Exos, VAT-Exos and SAT-Exos that were purified from DIO mice into the B6/J-Rab27a-Cas9-KO mouse models; PBS served as vehicle control. Based peak ion chromatography of the plasma metabolites in (a) positive ion mode and (b) negative ion mode; and (c) the coefficient of variation of the relative peak area. KEGG functional analysis of the detected metabolites in (d) positive ion mode and (e) negative ion mode. The PLS-DA analysis model of the plasma metabolites in B6/J-Rab27a-Cas9-KO mice after (f) EAT-Exos, (g) VAT-Exos and (h) SAT-Exos injection. Heatmap display of the differential metabolites in the B6/J-Rab27a-Cas9-KO mice after (i) EAT-Exos, (j) VAT-Exos and (k) SAT-Exos injections. EAT-Exos, exosomes derived from epididymal adipose tissues; VAT-Exos, exosomes derived from visceral adipose tissues; SAT-Exos, exosomes derived from subcutaneous adipose tissues of the high fat diet-induced obesity (DIO) mice. n = 3-4 mice in each group. Supplementary Figure 5 Based peak ion chromatography of the plasma lipid metabolites in the B6/J-Rab27a-Cas9-KO mice after SAT-Exos injection in (a) positive ion mode and (b) negative ion mode; and (c) the coefficient of variation of the relative peak area. (d) Principal component analysis (PCA) of the plasma metabolites in the mice. SAT-Exos, exosomes derived from subcutaneous adipose tissues of the high fat diet-induced obesity (DIO) mice. n = 3-4 mice in each group. Supplementary Figure 6 Based peak ion chromatography of the plasma lipid metabolites in (a) CD mice and (b) DIO mice in positive ion mode; and (c) CD mice and (d) DIO mice in negative ion mode. (e) Principal component analysis (PCA) of the differentiate lipid metabolites in DIO and CD mice. (f) Heatmap showing the differential lipid molecules in DIO and CD mice. DIO, high fat diet-induced obesity mice; CD, control diet fed mice. n = 3-4 mice in each group (PDF 3329 KB)
Supplementary Table 2
The number of differential metabolites in the B6/J-Rab27a-Cas9-KO mice after EAT-Exos, VAT-Exos or SAT-Exos injection. EAT-Exos, exosomes derived from epididymal adipose tissues; VAT-Exos, exosomes derived from visceral adipose tissues; SAT-Exos, exosomes derived from subcutaneous adipose tissues of the high fat diet-induced obesity (DIO) mice. n = 3-4 mice in each group (XLSX 11 KB)
Supplementary Table 3
The plasma metabolites in the B6/J-Rab27a-Cas9-KO mice after EAT-Exos injection detected in (a) positive ion mode and (b) negative ion mode. The plasma metabolites in the B6/J-Rab27a-Cas9-KO mice after VAT-Exos injection detected in (c) positive ion mode and (d) negative ion mode. The plasma metabolites in the B6/J-Rab27a-Cas9-KO mice after SAT-Exos injection detected in (e) positive ion mode and (f) negative ion mode. EAT-Exos, exosomes derived from epididymal adipose tissues; VAT-Exos, exosomes derived from visceral adipose tissues; SAT-Exos, exosomes derived from subcutaneous adipose tissues of the high fat diet-induced obesity (DIO) mice. n = 3-4 mice in each group (XLSX 270 KB)
Supplementary Table 4
The plasma lipid metabolites in different sub-classes detected in the B6/J-Rab27a-Cas9-KO mice after SAT-Exos injection. SAT-Exos, exosomes derived from subcutaneous adipose tissues of the high fat diet-induced obesity (DIO) mice. n = 3-4 mice in each group (XLSX 119 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, M., Zhang, F., Chen, B. et al. Omics approach to reveal the effects of obesity on the protein profiles of the exosomes derived from different adipose depots. Cell. Mol. Life Sci. 79, 570 (2022). https://doi.org/10.1007/s00018-022-04597-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04597-4