Abstract
Human monocarboxylate transporters (hMCTs/SLC16As) mediate the transport of monocarboxylic compounds across plasma membranes. Among the hMCTs, hMCT1 and hMCT4 are expressed in various tissues, and transport substrates involved in energy metabolism. Both transporters mediate l-lactate transport, but, although hMCT1 also transports l-5-oxoproline (l-OPro), this compound is minimally transported by hMCT4. Thus, we were interested in the molecular mechanism responsible for the difference in substrate specificity between hMCT1 and hMCT4. Therefore, we generated 3D structure models of hMCT1 and hMCT4 to identify amino acid residues involved in the substrate specificity of these transporters. We found that the substrate specificity of hMCT1 was regulated by residues involved in turnover number (M69) and substrate affinity (F367), and these residues were responsible for recognizing (directly or indirectly) the –NH– moiety of l-OPro. Furthermore, our homology model of hMCT1 predicted that M69 and F367 participate in hydrophobic interactions with another region of hMCT1, emphasizing its potentially important role in the binding and translocation cycle of l-OPro. Mutagenesis experiments supported this model, showing that efficient l-OPro transport required a hydrophobic, long linear structure at position 69 and a hydrophobic, γ-branched structure at position 367. Our work demonstrated that the amino acid residues, M69 and F367, are key molecular elements for the transport of l-OPro by hMCT1. These two residues may be involved in substrate recognition and/or substrate-induced conformational changes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human monocarboxylate transporters (hMCTs/SLC16As) are transport proteins of the major facilitator superfamily (MFS), and they mediate the absorption and distribution of monocarboxylic compounds across the plasma membrane in various tissues [1]. Among the hMCTs, hMCT1 and hMCT4 are responsible for the pH-related transport of l-lactate, a common substrate in both normal and cancer cells [1,2,3,4,5]. To facilitate investigations of the physiological roles of hMCT1 and hMCT4, selective substrates and inhibitors of each transporter have been developed. In the previous studies, several monocarboxylate derivatives have been created as selective inhibitors of human or rodent MCT1 [6,7,8,9,10]. Our recent studies demonstrate that l-5-oxoproline (l-OPro) is a selective hMCT1 substrate [11]. l-OPro is an intermediate metabolite in the glutathione cycle. In this cycle, the glutathione formed is degraded by γ-glutamyl cyclotransferase to yield l-OPro and cysteinyl glycine [12]. The physiological role of l-OPro as an analog or a reservoir of glutamate, or an osmoprotectant has been discussed in a recent study [13]. Moreover, several studies have shown that the serum concentration of l-OPro may serve as a biomarker for several diseases [14,15,16]. In addition, Thiesen et al. [17] showed that 3-hydroxycyclopent-1-enecarboxylic acid (HOCPCA; γ-hydroxybutyric acid analog) is a selective substrate for rat MCT1/2. Although our recent research has identified bindarit (2-[(1-benzyl-1H-indazol-3-yl)methoxy]-2-methylpropanoic acid) as a non-competitive, selective inhibitor of hMCT4 [18], a specific substrate for hMCT4 has not yet been identified. Thus, we were interested in the molecular mechanism responsible for the substrate specificity difference between hMCT1 and hMCT4.
Our previous studies showed that the intracellular loop 3 (TM6/7loop) of hMCT1 and hMCT4 contribute to the substrate affinity of the transporters, whereas their substrate specificity depends on other domains [5]. It has been reported that K142 and R143 residues in cytoplasmic loop 2 (TM4/5loop) are involved in the stereoselectivity of rat MCT1 [19]. Moreover, substrate specificity is affected by specific residues in transmembrane domain 10 (TM10). It is known that the F360C mutant of rodent MCT1 allows mevalonate to be transported [20, 21]. The most effective way to identify the residues involved in substrate specificity is to determine the co-crystal structures of transporters with their substrates [22,23,24,25,26,27,28]. However, the crystal structures of hMCT1 and hMCT4 are currently not available. In such cases, the homology modeling technique is valuable for predicting 3D structures of proteins. Nancolas et al. [29] recently generated molecular models of rat MCT1 to identify key binding site residues for AR-C155858, a selective inhibitor of rat MCT1/2. They also showed that the residues near R306, which is an essential residue for transport in TM8, are not always conserved between rat MCT1 and MCT4. Furthermore, as predicted by an hMCT8 homology model, the substrate specificity of hMCT8 and hMCT10, in substrate translocation pathways, is altered by the 2–3 amino acid differences between the two proteins [30]. In addition, several studies have also identified differences in the substrate specificity of transport proteins using the homology modeling technique [31,32,33,34]. Based on these studies, we speculated that the substrate specificity of hMCT1 and hMCT4 is regulated by amino acid residues involved in substrate translocation pathways. Therefore, we generated 3D structure models of hMCT1 and hMCT4 to identify amino acid residues involved in the substrate specificity of these transporters.
Materials and methods
Materials
Sodium l-[14C]lactate was purchased from PerkinElmer (Waltham, MA, USA). l-5-[3H]Oxoproline ([3H]pyroglutamic acid) was purchased from Moravek (Brea, CA, USA). Sodium l-lactate was purchased from Sigma-Aldrich (St. Louis, MO, USA). Cyclopentanecarboxylic acid and l-OPro (l-pyroglutamic acid) were purchased from Tokyo Chemical Industry (Tokyo, Japan). (R)-3-Oxocyclopentanecarboxylic acid was purchased from AstaTech (Bristol, PA, USA). All nucleotide primers were synthesized by Eurofins Genomics (Tokyo, Japan). Xenopus laevis frogs were purchased from Hokudo (Sapporo, Hokkaido, Japan). This study was approved by the National University Corporation Hokkaido University Animal Experimentation Committee. All compounds were of the highest purity available.
Plasmids and site-directed mutagenesis
pGH19-hMCT1 and pGH19-hMCT4 plasmids were obtained as described previously [35]. hMCT1 and hMCT4 mutants were generated using the QuikChange site-directed mutagenesis protocol (Agilent Technologies, Santa Clara, CA, USA). Sequences of the primers used for site-directed mutagenesis are provided in Table S1. All mutations were confirmed by DNA sequencing, which was performed by Hokkaido System Science (Sapporo, Hokkaido, Japan).
Expression of cRNA in Xenopus oocytes and immunohistochemistry
Oocyte preparation, in vitro transcription, and cRNA microinjection were performed as outlined elsewhere [35]. Water-injected oocytes were used as negative controls. Immunohistochemical analysis was performed as described elsewhere, with some modifications [4]. Oocytes were stained with a mouse anti-hMCT1 antibody (sc-365501; Santa Cruz Biotechnology, Dallas, TX, USA), which recognizes the intracellular loop 3 of hMCT1, or a rabbit anti-hMCT4 antibody (22787-1-AP; Proteintech Group, Rosemont, IL, USA), which recognizes the C-terminal cytoplasmic domain of hMCT4. The prepared oocytes were monitored under an FV10i confocal laser scanning microscope (Olympus Corporation, Tokyo, Japan).
Substrate transport assays
l-[14C]lactate (1 μM; 0.14 µCi/mL) or l-5-[3H]oxoproline (0.2 μM; 0.5 µCi/mL) uptake by oocytes was measured as detailed previously [5, 18]. In brief, oocytes were placed on 24-well plates and were incubated at 25 °C in a standard buffer (100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 10 mM 2-morpholinoethanesulfonic acid, pH 5.5), containing the radiotracers. The incubation time point (l-[14C]lactate, 10 min; l-[3H]Opro, 15 min) was chosen in the linear portion of the time-course curve, as determined in our previous work [4, 11, 35].
Sequence alignment
Multiple protein sequence alignments were performed between vertebrate MCT1 (human, NP_001159968; rat, NP_036848; chicken, NP_001006323; green anole, XP_003220493; western clawed frog, NP_001015931; zebrafish, NP_001243569 [MCT1a] and NP_956379 [MCT1b]) and MCT4 (human, NP_001035887; rat, NP_110461; chicken, NP_989994; green anole, XP_008102768; western clawed frog, NP_001007912; zebrafish, NP_997873). Alignment were generated using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) using default parameters [36] and were visualized using GeneDoc [37].
Homology modeling and domain analysis
3D structure models of hMCT1 and hMCT4 were generated by performing homology modeling using the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) [38]. The amino acid sequences of hMCT1 and hMCT4 were submitted to the Phyre2 server for modeling in the “intensive mode”. Detailed information about the entire homology modeling procedure has been described previously [38]. In brief, initially, the protein sequences were scanned with HHblits method [39] against a protein sequence database, where no pair of sequences exhibited > 20% identity. The resulting multiple-sequence alignment was utilized for the prediction of secondary structure with PSI-BLAST-based secondary structure prediction [40]. The results from the alignment and secondary structure prediction were combined into a query hidden Markov model (HMM). This query HMM was then scanned against a database containing HMMs of known protein structures (HHsearch) [41]. The top-scoring alignments from HHsearch were used to construct crude backbone-only models. The insertions and deletions in these models were handled using a library of fragments of lengths ranging 2–15 amino acids obtained from known protein structures. Furthermore, multi-template modeling was carried out using Poing [42], a simplified protein-folding simulator, to create a complete model of the query protein even when different regions or domains are modeled by separate templates, or when there are no templates at all (ab initio modeling). Finally, the amino acid side chains were added to the backbone using the R3 protocol [43] to generate the final Phyre2 model. During the modeling process, six templates (MFS transporters) were selected to model each protein, based on heuristics to maximize confidence levels, percentage identity, and alignment coverage (Fig. S1). PDB accession numbers 1PW4 (inward open) [44], 4IU8 (inward-facing occluded) [27], 4LDS (inward open) [45], 3WDO (outward open) [46], 4CL5 (inward open) [24], and 4J05 (inward-facing occluded) [26] were used for the hMCT1 model and 1PW4 (inward open) [44], 3WDO (outward open) [46], 4IU8 (inward-facing occluded) [27], 4CL5 (inward open) [24], 4ZP0 (inward open) [23], and 4J05 (inward-facing occluded) [26] were used for the hMCT4 model. The energy minimization of the final Phyre2 models was performed using the 3Drefine server (http://sysbio.rnet.missouri.edu/3Drefine/) [47]. The 3Drefine server performs protein structure refinement by making use of iterative optimization of hydrogen bonding network associated with atomic-level energy minimization on the optimized model by employing composite physics and knowledge-based force fields. Ramachandran plot analysis using RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) was performed to predict the stereochemical quality of the homology models [48]. The Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/) was searched for putative substrate translocation pore domains of MFS proteins (PSSM-ID: 119392), using CD search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) [49, 50]. We performed the multiple structure alignment for the models of hMCT1 and hMCT4 using Matt [51]. The models were visualized and analyzed using PyMOL 2.1.0 (Schrödinger, New York, NY, USA).
Molecular docking analysis
The 3D structures of l-lactate and l-OPro were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) [52]: PubChem CIDs 107689 (l-lactate) and 7405 (l-OPro). The energy minimization of each ligand was performed with UCSF Chimera 1.13.1 [53] using the following set of parameters: 1000 steepest descent steps with step size 0.02 Å and 1000 conjugate gradient steps with step size 0.02 Å. Furthermore, the hydrogen atoms and charges were added into the ligand structures as well as the energy-minimized hMCT1 model using the Dock Prep function in UCSF Chimera 1.13.1 with default parameters [54, 55]. Molecular docking analysis was carried out using the software AutoDock Vina [56]. The grid box parameters were set with center (X = 34, Y = 17, and Z = − 35) and size (X = 29, Y = 13, and Z = 18), and were large enough to cover M69, R313, and F367 residues. The default settings were used for all other parameters. All the ligands were docked separately against hMCT1 model. The top nine conformations of the generated docked complexes were selected after evaluating the binding free energy of each conformation.
Data analysis
To determine inhibition constants (Ki), the data were fitted to a two-parameter hyperbolic decay equation using SigmaPlot 14 (Systat Software, San Jose, CA, USA), as described previously [18]. Initial uptake clearances (CL = Vmax/Km) for l-lactate and l-OPro were determined from the ratio of the initial uptake rate over the substrate concentration, which is much less than the Km values of the substrates [4, 11, 35]. Because Ki = Km when an inhibitor is also transported by an identical mechanism to the compound designated as the substrate [57], turnover number (kcat) ratios of the transporters were estimated from the Ki and CL values, as follows:
All experimental data are expressed as the mean ± SE. Structure property predictions and calculations were performed using the Calculator Plugins of MarvinSketch 19.2.0 (2019; ChemAxon, Budapest, Hungary).
Statistical analysis
Group results were compared by one-way ANOVA, followed by the Holm–Sidak post hoc test with multiple comparisons or by unpaired t test, when appropriate. Data were analyzed using SigmaPlot 14, and p < 0.05 was considered statistically significant.
Results
Homology modeling and putative substrate translocation pathway of hMCT1 and hMCT4
Our previous reports have shown that l-OPro is a selective hMCT1 substrate that has inhibitory effects on hMCT1 (at 10 mM), but not on hMCT4 [11, 18]. To investigate the inhibitory properties of l-OPro on hMCT1 and hMCT4, we performed a dose–response experiment using l-OPro concentrations ranging from 0 to 100 mM (Fig. 1a). l-OPro inhibited hMCT1 with a Ki value of 5.9 ± 0.7 mM. However, no inhibitory effect was observed on hMCT4, even at the highest concentration tested (100 mM). These data indicated that l-OPro was selectively recognized by hMCT1. The previous reports on rat MCT1, hMCT4, and hMCT8 have identified an Arg residue located in TM8 that is essential for substrate transport [4, 21, 58]. However, the role of this Arg residue in hMCT1 (R313) has not been reported. Multiple protein sequence alignments of vertebrate MCT1 and MCT4 proteins showed that this Arg residue is conserved in hMCT1 (Fig. 1b). Substrate transport activity was drastically diminished in hMCT1-R313Q, but membrane protein expression was not affected (Fig. 1c, d), indicating that this Arg residue is essential for substrate transport, not only in rat MCT1, hMCT4, and hMCT8, but also in hMCT1.
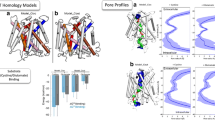
The conserved TM8 Arg residue is required for l-lactate and l-OPro transport by hMCT1. a The uptake of l-[14C]lactate was monitored in the presence of increasing doses of l-OPro (0–100 mM). The Ki value was determined using non-linear fitting, as described in the “Materials and methods” section. Transporter-specific uptake was calculated by subtracting the uptake in water-injected oocytes from the uptake in cRNA-injected oocytes. Values represent the mean ± SE from three independent experiments using a single batch of oocytes, each performed with 3–5 replicates. b Multiple protein sequence alignments were performed between vertebrate MCT1 and MCT4 (hsa, human; rno, rat; gga, chicken; acs, green anole; xtr, western clawed frog; dre, zebrafish). The alignments were generated using Clustal Omega, under default parameters and were visualized using GeneDoc. The conserved TM8 Arg residues of vertebrate MCT1 and MCT4 are marked in red. cl-[14C]Lactate and l-[3H]OPro uptake via hMCT1-WT and -R313Q compared to controls (water-injected oocytes). Values represent the mean ± SE from three independent experiments using a single batch of oocytes, each performed with 3–5 replicates. d Localization of hMCT1-WT and -R313Q in the oocyte membrane. Oocytes were treated with antibodies against hMCT1. e, f 3D structure models of hMCT1 (e) and hMCT4 (f) were generated by the homology modeling technique, and the TMs are numbered. Six templates were selected to model each protein: PDB accession numbers 1PW4, 4IU8, 4LDS, 3WDO, 4CL5, and 4J05 for the hMCT1 model and 1PW4, 3WDO, 4IU8, 4CL5, 4ZP0, and 4J05 for the hMCT4 model. g Homology model of hMCT1 superimposed upon the model of hMCT4. The putative substrate translocation pathways of the transporters are shown as sticks
To reconcile the observed differences in substrate specificity between hMCT1 and hMCT4, homology modeling and domain analysis were used to identify the differences in substrate translocation pathways. As reported by Quistgaard et al., all MFS transporters share the same structural fold [59]. Since hMCTs, including hMCT1 and hMCT4, are members of the MFS family, these transporters are anticipated to form similar structures to the experimentally solved 3D structures of MFS transporters [60]. Here, we generated 3D structure models of hMCT1 and hMCT4 using Phyre2 [38], and the models were further energy minimized using 3Drefine (Fig. 1e, f) [47]. Although the TM6 domain of the hMCT1 structure was not modeled as a complete α-helix, these homology models had 12 TM helices, which is typical of MFS transporters [60]. The stereochemical quality of the predicted models was evaluated by Ramachandran plot analysis using RAMPAGE (Fig. S2) [48]. This analysis demonstrated that most of the residues were sterically allowed (95% for hMCT1; 97% for hMCT4), suggesting that the obtained models are of good quality. Next, we identified the putative substrate translocation pathways of hMCT1 and hMCT4, which are each comprised of 51 amino acid residues, based on the Conserved Domain Database (Figs. 1g, S3) [49, 50]. All of these residues fell within the favored region in the Ramachandran plot analysis (Fig. S2).
M69 and F367 in hMCT1 are key molecular determinants of substrate specificity
To identify the amino acid residues involved in the substrate specificity of hMCT1 and hMCT4, we focused on non-conserved residues, which constitute the putative substrate translocation pathways. As shown in Table 1 and Fig. S3, 20 residues were found in each of hMCT1 and hMCT4. We, then, performed residue substitution experiments between the non-conserved residues of hMCT1 and hMCT4, and examined whether substrate specificity was altered by the mutations. We first assessed the transport activity of the substitution mutants of hMCT1, using l-lactate and l-OPro (Fig. 2a). l-Lactate transport activity was significantly reduced in F274 V, L281P, and F367Y mutants, while l-OPro transport activity was significantly reduced in M69L, L281P, F367Y, and T395L mutants. We also confirmed membrane localization of the hMCT1 mutants using immunohistochemistry (Fig. S4a). Appropriate expression in the oocyte plasma membrane was observed for all mutants, except L281P, which showed decreased membrane expression. These results indicated that M69, F274, F367, and T395 residues are involved in substrate translocation or that the mutated residues interfere with this process. However, the L281 residue appeared to participate in protein trafficking, since a proline residue at this position interfered with this process. To analyze changes in substrate specificity, normalized initial uptake clearance of l-OPro (CLl-OPro/CLl-Lactate) was calculated (Fig. 2b). Uptake clearance was markedly decreased in M69L and F367Y mutants, indicating that M69 and F367 residues play important roles in substrate specificity. We next examined the substrate transport activity of the 20 reciprocal mutants of hMCT4 (Fig. 2c). l-Lactate transport activity was significantly reduced in L67M, F130L, Q131N, V239F, P246L, V250S, G271A, G336S, L360T, and A364C mutants. No l-OPro transport activity was detected in any of the mutants tested, including hMCT4-L71M and -Y332F, which are the reciprocal mutants of hMCT1-M69L and -F367Y, respectively. Appropriate membrane localization was observed in all mutants, except for P246L, which exhibited reduced protein expression (Fig. S4b). These data suggested that the P246 residue and the equivalent residue in hMCT1, L281, are important for appropriate protein trafficking.
M69 and F367 are key molecular determinants of substrate specificity in hMCT1. al-[14C]Lactate and l-[3H]OPro uptake by the substitution mutants of hMCT1. ap < 0.05, dp < 0.001 versus WT (l-lactate), and Ap < 0.05, Cp < 0.005, Dp < 0.001 versus WT (l-OPro). b Normalized l-OPro initial uptake clearance (CLl-OPro/CLl-Lactate) for the substitution mutants of hMCT1. Transporter-specific uptake was calculated by subtracting the uptake in water-injected oocytes from the uptake in cRNA-injected oocytes. ap < 0.05, cp < 0.005, dp < 0.001 versus WT. cl-[14C]Lactate and l-[3H]OPro uptake by the substitution mutants of hMCT4. ap < 0.05; dp < 0.001 versus WT (l-lactate). Values represent the mean ± SE from 3 to 12 independent experiments using different batches of oocytes, each performed with 3–5 replicates
Substrate transport via multiple mutants of hMCT1 and hMCT4
On the basis of single mutation results, we constructed multiple mutants of hMCT1 (double, M69L/F367Y; novemdecuple, I40V/T41S/M65L/M69L/S76C/L128F/N129Q/T133I/G136N/M151A/F274V/S285V/S298A/A306G/F367Y/S371G/V391I/T395L/C399A) and hMCT4 (double, L71M/Y332F; undecuple, V42I/S43T/L67M/L71M/C78S/I135T/N138G/A153M/A263S/Y332F/I356V), and measured their transport activity using l-lactate and l-OPro (Fig. 3a). The l-lactate transport activity of hMCT1-M69L/F367Y remained ~ 50% of wild-type (WT) levels, while the l-OPro transport activity almost completely disappeared. However, the reciprocal hMCT4 double mutant (hMCT4-L71 M/Y332F) showed almost the same l-lactate transport activity as hMCT4-WT, but no l-OPro transport activity. These results were consistent with the observations in the single mutation study (Fig. 2a, c). Moreover, the novemdecuple mutant of hMCT1 and the undecuple mutant of hMCT4 showed no substrate transport activity (Fig. 3a). We then assessed protein expression levels and translocation of the multiple mutants to the plasma membrane (Fig. S5). Correct protein expression levels and localization were observed in all multiple mutants, except for the novemdecuple mutant of hMCT1, which showed decreased protein expression. These results indicated that the native residues where the mutations were introduced work cooperatively in regulating substrate transport in hMCT1 and hMCT4, since the single mutations, other than M69L and F367Y in hMCT1, had only minor effects on substrate transport activity (Fig. 2a, c). In addition, these native residues in hMCT1 work cooperatively to ensure membrane localization of the transporter, since the single mutations did not affect its localization (Fig. S4). We then examined changes in substrate specificity by calculating CLl-OPro/CLl-Lactate values (Fig. 3b). Uptake clearance was greatly diminished in hMCT1-M69L/F367Y, which is consistent with the findings in the single mutation assay (Fig. 2b).
Substrate transport by the multiple mutants of hMCT1 and hMCT4. al-[14C]Lactate and l-[3H]OPro uptake by the multiple mutants of hMCT1 and hMCT4. b Normalized l-OPro initial uptake clearance (CLl-OPro/CLl-Lactate) of the multiple mutants of hMCT1 and hMCT4. Transporter-specific uptake was calculated by subtracting the uptake in water-injected oocytes from the uptake in cRNA-injected oocytes. NC, not calculated; dp < 0.001 versus hMCT1-WT. Values represent the mean ± SE from 3 to 6 independent experiments using different batches of oocytes, each performed with 3–5 replicates
Characterization of the inhibitory kinetics of l-lactate transport via hMCT1 and hMCT4 mutants
We identified the residues related to substrate specificity, M69 and F367, in hMCT1 (L71 and Y332 residues in hMCT4). To further explore the function of these residues, we characterized the effect of substitution mutations on the inhibitory kinetics of hMCT1 and hMCT4. We first investigated the ability of l-lactate and l-OPro to inhibit l-[14C]lactate transport by hMCT1-M69L, -F367Y, and -M69L/F367Y (Fig. 4a–c). The M69L mutant had an approximately twofold increase in the Ki values for l-lactate and l-OPro, whereas the F367Y mutant had an approximately twofold increase in the Ki value for l-lactate and an approximately fourfold increase in the Ki value for l-OPro (Table 2). In addition, the M69L/F367Y double mutant had an approximately sixfold increase in the Ki value for l-lactate and an approximately 13-fold increase in the Ki value for l-OPro (Tables 2 and 3). These results indicated that M69 and F367 residues are involved in determining the substrate affinity of hMCT1. We also calculated the values for normalized l-OPro affinity (Ki, l-OPro/Ki, l-Lactate) and normalized l-OPro turnover number (kcat, l-OPro/kcat, l-Lactate), as indicators of differences in substrate specificity between hMCT1 and its mutants. Although a statistical comparison of the values was not performed, since they are ratios of mean Ki or kcat values, the data indicated that the normalized affinity of l-OPro for the F367Y mutant was higher than that for WT hMCT1, and the normalized l-OPro turnover number for the M69L mutant was lower than that for WT hMCT1 (Table 2). Furthermore, the normalized l-OPro affinity and turnover number for the M69L/F367Y double mutant were altered in the same manner as the single mutants (Table 3). These results suggested that the substrate specificity of hMCT1 was regulated by the residues involved in turnover number (M69) and substrate affinity (F367). Similarly, we investigated the inhibitory kinetics of the reciprocal mutants of hMCT4 (L71M, Y332F, and L71M/Y332F; Fig. 4d, e). Y332F and L71M/Y332F mutants had a significant, approximately 0.2-fold, decrease in the Ki value of l-lactate, which is consistent with the increase in Ki value seen in the reciprocal hMCT1 mutants (F367Y and M69L/F367Y). However, the Ki value of l-lactate in the L71M mutant was not significantly different from that in WT hMCT4 (Table 2). No inhibitory effect was observed with l-OPro in WT or mutant hMCT4 (Fig. 4e), which agreed with the finding that l-OPro transport activity was not observed in the mutants (Figs. 2c, 3a).
Characterization of the inhibitory kinetics of l-lactate transport by hMCT1 and hMCT4 mutants. a–el-[14C]Lactate uptake by hMCT1 (a–c) and hMCT4 (d, e) mutants in the presence of unlabeled l-lactate (a, d) or l-OPro (b, c, e). Data are expressed as % of control (absence of competitor) after the subtraction of uptake into water-injected oocytes. Values represent the mean ± SE from three independent experiments, each performed with 3–5 replicates. Each experiment shown in the panels a–e was performed using a single batch of oocytes. f 3D structure models of hMCT1 with the M69 and F367 residues (magenta) and their neighboring residues (green). g, h Interactions between the key residues involved in substrate specificity (M69 [g] and F367 [h]) and their neighboring residues (distances in Å). The helices are numbered as shown in Fig. 1e
We examined the homology model of hMCT1 to visualize possible interactions between the key residues involved in substrate specificity (M69 and F367) and their neighboring residues. The side chains of M69 and F367 were facing toward the putative substrate translocation pathway (Figs. 1g, 4f), and may form hydrophobic interactions (≤ 4.5 Å) with other aliphatic segments of hMCT1 (Fig. 4g, h). The side chain of M69 in TM2 appeared to form hydrophobic interactions with L66 and Y70 in TM2 and with G123 and L124 in TM4 (Fig. 4g). Four residues (D309 in TM8, and F363, G364, and G368 in TM10) appeared to participate in hydrophobic interactions with F367 in TM10 (Fig. 4h). The turnover changes seen in the M69L mutant and the substrate affinity changes seen in the F367Y mutant may be caused by disruption of the hydrophobic interactions between these residues and their neighboring residues.
The role of position 69 and 367 in the substrate specificity of hMCT1
To determine how the unique chemical properties of M69 and F367 residues affect substrate transport and specificity, we measured l-lactate and l-OPro uptake by a panel of hMCT1 mutants, with each possible amino acid substituted at positions 69 and 367 (M69X and F367X). The appropriate localization of the expressed protein to the oocyte plasma membrane was confirmed, except for M69P and F367P mutants, which showed decreased membrane expression (Fig. S6), suggesting that these mutations may cause protein misfolding. In M69X mutants, the introduction of aromatic or positively charged residues resulted in the loss of substrate transport activity (Fig. 5a, b). In particular, the substitution of M69 to lysine (sterically conserved, but more polar than methionine) impaired substrate transport ability, suggesting that a hydrophobic environment at this position is important for substrate transport. All the remaining mutants transported the substrates, but the mutant CLl-OPro/CLl-Lactate values were significantly decreased compared to the WT values (Fig. 5c). These observations indicated that the long linear structure of a Met side chain is needed at this position to transport l-OPro effectively. In F367X mutants, the clear trend was that only large aliphatic/aromatic residues enabled l-lactate transport (Fig. 5d, e), albeit at levels significantly below those measured in the WT protein. In addition, little substrate transport was observed when F367 was replaced with the sterically similar, but basic, His residue, emphasizing the importance of a hydrophobic moiety at this position to transport the substrates. These results suggested that other large hydrophobic functional groups may be substituted for the benzyl group of F367 and still preserve substrate transport. To examine changes in substrate specificity, CLl-OPro/CLl-Lactate values were calculated (Fig. 5f). Uptake clearance was significantly decreased in all mutants, but the F367L mutant had a relatively small effect. These data indicated that a γ-branched structure is required at position 367 for efficient l-OPro transport.
The role of position 69 and 367 in determining the substrate specificity of hMCT1. a, dl-[14C]Lactate and l-[3H]OPro transport activity of M69X (a) and F367X (d) mutants of hMCT1. cp < 0.005, dp < 0.001 versus WT (l-lactate), and Dp < 0.001 versus WT (l-OPro). b, e These data were taken from Fig. 5a and d. Data are expressed as % of control (hMCT1-WT) after the subtraction of uptake into water-injected oocytes. Different colors represent different amino acid characteristics: red, acidic; orange, amide; yellow, sulfur; green, aliphatic; cyan, hydroxy; blue, basic; magenta, aromatic; black, Gly/Pro. c, f Normalized l-OPro initial uptake clearance (CLl-OPro/CLl-Lactate) by the M69X (c) and F367X (f) mutants. Transporter-specific uptake was calculated by subtracting the uptake in water-injected oocytes from the uptake in cRNA-injected oocytes. NC, not calculated; bp < 0.01, dp < 0.001 versus WT. Values represent the mean ± SE from 3 to 6 independent experiments using different batches of oocytes, each performed with 3–5 replicates
Structure–affinity relationship of small monocarboxylates for hMCT1 mutants, M69L and F367Y
To examine the effect of M69L and F367Y mutations on the ligand-binding affinity of hMCT1, we evaluated its interaction with l-OPro analogs. We investigated the ability of cyclopentanecarboxylate (CPC) and (R)-3-oxocyclopentanecarboxylate (l-OCPC) to inhibit l-[14C]lactate transport via M69L and F367Y mutants (Fig. 6a, b). Although these compounds inhibited the transport activity of M69L and F367Y mutants, the Ki values of CPC and l-OCPC in the mutants did not significantly increase compared to those in the WT protein (Table 4). By comparing the Ki values of l-OCPC and l-OPro (Fig. 6c), the effect of replacing the nitrogen atom at position 1 with carbon was assessed. This replacement was shown to increase ligand recognition > 8-fold in the M69L and F367Y mutants. These observations suggested that M69 and F367 residues are responsible for recognizing (directly or indirectly) the –NH– moiety of l-OPro. Our in silico docking analysis revealed the potential interactions between l-OPro and the residues (Fig. 6d). Notably, one of the l-OPro conformers showed possible interactions between the amide nitrogen atom on l-OPro and aromatic carbon atoms on F367 residue. However, the Ki values in hMCT1-WT, -M69L, and -F367Y were similar between CPC and l-OCPC (< 2-fold; Fig. 6c), suggesting that an oxo group in l-OCPC has minimal involvement in the interaction with hMCT1. In addition, by comparing the Ki values of CPC and l-lactate, the effect of core structure rigidification and the replacement of oxygen with carbon in the β-position were assessed. This replacement decreased ligand recognition > 6.5-fold in hMCT1-WT. These data implied that M69 and F367 residues play a role in recognizing (directly or indirectly) the flexible polar structure of l-lactate. This suggestion was supported by the in silico simulations showing possible interactions between l-lactate and the residues (Fig. 6e).
Structure–affinity relationships of small monocarboxylates for hMCT1 mutants, M69L and F367Y. a, b l-[14C]Lactate uptake by hMCT1 single mutants in the presence of CPC (a) or l-OCPC (b). Data are expressed as % of control (absence of competitor) after the subtraction of uptake into water-injected oocytes. cKi values of hMCT1 single mutants from radioligand competition assays with four different inhibitors are shown as bars. The Ki values for each of the shown inhibitors were obtained from data presented in Tables 2 and 4. Values represent the mean ± SE from three independent experiments, each performed with 3–5 replicates. Each experiment in the panels a, b was performed using a single batch of oocytes. d, e Docking poses of l-OPro (d) and l-lactate (e) into the substrate selection region in the 3D structure models of hMCT1 (distances in Å). The helices are numbered as shown in Fig. 1e
Discussion
Our current work focused on the identification of specific residues that determine the substrate specificity of hMCT1 and hMCT4, by measuring transport activities of site-directed mutants. hMCT1 and hMCT4 are homologous proteins and are both efficient l-lactate transporters, but they differ in their substrate specificity. hMCT1 is capable of l-OPro transport, whereas hMCT4 is not. There is evidence of the importance of the TM4/5 loop and TM10 in determining the substrate specificity of rodent MCT1. Galić et al. have reported that K142 and R143 residues in the TM4/5 loop appear to be involved in the stereoselectivity of rat MCT1 [19]. Moreover, it is known that the F360C mutant of rodent MCT1 allows it to transport mevalonate [20, 21]. The F360 residue of rodent MCT1 corresponds to the F367 residue of hMCT1, which was found to be associated with the substrate specificity in this study. There is also evidence that certain residues in TM2, TM4, and TM5 (S121, Y184, and T207 in hMCT10, respectively) are important for the substrate specificity of hMCT8 and hMCT10 [30]. On the basis of these studies, we postulated that differences in amino acids lining the substrate translocation pathway would determine the substrate specificity of these transporters. To identify the residues that are essential for hMCT1-mediated l-OPro transport, we used site-directed mutagenesis to change individual amino acid residues in hMCT1 to the corresponding residues in hMCT4 and vice versa. During the course of these studies, we identified several mutants with reduced substrate transport activity. Some of these mutants, such as hMCT1-L281P, hMCT4-V239F, -V250S, and -G336S, have already been reported to have low activity in rat MCT1 and MCT4 (rat MCT1-L274P and rat MCT4-V243F, -V254S, and -G340S) [29]. In addition, Stäubli et al. [61] showed reduced creatine transport in hMCT12-S158P compared with WT hMCT12 in Xenopus laevis oocytes and human HEK293T cells. Since S158 in hMCT12 is equivalent to N129 in hMCT1 and Q131 in hMCT4, it is reasonable to expect that l-lactate transport activity would be reduced in hMCT4-Q131N. We also identified two amino acid residues in TM2 and TM10 (M69 and F367) that appeared to be the most important residues for l-OPro transport among the 20 different residues in the putative substrate translocation pathways of hMCT1 and hMCT4. Moreover, double mutation of M69 and F367 residues severely impaired hMCT1-mediated l-OPro transport. Since Johannes et al. [30] identified S121, Y184, and T207 residues in hMCT10 as substrate specificity determinants, we initially speculated that M65/L67, L128/F130, and M151/A153 in hMCT1/hMCT4 (equivalent to S121, Y184, and T207, respectively, in hMCT10) may be some of the residues involved in substrate specificity. Unexpectedly, our experiments revealed that these residues do not participate in substrate specificity, although l-lactate transport activity was reduced in hMCT4-L67M and -F130L. Our homology model of hMCT1 predicted that M69 and F367 participate in hydrophobic interactions with another region of hMCT1, emphasizing its potentially important role in the binding and translocation cycle of l-OPro. Our M69X and F367X substitution panel results supported this model. Efficient l-OPro transport required a hydrophobic, long linear structure at position 69 and a hydrophobic, γ-branched structure at position 367. Our data also suggested that M69 and F367 residues recognize the –NH– moiety of l-OPro and the flexible and polar structure of l-lactate. In addition, we previously demonstrated that CPC inhibits these transporters, while dl-OCPC selectively inhibits hMCT1, suggesting that the introduction of an oxo group at position 3 of CPC appears to be important for the determination of ligand selectivity [18]. In the present study, analysis of structure–affinity relationships suggested that the oxo group has minimal involvement in the interaction with hMCT1. Thus, the selective recognition of l-OPro by hMCT1 occurs, not because the introduction of the oxo group provides a favorable interaction between hMCT1 and l-OPro, but, rather, because the introduced oxo group reduces molecular recognition by hMCT4.
The question remains why the M69 and F367 mutants have altered substrate specificities. The M69 side chain is highly non-reactive and, thus, infrequently directly participates in substrate transport in general. The bulk of the Met side chain is proposed to fulfill a steric role in the determination of substrate specificity. Our homology model revealed that there were hydrophobic interactions between M69 and its neighboring residues (L66, Y70, G123, and L124) and that these interactions would be impaired in the mutants with a shortened side chain (e.g., M69G, M69A, M69C, and M69T). Indeed, the CLl-OPro/CLl-Lactate values for these mutants were significantly decreased compared to the values for hMCT1-WT. The potential role of this Met residue had not been studied for some time, but, recently, Nancolas et al. [29] investigated the importance of the residue in the interaction with AR-C155858 (a selective rat MCT1/2 inhibitor). They concluded that M69 is not involved in mediating binding of the inhibitor in rat MCT1, but we identified M69 as one of the substrate specificity determinants. These data suggest that the ligand recognition mechanism of mammalian MCT1 differs between the substrate and the inhibitor, even for the same selective hMCT1 ligand. The simple addition of phenolic oxygen at the para position of the phenyl group of F367 (F367Y) was sufficient to decrease the CLl-OPro/CLl-Lactate value. In addition, substitution of F367 to Val resulted in a decrease in substrate uptake, but a significant fraction of the transport activity remained, whereas substitution to Thr completely abolished the transport activity. Since Val and Thr have sterically similar structures and differ only in a hydroxy group, the reduction of substrate transport activity in the F367T mutant is likely caused by steric or electrostatic effects of the hydroxyl group of Thr. It is possible that the aromatic side chain of F367 is involved in hydrophobic interactions with other hydrophobic side chains or ligands. Based on the hMCT1 homology model, there are hydrophobic interactions between F367 and its surrounding residues (D309, F363, G364, and G368). It has been reported that D309 (D302 in rat MCT1), one of the amino acid residues surrounding F367, forms an ion pair with the Arg residue present in TM8 and plays a pivotal role in substrate translocation [21, 62]. Hence, we considered that this Phe residue may be important for the positioning of D309, allowing correct TM8 orientation. It appeared that the interactions between F367 and its neighboring residues would be maintained, as long as the side chain at this site is large and hydrophobic (e.g., F367I, F367L, F367M, F367V, F367W, and F367Y). In particular, the CLl-OPro/CLl-Lactate value of the F367L mutant was almost the same as that of hMCT1-WT, indicating the importance of the γ-branched hydrophobic structure at this position. Using mutagenesis techniques, this Phe residue and the equivalent Asp residue in hMCT8 have been identified as being required for efficient substrate transport and the F360C mutant of rodent MCT1 allows mevalonate to be transported, as mentioned above [20, 21, 29, 58, 63]. However, we demonstrated that F367C (F360C in rodent MCT1) was not capable of l-lactate or l-OPro transport, indicating that the influence of F367 on the substrate specificity of hMCT1 is different between these substrates and mevalonate.
In conclusion, we have identified two amino acid residues, M69 and F367, which are key molecular elements for the transport of l-OPro by hMCT1. However, we cannot exclude the possibility that other unidentified residues are also involved in determining the substrate specificity of hMCT1. These two residues are predicted to be involved in substrate recognition and/or substrate-induced conformational changes. This information will be valuable for the elucidation of the structure–function relationships of hMCTs.
Abbreviations
- hMCTs:
-
Human monocarboxylate transporters
- MFS:
-
Major facilitator superfamily
- l-OPro:
-
l-5-oxoproline
- TM:
-
Transmembrane domain
- HMM:
-
Hidden Markov model
- CPC:
-
Cyclopentanecarboxylate
- l-OCPC:
-
(R)-3-Oxocyclopentanecarboxylate
References
Fisel P, Schaeffeler E, Schwab M (2018) Clinical and functional relevance of the monocarboxylate transporter family in disease pathophysiology and drug therapy. Clin Transl Sci 11:352–364. https://doi.org/10.1111/cts.12551
Carneiro L, Pellerin L (2015) Monocarboxylate transporters: new players in body weight regulation. Obes Rev 16:55–66. https://doi.org/10.1111/obr.12256
Pinheiro C, Longatto-Filho A, Azevedo-Silva J et al (2012) Role of monocarboxylate transporters in human cancers: state of the art. J Bioenerg Biomembr 44:127–139. https://doi.org/10.1007/s10863-012-9428-1
Sasaki S, Kobayashi M, Futagi Y et al (2013) Crucial residue involved in L-lactate recognition by human monocarboxylate transporter 4 (hMCT4). PLoS One 8:e67690. https://doi.org/10.1371/journal.pone.0067690
Futagi Y, Sasaki S, Kobayashi M et al (2017) The flexible cytoplasmic loop 3 contributes to the substrate affinity of human monocarboxylate transporters. Biochim Biophys Acta Biomembr 1859:1790–1795. https://doi.org/10.1016/j.bbamem.2017.05.014
Polanski R, Hodgkinson CL, Fusi A et al (2014) Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin Cancer Res 20:926–937. https://doi.org/10.1158/1078-0432.CCR-13-2270
Gurrapu S, Jonnalagadda SK, Alam MA et al (2016) Coumarin carboxylic acids as monocarboxylate transporter 1 inhibitors: in vitro and in vivo studies as potential anticancer agents. Bioorganic Med Chem Lett 26:3282–3286. https://doi.org/10.1016/j.bmcl.2016.05.054
Quanz M, Bender E, Kopitz C et al (2018) Preclinical efficacy of the novel monocarboxylate transporter 1 inhibitor BAY-8002 and associated markers of resistance. Mol Cancer Ther 17:2285–2296. https://doi.org/10.1158/1535-7163.MCT-17-1253
Ovens MJ, Davies AJ, Wilson MC et al (2010) AR-C155858 is a potent inhibitor of monocarboxylate transporters MCT1 and MCT2 that binds to an intracellular site involving transmembrane helices 7–10. Biochem J 425:523–530. https://doi.org/10.1042/BJ20091515
Wang H, Yang C, Doherty JR et al (2014) Synthesis and structure-activity relationships of pteridine dione and trione monocarboxylate transporter 1 inhibitors. J Med Chem 57:7317–7324. https://doi.org/10.1021/jm500640x
Sasaki S, Futagi Y, Kobayashi M et al (2015) Functional characterization of 5-oxoproline transport via SLC16A1/MCT1. J Biol Chem 290:2303–2311. https://doi.org/10.1074/jbc.M114.581892
Bachhawat AK, Yadav S (2018) The glutathione cycle: glutathione metabolism beyond the γ-glutamyl cycle. IUBMB Life 70:585–592. https://doi.org/10.1002/iub.1756
Kumar A, Bachhawat AK (2012) Pyroglutamic acid: throwing light on a lightly studied metabolite. Curr Sci 102:288–297. https://www.jstor.org/stable/24083854
Chou J, Liu R, Yu J et al (2018) Fasting serum α-hydroxybutyrate and pyroglutamic acid as important metabolites for detecting isolated post-challenge diabetes based on organic acid profiles. J Chromatogr B Anal Technol Biomed Life Sci 1100–1101:6–16. https://doi.org/10.1016/j.jchromb.2018.09.004
Qi S, Xu D, Li Q et al (2017) Metabonomics screening of serum identifies pyroglutamate as a diagnostic biomarker for nonalcoholic steatohepatitis. Clin Chim Acta 473:89–95. https://doi.org/10.1016/j.cca.2017.08.022
Yu B, Zheng Y, Nettleton JA et al (2014) Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol 9:1410–1417. https://doi.org/10.2215/CJN.11971113
Thiesen L, Kehler J, Clausen RP et al (2015) In vitro and in vivo evidence for active brain uptake of the GHB analog HOCPCA by the monocarboxylate transporter subtype 1. J Pharmacol Exp Ther 354:166–174. https://doi.org/10.1124/jpet.115.224543
Futagi Y, Kobayashi M, Narumi K et al (2018) Identification of a selective inhibitor of human monocarboxylate transporter 4. Biochem Biophys Res Commun 495:427–432. https://doi.org/10.1016/j.bbrc.2017.10.025
Galić S, Schneider H-PP, Bröer A et al (2003) The loop between helix 4 and helix 5 in the monocarboxylate transporter (MCT1) is important for substrate selection and protein stability. Biochem J 376:413–422. https://doi.org/10.1042/BJ20030799
Garcia CK, Goldstein JL, Pathak RK et al (1994) Molecular characterization of a membrane transporter for lactate, pyruvate, and other monocarboxylates: implications for the Cori cycle. Cell 76:865–873. https://doi.org/10.1016/0092-8674(94)90361-1
Rahman B, Schneider HP, Bröer A et al (1999) Helix 8 and helix 10 are involved in substrate recognition in the rat monocarboxylate transporter MCT1. Biochemistry 38:11577–11584. https://doi.org/10.1021/bi990973f
Deng D, Sun P, Yan C et al (2015) Molecular basis of ligand recognition and transport by glucose transporters. Nature 526:391–396. https://doi.org/10.1038/nature14655
Zhao Y, Heng J, Zhao Y et al (2015) Substrate-bound structure of the E. coli multidrug resistance transporter MdfA. Cell Res 25:1060–1073. https://doi.org/10.1038/cr.2015.94
Parker JL, Newstead S (2014) Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507:68–72. https://doi.org/10.1038/nature13116
Sun J, Bankston JR, Payandeh J et al (2014) Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507:73–77. https://doi.org/10.1038/nature13074
Pedersen BP, Kumar H, Waight AB et al (2013) Crystal structure of a eukaryotic phosphate transporter. Nature 496:533–536. https://doi.org/10.1038/nature12042
Yan H, Huang W, Yan C et al (2013) Structure and mechanism of a nitrate transporter. Cell Rep 3:716–723. https://doi.org/10.1016/j.celrep.2013.03.007
Sun L, Zeng X, Yan C et al (2012) Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature 490:361–366. https://doi.org/10.1038/nature11524
Nancolas B, Sessions RB, Halestrap AP (2015) Identification of key binding site residues of MCT1 for AR-C155858 reveals the molecular basis of its isoform selectivity. Biochem J 466:177–188. https://doi.org/10.1042/BJ20141223
Johannes J, Braun D, Kinne A et al (2016) Few amino acid exchanges expand the substrate spectrum of monocarboxylate transporter 10. Mol Endocrinol 30:796–808. https://doi.org/10.1210/me.2016-1037
Edwards N, Anderson CMH, Conlon NJ et al (2018) Resculpting the binding pocket of APC superfamily LeuT-fold amino acid transporters. Cell Mol Life Sci 75:921–938. https://doi.org/10.1007/s00018-017-2677-8
Krypotou E, Kosti V, Amillis S et al (2012) Modeling, substrate docking, and mutational analysis identify residues essential for the function and specificity of a eukaryotic purine-cytosine NCS1 transporter. J Biol Chem 287:36792–36803. https://doi.org/10.1074/jbc.M112.400382
Monné M, Miniero DV, Daddabbo L et al (2012) Substrate specificity of the two mitochondrial ornithine carriers can be swapped by single mutation in substrate binding site. J Biol Chem 287:7925–7934. https://doi.org/10.1074/jbc.M111.324855
Gui C, Hagenbuch B (2008) Amino acid residues in transmembrane domain 10 of organic anion transporting polypeptide 1B3 are critical for cholecystokinin octapeptide transport. Biochemistry 47:9090–9097. https://doi.org/10.1021/bi8008455
Sasaki S, Futagi Y, Ideno M et al (2016) Effect of diclofenac on SLC16A3/MCT4 by the Caco-2 cell line. Drug Metab Pharmacokinet 31:218–223. https://doi.org/10.1016/j.dmpk.2016.03.004
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. Embnet News 4:1–4
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015-053
Remmert M, Biegert A, Hauser A, Söding J (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9:173–175. https://doi.org/10.1038/nmeth.1818
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. https://doi.org/10.1006/jmbi.1999.3091
Söding J (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960. https://doi.org/10.1093/bioinformatics/bti125
Jefferys BR, Kelley LA, Sternberg MJE (2010) Protein folding requires crowd control in a simulated cell. J Mol Biol 397:1329–1338. https://doi.org/10.1016/j.jmb.2010.01.074
Xie W, Sahinidis NV (2006) Residue-rotamer-reduction algorithm for the protein side-chain conformation problem. Bioinformatics 22:188–194. https://doi.org/10.1093/bioinformatics/bti763
Huang Y, Lemieux MJ, Song J et al (2003) Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301:616–620. https://doi.org/10.1126/science.1087619
Iancu CV, Zamoon J, Woo SB et al (2013) Crystal structure of a glucose/H + symporter and its mechanism of action. Proc Natl Acad Sci 110:17862–17867. https://doi.org/10.1073/pnas.1311485110
Jiang D, Zhao Y, Wang X et al (2013) Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc Natl Acad Sci 110:14664–14669. https://doi.org/10.1073/pnas.1308127110
Bhattacharya D, Nowotny J, Cao R, Cheng J (2016) 3Drefine: an interactive web server for efficient protein structure refinement. Nucleic Acids Res 44:W406–W409. https://doi.org/10.1093/nar/gkw336
Lovell SC, Davis IW, Arendall WB III et al (2003) Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins: structure. Funct Genet 50:437–450
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32:327–331. https://doi.org/10.1093/nar/gkh454
Marchler-Bauer A, Bo Y, Han L et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. https://doi.org/10.1093/nar/gkw1129
Menke M, Berger B, Cowen L (2008) Matt: local flexibility aids protein multiple structure alignment. PLoS Comput Biol 4:0088–0099. https://doi.org/10.1371/journal.pcbi.0040010
Kim S, Chen J, Cheng T et al (2019) PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 47:D1102–D1109. https://doi.org/10.1093/nar/gky1033
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Shapovalov MV, Dunbrack RL (2011) A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 19:844–858. https://doi.org/10.1016/j.str.2011.03.019
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25:247–260. https://doi.org/10.1016/j.jmgm.2005.12.005
Trott oleg O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc
Vivian D, Polli JE (2014) Mechanistic interpretation of conventional Michaelis-Menten parameters in a transporter system. Eur J Pharm Sci 64:44–52. https://doi.org/10.1016/j.ejps.2014.08.007
Groeneweg S, Friesema ECH, Kersseboom S et al (2014) The role of Arg445 and Asp498 in the human thyroid hormone transporter MCT8. Endocrinology 155:618–626. https://doi.org/10.1210/en.2013-1521
Quistgaard EM, Löw C, Guettou F, Nordlund P (2016) Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat Rev Mol Cell Biol 17:123–132. https://doi.org/10.1038/nrm.2015.25
Perland E, Fredriksson R (2017) Classification systems of secondary active transporters. Trends Pharmacol Sci 38:305–315. https://doi.org/10.1016/j.tips.2016.11.008
Stäubli A, Capatina N, Fuhrer Y et al (2017) Abnormal creatine transport of mutations in monocarboxylate transporter 12 (MCT12) found in patients with age-related cataract can be partially rescued by exogenous chaperone CD147. Hum Mol Genet 26:4203–4214. https://doi.org/10.1093/hmg/ddx310
Manoharan C, Wilson MC, Sessions RB, Halestrap AP (2006) The role of charged residues in the transmembrane helices of monocarboxylate transporter 1 and its ancillary protein basigin in determining plasma membrane expression and catalytic activity. Mol Membr Biol 23:486–498. https://doi.org/10.1080/09687860600841967
Islam MS, Namba N, Ohata Y et al (2018) Functional analysis of monocarboxylate transporter 8 mutations in Japanese Allan-Herndon-Dudley syndrome patients. Endocr J 2018:1–11. https://doi.org/10.1507/endocrj.EJ18-0251
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (JSPS; KAKENHI Grant Number JP17J00013). We would like to thank Editage (http://www.editage.jp) for English language editing.
Author information
Authors and Affiliations
Contributions
YF designed and performed the experiments, analyzed the results, and wrote the first draft of the manuscript. MK, KN, AF, and KI contributed to the writing of the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Futagi, Y., Kobayashi, M., Narumi, K. et al. Homology modeling and site-directed mutagenesis identify amino acid residues underlying the substrate selection mechanism of human monocarboxylate transporters 1 (hMCT1) and 4 (hMCT4). Cell. Mol. Life Sci. 76, 4905–4921 (2019). https://doi.org/10.1007/s00018-019-03151-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03151-z