Abstract
Trophoblasts are specialized epithelial cells that perform critical functions during blastocyst implantation and mediate maternal–fetal communication during pregnancy. However, our understanding of human trophoblast biology remains limited since access to first-trimester placental tissue is scarce, especially between the first and fourth weeks of development. Moreover, animal models inadequately recapitulate unique aspects of human placental physiology. In the mouse system, the isolation of self-renewing trophoblast stem cells has provided a valuable in vitro model system of placental development, but the derivation of analogous human trophoblast stem cells (hTSCs) has remained elusive until recently. Building on a landmark study reporting the isolation of bona fide hTSCs from blastocysts and first-trimester placental tissues in 2018, several groups have developed methods to derive hTSCs from pluripotent and somatic cell sources. Here we review the biological and molecular properties that define authentic hTSCs, the trophoblast potential of distinct pluripotent states, and methods for inducing hTSCs in somatic cells by direct reprogramming. The generation of hTSCs from pluripotent and somatic cells presents exciting opportunities to elucidate the molecular mechanisms of human placental development and the etiology of pregnancy-related diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The placenta is a complex organ system that mediates the exchange of nutrients, gases and waste products between the mother and the developing fetus. Human placentas are hemochorial, which means that trophoblast cells come into direct contact with maternal blood. Placental development occurs in two morphologically and temporally distinct stages, the pre-villous and villous stages. The pre-villous placenta emerges upon implantation of the embryo within the maternal decidua starting at 7–9 days postfertilization (dpf). Multinucleated primitive syncytium invades into the maternal decidua and is involved in the implantation process and forming the first conduit of nutrient and gas exchange with endometrial glands and maternal endothelial capillary beds [1,2,3]. Cytotrophoblasts (CTBs) derived from the outer trophectoderm (TE) layer of the blastocyst rapidly divide and form a shell, which surrounds the epiblast (EPI, will give rise to the fetus) and primitive endoderm (PrE, will become the yolk salk) [4, 5]. Extraembryonic mesoderm descends from the embryonic compartment around 14–16 dpf and stretches through this shell, aligning with CTBs that further differentiate into two functionally distinct terminally differentiated trophoblast cell types: syncytiotrophoblast (STB) and extravillous trophoblast (EVT) [6]. After the emergence of the villous core, which is supplied with fetal blood vessels and placental macrophages (Hofbauer cells), this structure is considered the villous placenta and is maintained throughout the remainder of pregnancy.
STBs emerge directly from the underlying CTBs and are in direct contact with maternal blood. While many trophoblast cell types in the first trimester express placental hormones, STBs are the main manufacturers of human chorionic gonadotropin (hCG), which communicates the presence of the fetus to the maternal system, and other signaling hormones responsible for altering maternal metabolism, including leptin, prolactin-growth hormone family, and various steroid hormones [7]. EVTs arise from the tips of villi that form a prominent column extending to the maternal endometrium. The base of these columns contains a proliferative pool of EVTs that eventually travel to the endometrium, complete partial epithelial-to-mesenchymal transition (EMT) [8], and invade multiple uterine structures [9]. Interstitial EVTs reside within the decidua and invade deeply into the myometrium where they interact with maternal immune cells, obtain nutrients from maternal glandular epithelial cells, and invade uterine veins [10]. Although glandular invasion is yet to be fully characterized, arterial invasion and remodeling are a well-defined feature of EVTs. EVTs remodel uterine spiral arteries by inducing apoptosis in the smooth muscle layer [11], expressing similar adhesive proteins as vascular endothelium [12], and repopulating the walls of arteries in order to establish a consistent supply of maternal blood to the fetus. Insufficient arterial remodeling by EVTs has been identified as a leading cause of severe forms of pre-eclampsia [13].
Owing to ethical and regulatory restrictions on studying the human placenta in vivo, and the scarcity of samples obtained through elective terminations, our understanding of human placental development remains limited. Primary placental cultures are particularly difficult to perform and immortalized trophoblast cell lines fail to recapitulate the emergence and development of trophoblast cells from an undifferentiated progenitor state [14,15,16]. Self-renewing trophoblast stem cells (TSCs) were first derived from E3.5 mouse blastocysts by Janet Rossant’s laboratory in 1998 [17]. Mouse TSCs can be propagated indefinitely in the presence of fibroblast growth factor 4 (FGF4), heparin, and fibroblast conditioned medium and differentiate into specialized trophoblast cell types by withdrawal of FGF4 and fibroblast conditioned medium, among other methods [18,19,20]. The derivation of equivalent TSCs in the human system remained challenging for many years, but in 2018 Takahiro Arima’s laboratory in Japan reported the successful derivation of self-renewing human TSCs (hTSCs) from first-trimester placental CTBs and blastocysts [21]. Importantly, these hTSC lines were capable of undergoing lineage-directed differentiation into specialized EVT and STB cell types [21].
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), have been reported to acquire trophoblast-like fates in response to bone morphogenetic protein 4 (BMP4) [22,23,24]. However, hPSCs derived under conventional conditions exhibit biological and molecular features consistent with a post-implantation epiblast (EPI) identity [25]. Based on the alignment with single cell expression data from monkey embryos, conventional – also known as “primed”—hPSCs most closely resemble the late post-implantation EPI just prior to gastrulation, which arises more than a week after segregation of the trophoblast lineage [26]. Furthermore, there remains significant debate whether BMP4 treatment of primed hPSCs may give rise to amnion and mesoderm fates in addition to trophoblast [27,28,29,30,31]. We and others have shown that naïve hPSCs, which display transcriptional and epigenetic properties of the pre-implantation embryo, can directly differentiate into self-renewing and bipotent hTSCs [32,33,34,35]. In addition, somatic cells can be reprogrammed into human induced TSCs (hiTSCs) by overexpression of defined transcription factors [34, 36]. Here, we review the key properties that define human trophoblasts, the isolation of hTSCs from human blastocysts and placental tissues, and recently developed methods to derive hTSCs from pluripotent and somatic cell sources.
Biological and molecular criteria for bona fide human trophoblast stem cells
The recent years have seen a surge of interest in methods for deriving trophoblast cells from human stem cell sources. How do we assess the trophoblast identity of these cells? Which trophoblast model is most suitable for investigating placental development or disease processes? These questions are not simply answered and the choice of one trophoblast model versus another requires diligent considerations [37].
Lee et al. proposed four criteria for identification of primary first-trimester CTBs [38]. These criteria include: (i) the expression of GATA3, TFAP2C, and KRT7 at the protein level in mononuclear CTBs; (ii) hypomethylation of the ELF5 promoter region; (iii) expression of microRNAs (miRNAs) from the imprinted chromosome 19 miRNA cluster (C19MC), which is almost exclusively expressed in the placenta [39]; and (iv) the absence of classical HLA receptors (HLA-A, -B, -C). These criteria were established by comparing isolated primary first-trimester CTBs with villous stroma (extraembryonic mesoderm-derived), choriocarcinoma cell lines, primed hESCs, trophoblast-like cells generated from primed hESCs, and an embryonic carcinoma cell line. All four of these trophoblast features were identified in villous CTBs and choriocarcinoma cells. On the other hand, primed hESCs, their trophoblast derivatives, and embryonic carcinoma cells exhibited only some of these features, while villous stroma exhibited none of them.
Although the four criteria proposed by Lee et al. are characteristic of villous CTBs, it remains unclear whether they also apply to the pre-villous post-implantation stage, which is extraordinarily difficult to access. Additional criteria are needed to stage-match CTBs across different timepoints of human post-implantation development. In addition, specialized trophoblast cell types have their own unique molecular properties and recent studies are just beginning to illuminate the complex interactions between EVTs and maternal immune cells, how they invade uterine glandular spaces, and the possibility that placental bed giant cells may arise by fusion of deeply invasive interstitial EVTs [40, 41]. To define bona fide hTSCs, we propose that the following additional features need to be considered as well: (i) the ability to undergo lineage-directed differentiation into EVT and STB lineages, which should include functional characterization of differentiated trophoblast cell types based on the hormone-producing syncytia and invasive potential; (ii) the capacity of hTSCs for long-term self-renewal beyond a certain number of replication cycles, termed the Hayflick limit [42], which defines a truly proliferative stem cell; (iii) rigorous transcriptional benchmarking to human trophoblast identities in vivo; and (iv) whenever possible, the use of primary tissue sections for morphological and molecular comparison, e.g. to assess the size of syncytia and marker expression.
The recent availability of single cell RNA-sequencing (scRNA-seq) datasets for several stages of early human development, including pre-implantation [43, 44], post-implantation [45, 46], and villous placental tissues [47,48,49] provides stringent criteria for evaluating trophoblast fate (see the companion review in this issue by Brian Cox). Still missing, however, is a comprehensive scRNA-seq analysis that traces human trophoblast development from the pre-implantation TE to post-implantation CTB and subsequent stages of placental development. This would help to address questions regarding the hierarchical relationship between trophoblast subpopulations leading to the emergence of the placenta.
Derivation of human trophoblast stem cells from blastocysts and placental tissues
The discovery of in vitro culture conditions for establishing authentic human trophoblast stem cells (hTSCs) has transformed our ability to model placental development [21]. To identify pathways required for hTSC derivation, Okae et al. performed RNA-seq analysis on CTBs, EVTs, and STBs isolated from first-trimester placental tissues. Since the Wingless/Integrated (WNT) and epidermal growth factor (EGF) signal transduction pathways were enriched in CTBs, they first tried to culture CTBs in the presence of the GSK3 inhibitor CHIR99021, which activates WNT signaling, and recombinant EGF. Because the cells did not adhere well, they screened additional inhibitors and growth factors that promote the expansion of epithelial stem cells, eventually arriving at a cocktail comprising the transforming growth factor β (TGF-β) inhibitors SB431542 and A83-01, the histone deacetylase (HDAC) inhibitor valproic acid, recombinant EGF, CHIR99021, and the Rho-associated kinase (ROCK) inhibitor Y27632 (termed SAVECY medium). Under these conditions, they were able to derive putative hTSCs from both primary first-trimester CTBs and TE outgrowths of attached blastocysts. These cells could be propagated for over 70 passages, and expressed trophoblast markers, such as GATA3, TP63, TEAD4, and C19MC miRNAs.
Okae et al. then examined whether their candidate hTSCs were capable of differentiating into functional placental cell types. Upon removal of the WNT-promoting factor, CHIR99021, they noticed that the cells differentiated into HLA-G-positive EVT-like cells. To further optimize their protocol for lineage-directed EVT differentiation, they also increased the concentration A83-01, provided an extracellular matrix (ECM) in the form of Matrigel, and added Neuregulin (NRG1), all of which have been shown to promote EVT differentiation in placental explant cultures [50,51,52]. These hTSC-derived EVTs completed key aspects of EMT specific to EVTs, displayed enhanced expression of HLA-G, and reduced expression of CTB markers CDH1 and ITGA6. In addition, they also showed that hTSCs could differentiate into STBs upon addition of Forskolin, a cyclic AMP (cAMP) agonist, which was used previously to promote the differentiation of primary trophoblast and choriocarcinoma cells towards an STB fate [53]. hTSC-derived STB cells expressed elevated CGB and SDC1, while downregulating markers of other trophoblast lineages, including CDH1, HLA-G, ITGA6, and TP63. Enhanced differentiation of STBs was observed when hTSCs were cultured in a low adhesion environment and supplemented with EGF. As an assessment of in vivo potential, subcutaneous injection of hTSCs into NOD-SCID mice resulted in invasion of dermal and subcutaneous tissues and expansion of all trophoblast cell types, although migratory EVT-like cells were few in number.
Transcriptional analysis of hTSC lines derived from blastocysts and CTBs revealed strong similarities between each other and to primary CTBs [21]. Likewise, hTSC-derived EVTs and STBs most closely resembled their respective in vivo counterparts. Okae et al. then used whole genome bisulfite sequencing (WGBS) to assess the DNA methylation landscape of hTSCs in relation to first-trimester CTBs. Overall, hTSCs were hypomethylated compared to primary CTBs (33.6% and 52.3%, respectively), but critical patterns of methylation were conserved, such as the retention of placenta-specific germline differentially methylated regions (gDMRs) [54] and the presence of large partially methylated domains [55]. A potential explanation for the apparent hypomethylation of in vitro derived hTSCs is that these culture conditions may enrich for proliferative CTBs, which tend to be less methylated than other CTB populations [56]. Specific examples of placenta-specific hypomethylation observed in hTSCs include the promoter regions of ELF5, INSL4, ZNF750, and DSCR4. Finally, hTSCs also maintained the expected intermediate methylation levels at gDMRs associated with placenta-specific imprinted genes. Altogether, these data suggest that the conditions for hTSC isolation developed by the Arima laboratory capture a reliable in vitro counterpart of human first-trimester CTBs.
The original report from Okae et al. stated that they were unable to derive hTSCs from term placenta. This suggested that bipotent proliferative CTBs, which are the likely source of hTSCs, may be lost during or after the second trimester of pregnancy. However, two recent reports indicate that it is possible to derive hTSCs from term placentas using modified protocols. Kessler and colleagues reported that non-integrating viral expression of five transcription factors (CDX2, ELF5, ETS2, TFAP2C, and TEAD4) can reprogram term villous CTBs into induced TSCs (iTSCs), which are capable of long-term self-renewal and display transcriptional similarity to hTSCs derived from first-trimester placental tissues [57]. In addition, Chen and colleagues reported that treatment with SAVECY media under hypoxic conditions facilitates the derivation of hTSCs from term placentas by targeting the GCM1-ΔNp63α antagonistic signaling axis [58]. The establishment of hTSC lines from term placentas enables the generation of trophoblast models from placental tissues with known pregnancy outcomes, but it is important to remember that term placenta is inherently programmed to stop functioning after 9 months due to accumulating DNA methylation and damage from reactive oxygen species, a decline in CTB proliferation, and fluctuations in placental hormone secretion during pregnancy and parturition [59,60,61,62]. Because of these accumulated phenomena over gestation, determining the cause of placental dysfunction is difficult without access to earlier stages of placental development.
Generation of human trophoblast stem cells from naïve pluripotent stem cells
Given limited access to human embryos, stem cell models have become an invaluable tool to study early human development. Self-renewing hESC lines were first derived from pluripotent cells in human blastocysts in 1998 [63]. However, experimentally observed molecular differences between mouse and human pluripotent stem cells raised the question whether there may be multiple pluripotent states. It is now well described that pluripotency exists in a continuum from the initial, preimplantation naïve pluripotent state to the primed pluripotent state, the last step before gastrulation [25]. These discrete hPSC states are isolated using different culture media. Whereas conventional hPSCs are commonly maintained in commercially available media containing high levels of FGF and Activin, naïve hPSCs can be established using small molecule inhibitor cocktails. In particular, the t2i/L/Gö [64] and 5i/L/A [65] cocktails, have been widely used to induce molecular signatures of naïve pluripotency in hPSCs, including transcriptional correspondence to the EPI compartment of the blastocyst [26, 66, 67], X chromosome reactivation in female cells [68, 69], and expression of blastocyst-specific cell surface markers [70, 71]. Detailed reviews on methods for inducing and maintaining naïve hPSCs and their properties are available elsewhere [72,73,74,75]. In addition, naïve hPSCs can also be reprogrammed directly from somatic cells [76,77,78,79,80] or derived de novo from human blastocysts [81, 82].
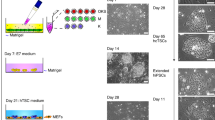
The TE vs. inner cell mass (ICM) cell fate decision is the first to occur in the mammalian embryo and is viewed as a strong barrier in mouse. Indeed, pluripotent cells in the mouse blastocyst have lost the ability to give rise to TE and mouse PSCs require genetic manipulation, such as overexpression of Cdx2 or downregulation of Oct4, to acquire TE fate [83, 84]. However, several lines of evidence suggested that naïve hPSCs may have an expanded fate potential compared to their murine counterparts. First, scRNA-seq profiling of human embryos has revealed more fluid lineage segregation compared to mouse embryos. The TE program is initiated by compaction in the human morula [85], but only becomes transcriptomically distinct from ICM cells 12 h later at the B2 blastocyst stage [86, 87]. CDX2 expression is acquired in TE cells at the B3 stage before it is specifically lost in polar TE cells, which subsequently gain NR2F2. This stage of blastocyst development also coincides with onset of PrE markers [86, 88]. Although these observations still require validation by time-lapse staging, overall lineage segregation appears to be more fluid than the paced, stepwise lineage segregation seen in mouse embryos [89,90,91]. This change of pace could explain retention of plasticity in human blastocyst lineages, as demonstrated by the ability of TE cells to form ICM upon re-aggregation [92]. Since naïve hPSCs correspond to the early human blastocyst based on the gene and transposon expression profiling [67, 69], they may conceivably retain developmental plasticity associated with this stage of human embryogenesis. Second, naïve hPSCs exhibit elevated expression of a subset of transcription factors and open chromatin sites associated with the human trophoblast lineage [69, 93], which raised the question whether they may have an enhanced potential for trophoblast differentiation. Indeed, we and others demonstrated that naïve cells can be directly converted into cells that closely resemble hTSCs upon application of the SAVECY media developed by Okae et al. [32,33,34,35] (Fig. 1). Some methodological differences were reported in these studies: the David and Pastor laboratories used naïve cells maintained in t2iLGöY [64], while Dong et al. primarily used naïve cells derived in 5i/L/A [65]. In addition, the David and Theunissen laboratories passaged the cells until homogeneous hTSC cultures were obtained [32, 34], while the Pastor laboratory performed FACS sorting based on the EpCAM and ITGA2 [33]. However, in all cases, cells derived from naïve hPSCs closely resemble primary hTSCs by morphology and surface marker profile.
Derivation of human trophoblast stem cells (hTSCs) from naïve human pluripotent stem cells (hPSCs). Naïve hPSCs can be directly converted into hTSCs by treatment with SAVECY media, which were originally developed to isolate primary hTSCs from blastocysts and first-trimester placental tissues [21]. The resulting hTSCs correspond to human post-implantation CTB-like cells [32,33,34]
The biological and molecular properties of naïve hPSC-derived hTSCs were evaluated by using a number of different assays. Bulk RNA-seq analyses revealed close transcriptional correspondence between naïve hPSC-derived hTSCs and primary hTSCs derived from blastocysts or first-trimester placental tissues by Okae et al. [32,33,34]. Compared with naïve and primed hPSCs, both types of hTSCs displayed significant induction of trophoblast markers, such as CCR7, ELF5, GATA3, KRT7, and TP63. In addition, the Pastor laboratory assessed the DNA methylation landscape of naïve hPSC-derived hTSCs, revealing the acquisition of a placenta-like methylome, including globally reduced DNA methylation levels and methylation patterns at individual CpG islands and gene promoters that resembled primary hTSCs [33]. This analysis also revealed that most placenta-specific imprinted genes were activated during differentiation of naïve hPSCs into hTSCs, although a few loci, including ZFAT and PROSER-A1, resisted demethylation both in the naïve hPSC state and upon differentiation into hTSCs. Importantly, all three studies also evaluated the differentiation potential of naïve hPSC-derived hTSCs towards specialized trophoblast fates, demonstrating that the cells were capable of giving rise to invasive EVTs and hormone-producing STBs using the methods for lineage-directed hTSC differentiation described by Okae et al. [32,33,34].
Based on the alignment with single cell expression data from human embryos, naïve hPSC-derived hTSCs most closely correspond to post-implantation trophoblast cells around 10–12 dpf [32, 34]. This suggests that the hTSC conditions developed by Okae et al. promote the differentiation of naïve hPSCs into a post-implantation CTB-like state and that it may be possible to capture a pre-implantation TE-like state from naïve cells under appropriate culture conditions. Indeed, recent work from the Smith laboratory demonstrated that naïve hPSCs transiently acquire a pre-implantation TE identity upon treatment with the MEK inhibitor PD0325901 and the TGFβ inhibitor A83-01 (PDA83) before the cells are switched to SAVECY media to attain a post-implantation CTB-like state [30] (Fig. 2). This work was independently corroborated by Takashima and colleagues, who supplemented the PDA83 cocktail with BMP4 and a Janus kinase (JAK) inhibitor [29] (Fig. 2). These naïve-like TE cells share common trophoblast signatures with post-implantation hTSCs, such as expression of GATA3, TFAP2C, and KRT7, demethylation of the ELF5 promoter region, and expression of C19MC miRNAs, but show increased expression of early TE markers, such as CDX2 and HAVCR1. Naïve TE-like cells can be distinguished by a unique cell surface profile, including expression of ENPEP and TACSTD2, which are also expressed in TE in human pre-implantation blastocysts, but reduced levels of HLA-ABC and SIGLEC6 as compared to hTSCs [94]. The absence of classical HLA molecules is one of the hallmarks of human trophoblast cells as proposed by Lee et al. [38], and therefore the increased expression of these antigens in hTSCs was unexpected. Takashima et al. proposed a simplified formulation for promoting the transition from naïve TE-like cells into a post-implantation CTB-like state by culture in the presence of A83-01, the GSK3 inhibitor CHIR99021, and EGF (ACE), which prevents aberrant HLA-ABC activation [94]. These conditions also support the derivation of hTSCs from primary chorionic villi of first-trimester placental tissues. By benchmarking their expression data to single cell studies in human and monkey embryos, these papers showed that the differentiation of naïve hPSCs into hTSCs via a pre-implantation TE intermediate recapitulates key steps during primate trophoblast development, offering an accessible model system of human trophoblast specification. Furthermore, the Smith laboratory demonstrated that EPI cells from human blastocysts harbor the intrinsic potential to generate TE [30], which reveals that the human EPI retains an expanded extraembryonic plasticity as compared to the mouse EPI, in accordance with in vitro studies using naïve hPSCs.
Recapitulating the trophoblast specification pathway from naïve hPSCs. Naïve hPSCs give rise to a transient pre-implantation TE state when cultured in BMP4, Activin/Nodal inhibitor A83-01, MEK inhibitor PD03, and a Janus Kinase inhibitor. TE cells can then be transitioned to stable post-implantation hTSCs in SAVECY or ACE, a reduced version of SAVECY [29, 30]
In addition to providing a source of 2D models of trophoblast development, naïve hPSCs can also be used to generate 3D organoids that encompass a diversity of trophoblast cell types. The isolation of trophoblast organoids was first reported by the Moffett and Knöfler laboratories in 2018 [95, 96], but required the isolation of CTBs from first-trimester placental tissue, which is not readily available in many jurisdictions due to ethical and practical restrictions. Given the transcriptional and epigenetic similarities between hTSCs and CTBs, we and others postulated that it should be possible to generate trophoblast organoids from naïve hPSCs via an hTSC intermediate. Indeed, when transferred to Matrigel droplets in the presence of trophoblast organoid medium (TOM), naïve hPSC-derived hTSCs self-organize into 3D organoids that exhibit a similar architecture as primary trophoblast organoids with an outer CTB layer and an inner STB compartment [29, 34, 97]. This marks a reversal of the architecture of the primary placental villi in which STBs are located towards the periphery. We recently performed single cell transcriptome profiling on stem-cell-derived trophoblast organoids (SC-TOs) generated from various hTSC lines, which revealed a highly concordant cellular distribution of progenitor and specialized trophoblast states [97]. This suggests that trophoblast organoid culture represents a powerful attractor state in which the influence of subtle epigenetic differences between hTSCs obtained from naïve hPSCs and those obtained from first-trimester placental tissues is mitigated. In addition, the proportion of EVTs can be enhanced by transfer to a WNT-reduced environment, similar to observations in primary trophoblast organoids [98]. We further showed that SC-TOs generated from naïve hPSCs recapitulate placental X chromosome inactivation patterns and model early placental susceptibility to emerging pathogens, including SARS-CoV-2 and Zika virus [97]. Like hTSCs in 2D culture, the CTB subpopulation within SC-TOs is most closely aligned with trophoblast progenitors in early post-implantation embryos and consequently these two culture systems exhibit many shared markers, such as GATA3, TFAP2C, KRT7, and C19MC miRNAs. However, the reduced expression of classical HLA molecules in trophoblast organoids and their 3D microenvironment present advantages for modeling placental organogenesis.
Is it possible to derive human trophoblast stem cells from primed and other pluripotent states?
The initial studies that directly compared the response of naïve and primed hPSCs to hTSC media all concluded that primed hPSCs fail to acquire an hTSC-like identity when directly transferred to the SAVECY media developed by Okae et al. [32,33,34]. In fact, primed hPSCs showed upregulation of neuroectoderm-related genes as opposed to trophoblast-related genes under these conditions [32]. Nevertheless, several other studies have reported that primed hPSCs are capable of generating hTSC-like cells under modified culture conditions. The generation of hTSCs from primed hPSCs was reported via micromesh culture [99] or by pre-treatment with BMP4, a combination of BMP4 and the WNT inhibitor IWP2, or the TGFβ inhibitor A83-01 before applying hTSC media (Fig. 3) [100,101,102,103,104]. The resulting cells share several global features with primary hTSCs, including gene expression, differentiation potential towards EVT and STB lineages, and increased chromatin accessibility at trophoblast-specific genes. These data indicate that it is possible to direct primed hPSCs towards an hTSC-like state. However, there may yet be subtle but significant differences between hTSC-like cells derived from naïve and primed states. For example, a recent study from the Arima laboratory reported that hTSCs derived from naïve, but not primed, hPSCs display complete activation of placentally imprinted C19MC miRNAs [105], which is one of the original trophoblast criteria proposed by Lee et al. [38]. Ectopic activation of C19MC miRNAs in primed hPSCs enhanced their potential to differentiate into proliferative hTSCs with full differentiation potential towards specialized trophoblast lineages [105]. Consistent with these findings, incomplete activation of C19MC miRNAs was also observed in two other studies that derived hTSCs from primed hPSCs [101, 103]. Therefore, an important objective in future studies will be to further compare the biological and molecular properties of hTSC-like cells isolated from isogenic naïve and primed states side-by-side.
A complicating factor in these studies is that trophoblast cells have substantially overlapping transcriptional signatures with amnion, which is another extraembryonic lineage that arises from the primate EPI following implantation [106]. Both the Pastor and Smith laboratories noted upregulation of amnion markers in hTSC-like cells generated from extended pluripotent stem cells (EPSCs), which lack clear alignment with a developmental equivalent in vivo [33, 107, 108]. On the other hand, the David laboratory reported that hTSCs can be derived from a different type of EPSC following extended culture in SAVECY media [34]. As noted earlier, there also remains a significant debate whether BMP4 treatment of primed hPSCs may give rise to amnion [27,28,29,30]. Therefore, careful analysis of amnion and trophoblast markers will be essential when assessing the quality of putative hTSCs and a recent scRNA-seq analysis of amnion development in primate embryos provides a helpful set of distinctive markers [109]. Adding further complexity, this study also demonstrated that early amnion differentiation from EPI cells follows a TE-like trajectory, including the activation of a broad range of STB-associated markers. Consequently, it may be instructive to reassess whether primed hPSCs or EPSCs acquire an amnion fate before being redirected towards an hTSC-like identity.
Direct reprogramming of human somatic cells into induced trophoblast stem cells
The derivation of hTSCs offers an in vitro model system of placental organogenesis and dysfunction during differentiation of villous CTB progenitors into specialized trophoblast cell types. However, human stem cell models are limited in terms of their genetic background, since they are mainly generated from embryos, or in the case of hTSCs, first-trimester placental tissues [21]. The David and Polo laboratories have developed methods to directly reprogram patient samples into an array of stem cells. Their initial focus was to directly reprogram somatic cells into naïve iPSCs, assessing multiple culture conditions in parallel. They showed that somatic cells could be directly reprogrammed using the canonical OCT4, SOX2, KLF4, and c-Myc (OSKM) transcription factor cocktail and two naïve-specific culture media, t2iLGö and 5iLAF, yielding cells that closely corresponded to the human pre-implantation EPI based on the transcriptional and epigenetic criteria [76, 77].
The David laboratory noticed that cells co-expressing the EPI marker NANOG and the TE marker GATA3 emerged transiently at an early stage of naïve reprogramming in reprogramming intermediates that retained high transgene expression levels [34]. The Polo laboratory came to the same conclusion by mapping the reprogramming routes from somatic cells to primed and naïve iPSCs [36]. These observations were concomitant with the publication of SAVECY media that support hTSC derivation and growth [21], prompting both laboratories to fine tune their reprogramming conditions to generate hiTSCs. Technically, the protocols established by both groups are similar, with minor differences (described in soon-to-be-published Nature Protocol papers). Both groups demonstrated that the hiTSCs were bona fide hTSCs: they reported in vivo differentiation into trophoblast-like tissues, the ability to differentiate into hCG-secreting syncytia and EVTs, and high expression levels of C19MC miRNAs as compared to both fibroblasts and iPSCs, a unique feature of primary trophoblast [38]. Building on their scRNA-seq analysis, Liu and colleagues [36] identified GATA2 and TFAP2C as important regulators during somatic cell reprogramming into naïve iPSCs. Indeed, naïve reprogramming was greatly impaired when TFAP2C was knocked down, while reprogramming to primed pluripotency was less impacted. Moreover, GATA2 knockdown resulted in impaired reprogramming to both naïve and primed states, suggesting an earlier role of GATA2 during reprogramming. Further analysis of the scRNA-seq dataset revealed that during naïve reprogramming under both 5iLAF and t2iLGö conditions some subpopulations of cells were enriched in TE signatures (Fig. 4). Building on their expertise in human peri-implantation development [86], Castel and colleagues matched hiTSCs with 8 dpf trophoblast cells, as demonstrated by precise gene-set enrichment of developmental stages and notably the presence of NR2F2, a transcription factor that is expressed upon progression of CDX2-positive TE cells into CDX2-negative trophoblast cells [88].
Direct reprogramming of somatic cells into human induced TSCs (hiTSCs). Somatic reprogramming of fibroblasts using the Yamanaka factors, OCT4, SOX2, KLF4, and c-MYC (OSKM), presents an alternative route for accessing the hTSC state. Reprogramming intermediates are treated with t2iLGöY or 5iLAF naïve conversion medium, giving rise to a transient mixture of cells that express TE markers. The treatment with SAVECY medium further differentiates these cells into a post-implantation CTB-like hTSC state [34, 36]
Concluding remarks
Since the elucidation of culture conditions for derivation of self-renewing hTSCs from blastocysts and first-trimester placental tissues by Okae et al. [21], a number of groups have explored alternative avenues for accessing the hTSC state. Here we reviewed three sources for creating hTSCs, each of which offers their own relative advantages and disadvantages for basic and applied research. First, the derivation of hTSCs from blastocysts and first-trimester placental tissues continues to provide a gold standard for the field, although ethical, legal, and practical constraints on accessing such tissues present a barrier for many researchers. The recent success in deriving hTSCs from placental tissues at term may offer a more accessible option for deriving primary hTSCs [57, 58]. Second, the generation of hTSCs from naïve hPSCs offers a renewable source of hTSCs and 3D trophoblast organoids that can be applied to a broad spectrum of hPSC lines, including patient-specific iPSCs [29, 30, 32,33,34, 97]. Furthermore, the ability to reconstitute the trophoblast lineage from naïve hPSCs, which correspond to pluripotent cells in the pre-implantation embryo, presents exciting possibilities to investigate the genetic and epigenetic mechanisms of human trophoblast specification in vitro. A potential limitation of the use of naïve hPSCs is the fact that they undergo erasure of parent-specific imprinting during extended culture [69, 110]. Nevertheless, most placentally imprinted genes are activated during the naïve-to-hTSC transition [33, 97]. Third, direct reprogramming of somatic cells into hiTSCs presents a more efficient route towards the creation of patient-specific hTSCs for modeling placental diseases but may be less suitable for modeling the process of trophoblast specification during human pre-implantation development.
Several recent studies indicate that it is possible to generate hTSC-like cells directly from primed hPSCs under modified culture conditions. A short pulse of BMP4, BMP4 and IWP2, or A83-01 can redirect primed hESCs away from a neural fate and towards a trophoblast identity [99,100,101,102,103,104]. However, there may be subtle but significant differences between hTSC-like cells derived from naïve and primed cells, such as the expression level of placentally imprinted C19 miRNAs [105], which is one of the original trophoblast criteria proposed by Lee et al. [38]. It is important to bear in mind that extensive single cell expression profiling of primate embryogenesis has not revealed evidence that post-implantation EPI cells contribute to the trophoblast lineage [26, 45, 111]. Therefore, we surmise that the generation of hTSC-like cells from primed hPSCs may represent a culture-induced trans-differentiation event. According to the recent work from Rugg-Gunn and colleagues, primate EPI cells coopt the transcriptional program of TE specification to initiate the amniotic cavity just after implantation [109]. Cooptation of the pre-existing TE differentiation program during the early wave of amnion differentiation may present a window of opportunity for a subset of EPI cells to be redirected towards an hTSC-like identity under pressure from strong external stimuli. It has also been suggested that amnion may provide an independent source of STB-like cells [112], but this hypothesis requires validation using lineage tracing.
The SAVECY media developed by the Arima laboratory have had a transformative impact on the field of trophoblast biology, but the culture conditions for hTSC derivation and differentiation require further refinement. Current hTSCs are hypomethylated and exhibit increased expression of HLA class I surface molecules compared to CTBs in vivo [21]. Furthermore, it remains unclear why hTSC derivation requires the use of two distinct TGFβ inhibitors, A83-01 and SB435412. In fact, the ACE formulation developed by the Takashima laboratory uses only one of these inhibitors and yields hTSCs with reduced HLA-ABC expression [94], as does transfer to trophoblast organoid media [37, 97]. Additional refinements to these conditions and those used for lineage-directed differentiation may enable the isolation of hTSCs that more readily transition into functional EVT and STB cells. Alongside this effort, a more complete in vivo reference for TE and CTB progenitors is needed to stage-match hTSCs derived under various conditions to their counterparts in the human embryo and placenta. New embryo models, such as blastoids [88, 113, 114], offer an opportunity to better understand TE fate progression from its initiation at the morula stage [85] until the appearance of the first STB and EVT cells [45].
While the culture conditions developed by Okae et al. capture hTSCs in a post-implantation trophoblast identity, a pre-implantation TE state can be transiently accessed by treating naïve hPSCs with MEK and TGFβ inhibitors in monolayer culture [29, 30] or by promoting their self-organization into blastoids. An important question for future research will be to investigate whether culture conditions can be devised to capture a self-renewing pre-implantation TE state in human cells. Mouse TSCs, in fact, more closely resemble the pre-implantation TE at the level of marker expression and based on their ability to colonize the placenta following injection into E3.5 mouse embryos [17], although significant heterogeneity has been reported within mouse TSC culture [115, 116]. Rivron and colleagues recently reported progress in capturing mouse TSCs with more specific features of pre-implantation polar TE by applying inductive signals originating from the inner embryonic cells of the blastocyst [117]. Conceivably, a similar approach may be effective for stabilizing a self-renewing pre-implantation TE state from naïve hPSCs. As an alternative approach, Mischler et al. reported that primed hPSCs can give rise to CDX2-positive hTSC-like cells in the presence of a sphingosine-1 phosphate agonist, a GSK3 inhibitor, a TGFβ inhibitor, and recombinant FGF10 [100], but it remains to be determined whether these cells correspond to TE cells in the human pre-implantation embryo.
What are the potential biomedical applications of hTSCs derived from pluripotent and somatic sources? The development of the placenta remains an understudied aspect of human embryology, but placental complications during the first trimester are associated with pregnancy complications such as preeclampsia, miscarriage, and fetal growth restriction [118]. In addition, there is increasing recognition that variations in the supply of nutrients to the developing fetus can manifest in disease during postnatal life, such as cardiovascular disease [119]. Therefore, it would be instructive to generate hTSCs from patients who suffered pre-eclampsia or fetal growth restriction using the various approaches discussed in this review and rigorously evaluate their phenotype relative to healthy controls based on the molecular profiling (transcriptome, DNA methylome, and chromatin accessibility) and differentiation towards specialized trophoblast fates using 2D monolayer and 3D organoid culture. More advanced phenotyping could involve the use of co-culture assays between trophoblasts and human endometrial cells to model the process of trophoblast implantation and invasion, as recently demonstrated using blastoids and stem-cell-derived trophoblast organoids [88, 97]. These studies could identify molecular targets for early detection and potential pharmacological intervention. In addition, an improved understanding of the interplay between placental genes and endometrial environmental cues that are essential for hTSC specification and differentiation may aid in understanding the genetic and environmental basis of placental pathologies leading to fetal growth restriction, preterm birth, and recurrent pregnancy loss [120,121,122], while genes that suppress the proliferation and invasion of hTSCs may provide candidates for treatment of choriocarcinoma, a highly malignant tumor of trophoblastic origin [123]. From this standpoint, recent candidate-based and genome-wide approaches to identify essential and growth-restricting genes in hTSCs present an important foundation for future studies [124,125,126,127,128]. We anticipate that the ensuing decade will witness unprecedented advances in our understanding of placental development and women’s health through the concerted efforts of reproductive scientists, stem cell biologists, clinicians, and biomedical engineers.
Availability of data and material
Not applicable.
References
Hertig AT, Rock J, Adams EC (1956) A description of 34 human ova within the first 17 days of development. Am J Anat 98(3):435–493
Enders AC (1976) Cytology of human early implantation. Res Reprod 8(5):1–2
Enders AC (1989) Trophoblast differentiation during the transition from trophoblastic plate to lacunar stage of implantation in the rhesus monkey and human. Am J Anat 186(1):85–98
Enders AC (1995) Transition from lacunar to villous stage of implantation in the macaque, including establishment of the trophoblastic shell. Acta Anat (Basel) 152(3):151–169
Boyd JHW (1970) The human placenta. W. Heffer & Sons Ltd., Cambridge
Kingdom J et al (2000) Development of the placental villous tree and its consequences for fetal growth. Eur J Obstet Gynecol Reprod Biol 92(1):35–43
Lacroix MC et al (2002) Human placental growth hormone—a review. Placenta 23(Suppl A):S87–S94
Davies JE et al (2016) Epithelial-mesenchymal transition during extravillous trophoblast differentiation. Cell Adh Migr 10(3):310–321
Lee CQE et al (2018) Integrin alpha2 marks a niche of trophoblast progenitor cells in first trimester human placenta. Development 145(16):dev162305
Moser G et al (2017) Extravillous trophoblasts invade more than uterine arteries: evidence for the invasion of uterine veins. Histochem Cell Biol 147(3):353–366
Red-Horse K et al (2006) Cytotrophoblast induction of arterial apoptosis and lymphangiogenesis in an in vivo model of human placentation. J Clin Invest 116(10):2643–2652
Zhou Y et al (1997) Human cytotrophoblasts adopt a vascular phenotype as they differentiate. A strategy for successful endovascular invasion? J Clin Invest 99(9):2139–2151
Fisher SJ (2015) Why is placentation abnormal in preeclampsia? Am J Obstet Gynecol 213(4 Suppl):S115–S122
King A, Thomas L, Bischof P (2000) Cell culture models of trophoblast II: trophoblast cell lines–a workshop report. Placenta 21(Suppl A):S113–S119
Ringler GE, Strauss JF 3rd (1990) In vitro systems for the study of human placental endocrine function. Endocr Rev 11(1):105–123
Genbacev O et al (2013) Human trophoblast progenitors: where do they reside? Semin Reprod Med 31(1):56–61
Tanaka S et al (1998) Promotion of trophoblast stem cell proliferation by FGF4. Science 282(5396):2072–2075
Hemberger M, Hughes M, Cross JC (2004) Trophoblast stem cells differentiate in vitro into invasive trophoblast giant cells. Dev Biol 271(2):362–371
Latos PA, Hemberger M (2014) Review: the transcriptional and signalling networks of mouse trophoblast stem cells. Placenta 35(Suppl):S81–S85
Natale DR et al (2009) Activin promotes differentiation of cultured mouse trophoblast stem cells towards a labyrinth cell fate. Dev Biol 335(1):120–131
Okae H et al (2018) Derivation of human trophoblast stem cells. Cell Stem Cell 22(1):50-63 e6
Xu RH et al (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20(12):1261–1264
Horii M et al (2016) Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc Natl Acad Sci USA 113(27):E3882–E3891
Amita M et al (2013) Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA 110(13):E1212–E1221
Nichols J, Smith A (2009) Naive and primed pluripotent states. Cell Stem Cell 4(6):487–492
Nakamura T et al (2016) A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537(7618):57–62
Bernardo AS et al (2011) BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9(2):144–155
Roberts RM et al (2014) Differentiation of trophoblast cells from human embryonic stem cells: to be or not to be? Reproduction 147(5):D1-12
Io S et al (2021) Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell 60:S57
Guo G et al (2021) Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 105:733
Seetharam AS et al (2022) The product of BMP-directed differentiation protocols for human primed pluripotent stem cells is placental trophoblast and not amnion. Stem Cell Reports 17:1289–1302
Dong C et al (2020) Derivation of trophoblast stem cells from naive human pluripotent stem cells. Elife. https://doi.org/10.7554/eLife.52504
Cinkornpumin JK et al (2020) Naive human embryonic stem cells can give rise to cells with a trophoblast-like transcriptome and methylome. Stem Cell Reports 15(1):198–213
Castel G et al (2020) Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep 33(8):108419
Dong C, Theunissen TW (2022) Generating trophoblast stem cells from human naive pluripotent stem cells. Methods Mol Biol 2416:91–104
Liu X et al (2020) Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 586(7827):101–107
Sheridan MA et al (2021) Characterization of primary models of human trophoblast. Development. https://doi.org/10.1242/dev.199749
Lee CQ et al (2016) What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Reports 6(2):257–272
Bortolin-Cavaille ML et al (2009) C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res 37(10):3464–3473
Jones CJP, Aplin JD (2021) A re-examination of the origins of placental bed giant cells. Placenta 114:39–41
Morey R et al (2021) Transcriptomic drivers of differentiation, maturation, and polyploidy in human extravillous trophoblast. Front Cell Dev Biol 9:702046
Hayflick L, Moorhead PS (1961) The serial cultivation of human diploid cell strains. Exp Cell Res 25:585–621
Petropoulos S et al (2016) Single-cell RNA-Seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 165(4):1012–1026
Meistermann D et al (2021) Integrated pseudotime analysis of human pre-implantation embryo single-cell transcriptomes reveals the dynamics of lineage specification. Cell Stem Cell 28(9):1625-1640 e6
Xiang L et al (2020) A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 577(7791):537–542
Tyser RCV et al (2020) A spatially resolved single cell atlas of human gastrulation. bioRxiv p. 2020.07.21.213512
Vento-Tormo R et al (2018) Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563(7731):347–353
Liu Y et al (2018) Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res 28(8):819–832
Marsh B et al (2022) Regionally distinct trophoblast regulate barrier function and invasion in the human placenta. Elife. https://doi.org/10.7554/eLife.78829
Fock V et al (2015) Neuregulin-1-mediated ErbB2-ErbB3 signalling protects human trophoblasts against apoptosis to preserve differentiation. J Cell Sci 128(23):4306–4316
Aplin JD et al (1999) Development of cytotrophoblast columns from explanted first-trimester human placental villi: role of fibronectin and integrin alpha5beta1. Biol Reprod 60(4):828–838
Prossler J et al (2014) The relationship between TGFbeta, low oxygen and the outgrowth of extravillous trophoblasts from anchoring villi during the first trimester of pregnancy. Cytokine 68(1):9–15
Chang CW et al (2005) Stimulation of GCMa transcriptional activity by cyclic AMP/protein kinase A signaling is attributed to CBP-mediated acetylation of GCMa. Mol Cell Biol 25(19):8401–8414
Court F et al (2014) Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res 24(4):554–569
Schroeder DI et al (2013) The human placenta methylome. Proc Natl Acad Sci USA 110(15):6037–6042
Gamage T et al (2018) The role of DNA methylation in human trophoblast differentiation. Epigenetics 13(12):1154–1173
Bai T et al (2021) Establishment of human induced trophoblast stem-like cells from term villous cytotrophoblasts. Stem Cell Res 56:102507
Wang LJ et al (2022) Functional antagonism between DeltaNp63alpha and GCM1 regulates human trophoblast stemness and differentiation. Nat Commun 13(1):1626
Wilson RL et al (2019) Characterization of 5-methylcytosine and 5-hydroxymethylcytosine in human placenta cell types across gestation. Epigenetics 14(7):660–671
Wilson SL, Liu Y, Robinson WP (2016) Placental telomere length decline with gestational age differs by sex and TERT, DNMT1, and DNMT3A DNA methylation. Placenta 48:26–33
Hemberger M et al (2010) ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum Mol Genet 19(12):2456–2467
Costa MA (2016) The endocrine function of human placenta: an overview. Reprod Biomed Online 32(1):14–43
Thomson JA et al (1998) Embryonic stem cell lines derived from human blastocysts. Science 282(5391):1145–1147
Takashima Y et al (2014) Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158(6):1254–1269
Theunissen TW et al (2014) Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15(4):471–487
Huang K, Maruyama T, Fan G (2014) The naive state of human pluripotent stem cells: a synthesis of stem cell and preimplantation embryo transcriptome analyses. Cell Stem Cell 15(4):410–415
Stirparo GG et al (2018) Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development. https://doi.org/10.1242/dev.158501
Sahakyan A et al (2017) Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 20(1):87–101
Theunissen TW et al (2016) Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19(4):502–515
Collier AJ et al (2017) Comprehensive Cell Surface Protein Profiling Identifies Specific Markers of Human Naive and Primed Pluripotent States. Cell Stem Cell 20(6):874-890 e7
Bredenkamp N et al (2019) The cell-surface marker Sushi Containing Domain 2 facilitates establishment of human naive pluripotent stem cells. Stem Cell Reports 12:1212–1222
Fischer LA, Khan SA, Theunissen TW (2022) Induction of human naive pluripotency using 5i/L/A medium. Methods Mol Biol 2416:13–28
Rugg-Gunn PJ (2022) Induction of human naive pluripotency using chemical resetting. Methods Mol Biol 2416:29–37
Rostovskaya M (2022) Maintenance of human naive pluripotent stem cells. Methods Mol Biol 2416:73–90
Dong C, Fischer LA, Theunissen TW (2019) Recent insights into the naive state of human pluripotency and its applications. Exp Cell Res 385(1):111645
Liu X et al (2017) Comprehensive characterization of distinct states of human naive pluripotency generated by reprogramming. Nat Methods 14:1055
Kilens S et al (2018) Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat Commun 9(1):360
Giulitti S et al (2019) Direct generation of human naive induced pluripotent stem cells from somatic cells in microfluidics. Nat Cell Biol 21(2):275–286
Onfray C et al (2022) Induction of human naive pluripotent stem cells from somatic cells. Methods Mol Biol 2416:39–51
Zorzan I et al (2022) Using microfluidics to generate human naive and primed pluripotent stem cells. Methods Mol Biol 2416:53–71
Guo G et al (2016) Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports 6(4):437–446
Strawbridge SE et al (2022) Deriving human naive embryonic stem cell lines from donated supernumerary embryos using physical distancing and signal inhibition. Methods Mol Biol 2416:1–12
Niwa H et al (2005) Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123(5):917–929
Gardner RL (1983) Origin and differentiation of extraembryonic tissues in the mouse. Int Rev Exp Pathol 24:63–133
Gerri C et al (2020) Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature 587(7834):443–447
Meistermann D et al (2021) Integrated pseudotime analysis of human pre-implantation embryo single-cell transcriptomes reveals the dynamics of lineage specification. Cell Stem Cell. https://doi.org/10.1016/j.stem.2021.04.027
Radley A, Smith AG, Dunn SJ (2022) Functional feature selection reveals the inner cell mass in human pre-implantation embryo single cell RNA sequencing data. bioRxiv
Kagawa H et al (2022) Human blastoids model blastocyst development and implantation. Nature 601(7894):600–605
Messerschmidt DM, Kemler R (2010) Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev Biol 344(1):129–137
Chazaud C et al (2006) Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell 10(5):615–624
Ralston A, Rossant J (2008) Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol 313(2):614–629
De Paepe C et al (2013) Human trophectoderm cells are not yet committed. Hum Reprod 28(3):740–749
Pontis J et al (2019) Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell 24(5):724-735 e5
Io S et al (2021) Capturing human trophoblast development with naive pluripotent stem cells in vitro. Cell Stem Cell 28(6):1023-1039 e13
Haider S et al (2018) Self-renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem Cell Reports 11(2):537–551
Turco MY et al (2018) Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 564(7735):263–267
Karvas RM et al (2022) Stem-cell-derived trophoblast organoids model human placental development and susceptibility to emerging pathogens. Cell Stem Cell 29(5):810-825 e8
Sheridan MA et al (2020) Establishment and differentiation of long-term trophoblast organoid cultures from the human placenta. Nat Protoc 15(10):3441–3463
Li Z, Kurosawa O, Iwata H (2019) Establishment of human trophoblast stem cells from human induced pluripotent stem cell-derived cystic cells under micromesh culture. Stem Cell Res Ther 10(1):245
Mischler A et al (2021) Two distinct trophectoderm lineage stem cells from human pluripotent stem cells. J Biol Chem 296:100386
Wei Y et al (2021) Efficient derivation of human trophoblast stem cells from primed pluripotent stem cells. Sci Adv. https://doi.org/10.1126/sciadv.abf4416
Jang YJ et al (2022) Induction of human trophoblast stem-like cells from primed pluripotent stem cells. Proc Natl Acad Sci USA 119(20):e2115709119
Soncin F et al (2022) Derivation of functional trophoblast stem cells from primed human pluripotent stem cells. Stem Cell Reports
Viukov S et al (2022) Human primed and naïve PSCs are both competent in differentiating into bona fide trophoblast stem cells. bioRxiv
Kobayashi N et al (2022) The microRNA cluster C19MC confers differentiation potential into trophoblast lineages upon human pluripotent stem cells. Nat Commun. https://doi.org/10.1038/s41467-022-30775-w
Boroviak T, Nichols J (2017) Primate embryogenesis predicts the hallmarks of human naïve pluripotency. Development 144(2):175–186
Posfai E et al (2021) Evaluating totipotency using criteria of increasing stringency. Nat Cell Biol 23(1):49–60
Guo G et al (2021) Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 28(6):1040-1056 e6
Rostovskaya M et al (2022) Amniogenesis occurs in two independent waves in primates. Cell Stem Cell 29(5):744-759 e6
Pastor WA et al (2016) Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell 18(3):323–329
Zhou F et al (2019) Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 572:660–664
Ohgushi M, Eiraku M (2021) Cell-autonomous differentiation of human primed embryonic stem cells into trophoblastic syncytia through the nascent amnion-like cell state. bioRxiv
Yu L et al (2021) Blastocyst-like structures generated from human pluripotent stem cells. Nature 591:620–626
Yanagida A et al (2021) Naive stem cell blastocyst model captures human embryo lineage segregation. Cell Stem Cell 28(6):1016-1022 e4
Motomura K et al (2016) Cellular dynamics of mouse trophoblast stem cells: identification of a persistent stem cell type. Biol Reprod 94(6):122
Kuales G et al (2015) A resource for the transcriptional signature of bona fide trophoblast stem cells and analysis of their embryonic persistence. Stem Cells Int 2015:218518
Frias-Aldeguer J et al (2020) Embryonic signals perpetuate polar-like trophoblast stem cells and pattern the blastocyst axis. BioRxiv
Smith GC (2010) First-trimester determination of complications of late pregnancy. JAMA 303(6):561–562
Barker DJ, Thornburg KL (2013) Placental programming of chronic diseases, cancer and lifespan: a review. Placenta 34(10):841–845
Audette MC, Kingdom JC (2018) Screening for fetal growth restriction and placental insufficiency. Semin Fetal Neonatal Med 23(2):119–125
Burton GJ, Jauniaux E (2018) Pathophysiology of placental-derived fetal growth restriction. Am J Obstet Gynecol 218(2S):S745–S761
Dimitriadis E et al (2020) Recurrent pregnancy loss. Nat Rev Dis Primers 6(1):98
Cheung AN et al (2009) Pathogenesis of choriocarcinoma: clinical, genetic and stem cell perspectives. Future Oncol 5(2):217–231
Saha B et al (2020) TEAD4 ensures postimplantation development by promoting trophoblast self-renewal: An implication in early human pregnancy loss. Proc Natl Acad Sci USA 117(30):17864–17875
Perez-Garcia V et al (2021) BAP1/ASXL complex modulation regulates epithelial-mesenchymal transition during trophoblast differentiation and invasion. Elife. https://doi.org/10.7554/eLife.63254
Shannon MJ et al (2022) Cell trajectory modeling identifies a primitive trophoblast state defined by BCAM enrichment. Development. https://doi.org/10.1242/dev.199840
Dong C et al (2022) A genome-wide CRISPR-Cas9 knockout screen identifies essential and growth-restricting genes in human trophoblast stem cells. Nat Commun 13(1):2548
Chen Y et al (2022) An integrated atlas of human placental development delineates essential regulators of trophoblast stem cells. Development. https://doi.org/10.1242/dev.200171
Acknowledgements
We thank Chen Dong for critical reading of the manuscript.
Funding
Research in the Theunissen laboratory is supported by the NIH Director's New Innovator Award (DP2 GM137418) and grants from the Shipley Foundation Program for Innovation in Stem Cell Science, the Edward Mallinckrodt, Jr. Foundation, and the Washington University Children's Discovery Institute. R.M.K. is supported by a Training in Regenerative Medicine training grant (T32 EB028092) from the NIH. Federal NIH/NIGMS funds were not used to develop integrated 3D models of human embryonic development.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
R.M.K. and T.W.T. are co-inventors on a patent application related to trophoblast organoids. L.D. is inventor of the patent WS 2019-093-04 related to induction of hiTSCs.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Karvas, R.M., David, L. & Theunissen, T.W. Accessing the human trophoblast stem cell state from pluripotent and somatic cells. Cell. Mol. Life Sci. 79, 604 (2022). https://doi.org/10.1007/s00018-022-04549-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04549-y