Abstract
MicroRNAs (miRNAs) are a class of endogenous small noncoding RNAs that participate in a majority of biological processes via regulating target gene expression. The post-transcriptional repression through miRNA seed region binding to 3′ UTR of target mRNA is considered as the canonical mode of miRNA-mediated gene regulation. However, emerging evidence suggests that other regulatory modes exist beyond the canonical mechanism. In particular, the function of intranuclear miRNA in gene transcriptional regulation is gradually revealed, with evidence showing their contribution to gene silencing or activating. Therefore, miRNA-mediated regulation of gene transcription not only expands our understanding of the molecular mechanism underlying miRNA regulatory function, but also provides new evidence to explain its ability in the sophisticated regulation of many bioprocesses. In this review, mechanisms of miRNA-mediated gene transcriptional and post-transcriptional regulation are summarized, and the synergistic effects among these actions which form a regulatory network of a miRNA on its target are particularly elaborated. With these discussions, we aim to emphasize the importance of miRNA regulatory network on target gene regulation and further highlight the potential application of the network mode in the achievement of a more effective and stable modulation of the target gene expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a class of single-strand non-coding RNAs about 22 nucleotides that are highly conserved evolutionarily and play important regulatory roles in a vast range of cellular processes in eukaryotic cells [1]. Most miRNAs are transcribed by RNA polymerase II (Pol II) in the nucleus, and then the transcripts referred to as pri-miRNAs are cleaved by Drosha and its essential cofactor DGCR8 [2]. After cleavage, hairpin-shaped precursor miRNAs (pre-miRNAs) with the length of 60–70 nucleotides(nt) are generated [3,4,5] and further exported to the cytoplasm by exportin-5 (XPO5) [6,7,8]. Dicer1, a member of the RNase III family, is responsible for the maturation of miRNAs in the cytoplasm, where it binds to the end of pre-miRNAs and produces approximately 22-nt miRNAs duplex [9]. Finally, the functional strand of miRNA duplex is incorporated into a miRNA-induced silencing complex (miRISC) assembled by Dicer1, transactivation-responsive RNA-binding protein (TRBP; also known as TARBP2) and Argonaute proteins (AGO1, AGO2, AGO3 or AGO4) to exert its post-transcriptional regulatory function [10,11,12].
It is generally accepted that miRNAs can directly mediate post-transcriptional gene silencing (PTGS) in the cytoplasm through as short as the 6nt seed sequence of miRNAs and the complementary sequences in the 3′-untranslated region (UTR) of target mRNA [13, 14]. Most studied miRNAs may follow the principle concluded by van den Berg et al. that complementarity degree between miRNA and its target mRNA determines the way of gene suppression. Exact matching has been proved to induce mRNA cleavage, whereas partial matching leads to translational inhibition [15]. However, the exact mechanism remains controversial and needs more evidence to be clarified.
Since miRNAs are responsible for the subtle and precise regulation of gene expression, several scientific issues about their function and mechanism are put forward. For example, is the canonical regulation of target by miRNA competent in the sophisticated regulation of gene expression? Are there any other types of regulation also contributing to the miRNAs-mediated regulation of gene expression besides canonical way? And whether seed sequence of miRNA is the only effective region in target recognition of miRNA? Up to now, more and more evidence shows that mature miRNAs can be detected in the nucleus. Although the function of nuclear miRNA has not been fully clarified, the majority of studies demonstrate that nuclear miRNA could induce transcriptional gene silencing (TGS) or transcriptional gene activating (TGA) [16,17,18,19,20,21,22,23]. Additionally, the imperfect matches between miRNAs seed sequence and their target mRNAs indicate the possibility that near-seed or non-seed region of miRNAs may play roles in gene regulation, which have been proved by a series of reports [14, 24,25,26,27]. As the mechanism of miRNA-mediated gene regulation has been identified to be far more intricate than the canonical mode, a blueprint of huge gene regulation network mediated by either different binding sites of miRNA on target gene or different functional regions in miRNA is gradually revealed.
In this review, we summarize the diverse mechanisms of miRNA-mediated gene transcriptional and post-transcriptional regulation in mammalian cells and elaborate the synergistic action among these regulations. With these examples, we aim to elicit that the multipoint actions of miRNAs and its regulatory network are involved in the sophisticated regulation of gene expression and further shed new light on the potential application of this network regulation by miRNAs in providing new insights for more effective and stable modulation of the target genes expression.
Transcriptional gene regulation by miRNA
Cellular distribution and nucleocytoplasmic shuttling of miRNA
Generally, miRNAs are involved in many processes as regulators through post-transcriptional regulation in the cytoplasm, and their subcellular distributions are thus very important to their potential functions [14]. It has been already proved that miRISC within the cytoplasm of the eukaryotic cell locates in Processing body (P-body) which consists of many enzymes involved in mRNA turnover, indicating that P-body is a pivotal place for the post-transcriptional silencing by miRNA [28]. Besides P-body, miRNAs have been observed in other cytoplasmic organelles, such as mitochondria [29, 30], endoplasmic reticulum, Golgi, lysosome, etc., and play a series of roles in regulating cellular function [31]. For instance, muscle-specific miR-1 which was specifically induced during myogenesis can enter mitochondria, where it stimulates translation of multiple specific mtDNA-encoded transcripts within mitochondria and AGO2 was required in this process [30].
At first, it was thought that the subcellular distributions of miRNAs were mainly in the cytoplasm, consistent with the discovery that the main function of miRNAs was to form miRISC and induce post-transcriptional regulation. However, researchers find that some mature miRNAs were also detected in the nucleus. In 2004, Meister and his colleagues detected the first miRNA (miR-21) in the nucleus and about 20% of mature miR-21 extracted from Hela cells were distributed in the nucleus [32]. Hwang et al. reported that human miR-29b was predominantly localized to the nucleus in mitotic cells [33]. And further research indicated that miRISC-related components including AGO2, Dicer1, TRBP, and TRNC6A/GW182 complexes were all found in the mammalian nucleus and miRNAs in complex with these factors can form miRISC which enables them to play the role of silencing nuclear RNA and inducing site-specific cleavage in the nucleus [34].
Since miRNAs in the nucleus also have the functions in RNA silencing (RNAi), their source arouses researchers’ interest. Mature miRNAs might be either transported back to the nucleus after being processed in the cytoplasm or directly produced in the nucleus. The shuttling of different RNA species including miRNAs between cytoplasm and nucleus is complicated and multiple mechanisms exist. Since small tRNAs, which are smaller than miRNAs, cannot pass the nuclear pore complex (NPC) without active transportation [35], it is unlikely that miRNAs can freely diffuse through the NPC and transfer from cytoplasm to nucleus. There might be transporters which mediate the transportation of mature miRNAs between cytoplasm and nucleus. Importin 8 (IPO8), a member of the karyopherin β family, is discovered to play a vital role in mediating the cytoplasm-to-nucleus transport of mature miRNAs and this process requires Ago2 complex [36]. Besides transporters, the sequence of miRNA, especially 3′ terminal sequence, is also a factor in determining its subcellular localization. Hwang et al. deduced that an AGNGUN element at the 3′ terminus of miR-29b was essential for its nucleus localization through mutagenesis studies [33]. Jeffries et al. found that 7 from 21 miRNAs highly expressed in the nucleus had an ASUS (S=C or G) motif in their 3′ terminus in neural stem cells [37]. However, a series of evidence also supports the hypothesis that nuclear mature miRNAs might be directly processed in the nucleus. In 2010, Brameier et al. reported that H/ACA box and C/D box small nucleolar RNAs (snoRNAs) in the nucleus can give rise to numerous small RNA fractions with lengths of ≥ 18 nt, some of which have miRNA-like functions, such as locating in the stem regions of predicted RNA structures and effectively silencing gene expression [38]. In another study about nucleolus miRNA in rat myoblasts, Politz et al. described that some miRNA precursors are present in the nucleolus, indicating the possibility of nuclear maturation of the miRNAs. They also found that miR-664 is highly abundant in the nucleolus and completely homologous to a fraction of snoRNA SNORA36B/ACA36b. This is consistent with Brameier’s finding of snoRNAs-derived small RNAs [39].
However, there remain several issues to be clarified. First, although some miRNAs can enter the nucleus through karyopherins, the molecular mechanism is still unclear. That is to say, the common transportation rules of miRNAs between nucleus and cytoplasm need to be further explicated. Recently, Pitchiaya et al. have developed single-molecule fluorescence-based tools to dissect the subcellular trafficking of miRNAs, which might be a helpful approach for the study of miRNA nucleocytoplasmic shuttling [40]. Second, as a novel and additional working mode, the exact mechanism and function of intranuclear miRNAs are not fully explained. With in-depth study, the inhibitory or activating roles of intranuclear miRNAs in gene transcription has gradually been discovered.
Transcriptional gene silencing (TGS)
It is well known that miRNA mainly play regulatory roles in the cytoplasm at the post-transcriptional level. However, lots of mature miRNAs have been found in the nucleus as discussed above and it is becoming evident that these miRNAs can also induce transcriptional gene silencing (TGS) [16, 22, 23, 41]. Since miRNAs usually function through sequence complementarity, there is also evidence showing putative binding sites of miRNAs in gene promoter regions with perfect or partial sequence complementarity, which enables nuclear miRNA to regulate gene transcription. For example, miR-320, a conserved miRNA encoded within the promoter region of POLR3D in the antisense orientation, inhibits its host gene transcription. The authors also expanded their study to search for miRNAs targeting gene promoter region in trans. By searching for miRNA target sites in 200 bp upstream of transcriptional start sites with bioinformatics analysis, they identified numerous miRNAs that potentially function in this way [16].
For most of reported TGS, miRNAs interact with their complementary promoter-associated RNA(s) or have putative binding sites in the promoter region, resulting in a decrease of RNA Pol-II activity and recruiting of corepressor or stabilization of a pre-existing repressor complex, and consequently leading to the target gene transcriptional silencing [17, 19, 20, 42, 43]. And in these situations, epigenetic modifications, such as DNA methylation, modification of H3K27me3 and H3K4me3, usually participate and might play a relatively prominent role [18, 44]. For example, miR-10a targets the promoter region of HOXD4 gene by its 3′ end sequence and inhibits HOXD4 expression at the transcriptional level, which requires the presence of Dicer, Ago1, and Ago3. This function of miR-10a is not only related to promoter-associated noncoding RNAs, but also associated with de novo DNA methylation in HOXD4 promoter and trimethylation of H3K27me3 [17]. Moreover, our recent work first discovered a new mechanism of miRNA in the regulation of gene transcription, where the non-seed sequence of miR-552-3p could directly bind to the loop hairpin in the cruciform structure of CYP2E1 promoter, prevent the interaction between the promoter and its transcription activator SMARCE1, and finally suppresses CYP2E1 transcription [23].
Besides protein-coding genes, nuclear miRNAs also regulate the biogenesis of non-coding RNAs at the transcriptional level. MALAT-1 is a conserved non-coding RNA, and it has been reported that miR-9 can bind to Ago2 in the nucleus and regulate the transcription of MALAT-1 [45]. Moreover, nuclear miRNAs can also interact with other pri-miRNAs and regulate the biogenesis of respective miRNAs. MiR-709 mainly locates in the nucleus and has 19nt completely complementary with the sequence of pri-miR-15a/16-1. MiR-709 suppresses the processing of pre-miR-15a/16-1 from pri-miR-15a/16-1 and finally decreases the level of mature miR-15a/16-1, which leads to cell apoptosis [46].
Transcriptional gene activating (TGA)
Besides transcriptional gene silencing, numerous studies also demonstrate the different function of nuclear miRNA in transcriptional gene activation. This mode of regulation was first reported in 2008, in which miR-373 can enhance E-cadherin and CSDC2 transcription and it depends on the miRNA target sites in the promoters of both genes [47]. In addition, miR-744 activates mouse cyclin B1 (Ccnb1) transcription by recruiting Ago1 to its promoter region. All these findings indicate that nuclear miRNAs could induce transcriptional gene activating (TGA) [48].
During the past 10 years, several hypotheses have been proposed based on experimental results, helping us better understand miRNA-mediated TGA. One possible mechanism is that miRNAs could bind to the enhancer. Some miRNAs have been found enriched in the enhancer region and activate gene transcription via chromatin remodeling [49,50,51,52]. For example, miR-24-1 which locates at the enhancer region could promote enhancer activation and finally induce the expression of its neighboring genes FBP1 and FANCC [41]. Another alternative hypothesis of miRNA-mediated TGA is generated from bidirectional transcription of the human genome. As reported, the transcription of E-cadherin can be modulated by its antisense transcript, and significant suppression of E-cadherin antisense transcript induced by miR-373 is correlated with the up-regulatory role of miR-373 in E-cadherin sense/mRNA transcript expression [53]. However, since this type of regulation through targeting complementary sequence within antisense transcripts including the long non-coding RNAs (lncRNAs) and promoter-associated RNAs (pRNAs) transcribed from the antisense strand of genome mostly came from siRNAs testing [53, 54], more observation of miRNA-mediated TGA by this mode should be discovered to validate this hypothesis. Additionally, miRNAs could induce TGA by recruiting a protein complex with transcriptional activators to the gene promoter. For example, miR-589 could bind to the cyclooxygenase-2 (COX-2) promoter and activate gene transcription which required the involvement of RNAi factors, Ago2 and GW182, as a scaffold [55]. The last but not least, nuclear miRNAs could also form a functional positive feedback loop on transcription of its host gene. For example, miR-483-5p which is embedded within the IGF2 gene could up-regulate IGF2 expression transcriptionally, resulting in increased tumorigenesis in vivo [56].
Post-transcriptional gene regulation by miRNA
For most miRNAs, matching with 3′ UTR of the mRNA is required for functional inhibition of the target genes. But with the in-depth study, the researches about the recognition sites by miRNAs have been expanded to the coding regions [57,58,59,60] or 5′ UTR [61] of targets. As reported, some miRNAs target the coding sequence of mRNA via the whole sequence, which is similar to siRNAs, and finally suppress mRNAs translation or stabilization. The interaction between the seed sequence of miRNAs and coding regions of mRNAs was discovered as well. For example, there are many miR-181a seed-matched sites within coding sequences of zinc finger genes (ZNFs), which is the mechanism of miR-181a-mediated downregulation of a large number of ZNFs [57]. As for 5′ UTRs, they may also interact with miRNA even though they usually have high GC content which tends to form secondary structure. For instance, miR-24 can inhibit Jab1 translation by directly binding to its 3′ UTR and 5′ UTR [62]. MiR-US25-1 from human cytomegalovirus (HCMV) binds to target sites primarily within 5′ UTRs of many genes associated with cell cycle control, demonstrating that a viral miRNA mediates translational repression of multiple cellular genes by targeting mRNA 5′ UTRs [63]. Interestingly, miRNAs targeting the 5′ UTRs can also trigger positive regulation on gene expression, in which some miRNAs bind to the 5′ UTR of target mRNAs to stabilize the target mRNAs or activate translation. For example, miR-122 can bind to the 5′ UTR of the positive-strand of HCV RNA genome to slow down the decay of the viral genome in infected cells, which led to the stimulation of viral protein expression and promotion of viral replication [64]. MiR-10a has been reported to interact with the 5′ UTR of mRNAs encoding ribosomal proteins to enhance their translation, which is the mechanism of miR-10a-induced global protein synthesis [65]. Mouse microRNA, miR-196b, can specifically target the 5′ UTR of the long insulin2 splice isoform to displace the RNA-binding protein HuD which represses insulin translation and as a result increase insulin translation [61]. Apparently, the interaction of miRNAs and 5′ UTR tends to induce an activation of translation rather than a repression, which provides a potential therapeutic strategy for rare disease caused by genetic defect.
A possible role of multiple modes of miRNA action in their sophisticated regulation of gene expression
The regulation of gene expression is a complicated multistep process. To produce a functional protein is subject to several factors, starting from transcription to translation and then post-translational modification, in which miRNAs participate in most processes and play pivotal roles in this highly sophisticated regulatory system. Besides post-transcriptional regulation, the discoveries of transcriptional regulation have greatly enriched the modes of miRNA action in fine-tuning protein expression, including interaction with promoter or regulation of epigenetic modifications to directly regulate promoter activity, and modulation of the transcriptional factor to indirectly regulate promoter activity, etc. Recently, the multi-omics techniques are applied to determine the gene regulatory function of a given miRNA as exhaustively as possible, by which more and more non-canonical regulations might be revealed. For example, the change of miR-223 could induce transcriptomic and proteomic changes in monocyte cells. However, the changes between transcript and protein levels were not always consistent and only some of the deregulated proteins had binding sites for miR-223, which indicates non-canonical regulations might exist between miR-223 and its targets [66, 67].
Additionally, the function of miRNA non-seed sequence also contributes to functional miRNA target recognition and this non-canonical interaction between miRNA and its target is extensive in mammalian cells, which expands our understanding in the manner of miRNA non-canonical regulation. Seed sequence, usually considered to be 6–8 ribonucleotide long at the 5′ end of the miRNA, is a key region of miRNA. However, there are also many imperfect matches between miRNAs seed sequence and their target mRNAs, which might be compensated by supplemental components in near-seed region sites to exert function in PTGS [68,69,70]. The observation that only 73% of the Ago-mRNA interactions can be explained by seed matches for Ago-bound miRNAs in mouse brain and the rest 27% have no predicted seed matches gives a hint of miRNA non-seed sequence playing a role in target recognition [71, 72]. And this probability was proved in succession by several reports which demonstrated miRNA 3′ end or central region can interact with the different region of target mRNA (3′ UTR, 5′ UTR, and coding region) or promoter region of the target gene [14, 24,25,26,27].
Therefore, endogenous miRNAs can control or fine-tune gene expression via a variety of ways and different regions, which enables miRNAs for a subtle and precise regulation in the modulation of gene expression.
Network regulation of a miRNA on its target
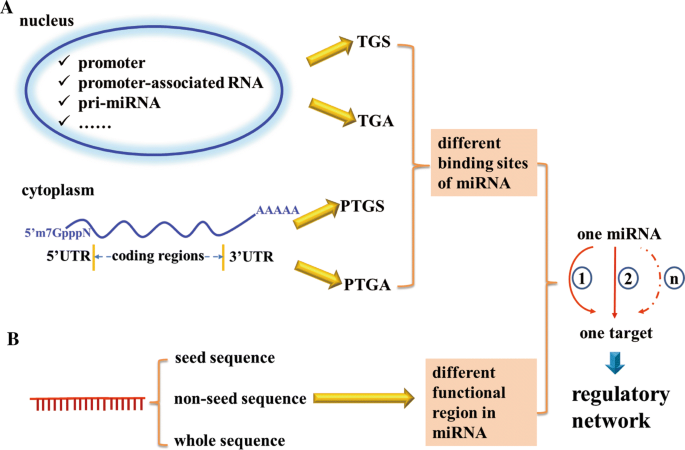
MiRNAs participate in a vast range of biological processes via the subtle and precise regulation of gene expression. Since the mechanisms of miRNAs regulating gene expression have been identified to be far more intricate than the canonical mode, a blueprint of huge gene regulation network mediated by either different binding sites of miRNA or different functional regions in miRNA is gradually revealed. On one hand, miRNAs not only target 3′ UTR of the mRNA, but also interact with other parts of the mRNA, such as 5′ UTR or coding sequence, and even gene promoter. On the other hand, besides the generally accepted function of miRNA targeting 3′ UTR of the mRNA via its seed region, several studies have bioinformatically predicted or functionally validated that the non-seed sequence of miRNAs could target mRNA transcripts or gene promoter as well. Based on these complicated mechanisms, single miRNA could impact on a series of pathways or axes to regulate one target gene expression (Fig. 1).
Multiple modes of miRNA action. The regulation by miRNA on its target can be achieved by a interacting with different binding sites including promoter, promoter-associated RNA, pri-miRNA, etc. in the nucleus to modulate target gene transcription or 5′ UTR, coding region, 3′ UTR of mRNA in the cytoplasm to modulate target gene translation. b The different functional region in miRNA including seed sequence, non-seed sequence or whole sequence. All these modes of miRNA action enable the network regulation between a miRNA and a target gene
Co-regulation by 3′ UTR and promoter
Our previous results first discovered the dual-inhibition of miRNA via PTGS and TGS [23]. MiR-552-3p was found to inhibit human cytochrome P450 (CYP) 2E1 translation via canonical regulatory mode, in which its seed sequence partially matches with CYP2E1 3′ UTR and the mutants in seed sequence suppressed the luciferase activity of the reporter containing 3′ UTR of CYP2E1. Interestingly, the non-seed sequence of miR-552, which is complementary to a loop hairpin of the cruciform structure in the CYP2E1 promoter, inhibits the binding of SMARCE1 and RNA polymerase II to the promoter and directs the TGS on CYP2E1. Moreover, this dual-inhibition is more effective in gene regulation, evidenced by experiments with wild-type, seed region mutant and non-seed region mutant of miR-552-3p.
Wang et al. discovered another case of dual-inhibition with PTGS and TGS. They predicted and validated that miR-215-5p inhibited the expression of PCDH9 at both mRNA and protein levels by targeting its promoter and 3′ UTR, respectively. Differently, the whole sequence of miR-215-5p is involved in recognition of the PCDH9 promoter. In this study, they proved the inhibitory effect of miR-215-5p on recombinant PCDH9 containing its own promoter and 3′ UTR more effective than that containing the promoter or 3′ UTR alone, which indicates the benefit of synergetic suppression by miRNA [73].
Co-regulation by 3′ UTR and transcriptional factor
Transcription factor and miRNA both are pivotal players participated in the regulation of gene expression in higher eukaryotes. To date, numerous reports have already identified the network between miRNAs and transcription factors [74]. For example, we demonstrated miR-491-3p downregulated ABCB1, a key factor in multidrug resistance, not only through its canonical regulatory role by directly targeting the ABCB1 3′-UTR, but also by targeting the 3′-UTR of Sp3, a transcription factor of ABCB1, to indirectly regulate the transcription of ABCB1. This dual inhibitory pathway finally leads to increased sensitivity of hepatoma cells to chemotherapeutic drugs [75].
The similar dual regulation is also discovered in another work from our lab. It is worth pointing out that we provide evidence again showing that a miRNA regulate the same target by matching to different sequences through its different regions. In our study, miR-1254 inhibits heme oxygenase-1 (HO-1) translation, dependent on its seed sequence complementary to HO-1 3′ UTR, and suppress HO-1 transcription, dependent on its non-seed sequence targeting TFAP2A which is transcription factor of HO-1 [26]. This coordination of miRNAs different regions achieves a more effective and stable inhibition effect on the target gene, and consequently inhibits tumor growth of NSCLC.
Co-regulation by 3′ UTR and signal cascades
The dual inhibition of miRNA is also achieved by the way of regulation of target gene 3′ UTR and the key factors in signal cascades associated with the target gene. The research of Sokolova et al. expounded on this type of dual regulation. First, miR-20a downregulates CDKN1A expression through direct binding to the 3′-UTR of CDKN1A mRNA. Second, miR-20a can subvert the TGF-β-mediated c-myc (an inhibitor of the CDKN1A promoter) repression via interference with the expression of factors in Smad/E2F-based core repressor complex to indirectly reduce CDKN1A promoter activity [76]. Another study in our lab also proved that this kind of dual regulation does exist in the sophisticated modulation of gene expression. We observe that miR-338-5p can downregulate ABCB1 by directly interacting with the ABCB1 3′ UTR and target the EGFR/ERK1/2 pathway to inhibit the expression of ABCB1 by pairing with EGFR 3′ UTR, resulting in increased sensitivity of hepatoma cells to DOX [77].
Co-regulation by 5′-UTR and 3′-UTR
Most synergetic effects usually occur between transcriptional and translational levels, however, some researches have also reported that a miRNA can target both 3′ UTR and 5′ UTR of mRNA, and achieve co-regulation on its target. Hsa-miR-34a was reported to target AXIN2 through both UTRs, in which the translation of mRNA luciferase reporter with both 3′ UTR and 5′ UTR of AXIN2 was more sensitive to miR-34a mimics or inhibitors (blocking endogenous miR-34a), compared to the reporter containing only 3′ UTR or 5′ UTR. These results suggest that both the 5′ UTR and the 3′ UTR of AXIN2 are functional target sites for miR-34a in the cells. Of course, this dual translational regulation is not restricted to miR-34a/AXIN2, and the similar regulation can be detected in miR-34a/WNT1 and has-miR-605/SEC24D [78], indicating that this might not be rare in higher eukaryotes.
Significance of dual regulation for miRNA’s sophisticated regulation of gene expression
In fact, miRNA’s fine control of the target genes expression depends on not only the diverse mechanisms, but also the complicated regulatory network. It is well known that a single miRNA can target several genes and one gene can also be targeted by multiple miRNAs. For example, miR-135b can regulate multiple important components in human Hippo pathway, including LATS2, BTRC, NDR2, and LZTS [79], while three miRNAs, miR-101, miR-129-5p, and miR-221, were identified to specifically target the 3′ UTR of FMR1 and regulate its expression [80]. Recently, emerging evidence pointed out a new network mode, the so-called “dual regulation” between one miRNA and one gene (Fig. 2). As summarized above, this network consists of coordination between 3′ UTR and promoter, 3′ UTR and transcriptional factor, 3′ UTR and signal cascades, 3′ UTR and 5′ UTR. This regulatory network within one miRNA and one target display the specificity in target recognition of miRNA and effectiveness in miRNA-mediated gene expression.
Different patterns of dual regulation on a target gene by miRNA. Currently, dual regulation of miRNA consists of coordination between 3′ UTR and promoter (a), in which miRNAs can directly regulate target gene transcription in nucleus and directly regulate target gene translation in cytoplasm; 3′ UTR and transcriptional factor (b), in which miRNAs can indirectly regulate target gene transcription in the nucleus by controlling the levels of transcriptional factors of the target in cytoplasm and directly regulate target gene translation in cytoplasm; 3′ UTR and signal cascades (c), in which miRNAs can indirectly regulate target gene mRNA or protein levels by modulating related signal pathways in the cytoplasm and directly regulate target gene translation in cytoplasm; 3′ UTR and 5′ UTR (d), in which miRNAs can directly regulate target gene translation in cytoplasm by interacting with the 3′ UTR and 5′ UTR of target mRNA
With the further research in this field, we believe more and more findings will prove that the dual regulation of miRNA on one target is universal and facilitate us to gradual deepen the recognition of the complexity of miRNA/target interactions and gene expression regulation network.
Potential application of the dual regulation in the development of small nucleic acid drug
Nucleic acid drugs are another emerging wave of modern medicine after small molecules and antibody drugs, in which obvious progress is found in miRNAs-related drugs including miRNAs itself and antisense oligonucleotides (ASO, antagomiR). MicroRNAs play important regulatory roles in a vast range of biological processes and are involved in the pathogenesis of most diseases. Compared with small molecule and biological molecule, miRNAs have their own unique advantages in druggability. Moreover, the discoveries of dual regulation of miRNA on one target might further help to enhance competitive advantage for its characteristics in the achievement of a productive and stable inhibition on the target with a smaller dose, which requires more preclinical and clinical trials to confirm.
Conclusions
As summarized above, miRNA can regulate gene transcription or translation in different manners via seed sequence, non-seed sequence or whole sequence, which provide a feasibility for synergism of dual or multi-regulation on one target gene by one miRNA. Since the dual regulation of miRNA on its target usually occurs at different levels, the spatiotemporal relationship between these two separated regulations needs to be figured out in the future research of dual-regulation. As well known, endogenous miRNAs usually have mild effects on their targets, however, dual-regulation of miRNA could form complex networks and finally produce additive effects at cellular and physiological levels. It is worth knowing that the precise modeling of miRNAs is the most important thing during the whole process. That is to say, does dual-regulation of miRNA happen simultaneously or successively? Is there a space–time specificity of this dual-regulation under different cellular or physiological conditions? Which regulation attributes more during disease progression? To address these questions, we need to have a better understanding of recognition rules between miRNA and its target and the functions of miRNA’s different region. Fortunately, technologies to identify miRNA targets have been improved a lot in these years. Transcriptome-wide and proteome-wide analyses along with RNA-sequencing and mathematical modeling approaches could help us to have an integrative comprehension of the whole regulatory network-biology of miRNA [81].
In addition, as for the miRNA with dual regulation both at the transcriptional level in nucleus and translation level in the cytoplasm, the nucleocytoplasmic shuttling of this miRNA should also be clarified. To understand the mechanisms of sublocation, cellular function and even dysfunction in disease of miRNA, we may need super-resolution imaging tools with single-molecule sensitivity and dynamic imaging capability to allow direct visualization of molecular interactions in cells [40].
In conclusion, the exploration of these scientific issues will help us to deeply understand the subtle regulation of miRNA and guide the development of miRNA-related nucleic acid drugs.
Abbreviations
- AGO:
-
Argonaute proteins
- ASO:
-
Antisense oligonucleotides
- CYP2E1:
-
Cytochrome P450 (CYP) 2E1
- DGCR8:
-
DiGeorge syndrome chromosomal [or critical] region 8
- DOX:
-
Doxorubicin
- HCV:
-
Hepatitis C virus
- IPO8:
-
Importin 8
- miRISC:
-
MicroRNA-induced silencing complex
- miRNA:
-
MicroRNA
- nt:
-
Nucleotides
- NPC:
-
Nuclear pore complex
- snoRNA:
-
Small nucleolar RNA
- SMARCE1:
-
SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1
- PTGS:
-
Post-transcriptional gene silencing
- pre-miRNAs:
-
Precursor miRNAs
- P-body:
-
Processing body
- Pol II:
-
Polymerase II
- RNAi:
-
RNA silencing
- TRBP:
-
Transactivation-responsive RNA-binding protein
- TGS:
-
Transcriptional gene silencing
- TGA:
-
Transcriptional gene activating
- trnc6a:
-
Trinucleotide repeat-containing gene 6A
- UTR:
-
Untranslated region
- XPO5:
-
Exportin-5
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23(20):4051–4060. https://doi.org/10.1038/sj.emboj.7600385
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956):415–419. https://doi.org/10.1038/nature01957
Carmell MA, Hannon GJ (2004) RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol 11(3):214–218. https://doi.org/10.1038/nsmb729
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432(7014):235–240. https://doi.org/10.1038/nature03120
Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17(24):3011–3016. https://doi.org/10.1101/gad.1158803
Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303(5654):95–98. https://doi.org/10.1126/science.1090599
Katahira J, Yoneda Y (2011) Nucleocytoplasmic transport of microRNAs and related small RNAs. Traffic 12(11):1468–1474. https://doi.org/10.1111/j.1600-0854.2011.01211.x
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818):363–366. https://doi.org/10.1038/35053110
Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115(2):209–216
Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436(7051):740–744. https://doi.org/10.1038/nature03868
Ameres SL, Zamore PD (2013) Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14(8):475–488. https://doi.org/10.1038/nrm3611
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115(7):787–798
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. https://doi.org/10.1016/j.cell.2009.01.002
van den Berg A, Mols J, Han J (2008) RISC-target interaction: cleavage and translational suppression. Biochim Biophys Acta 1779(11):668–677. https://doi.org/10.1016/j.bbagrm.2008.07.005
Kim DH, Saetrom P, Snove O Jr, Rossi JJ (2008) MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA 105(42):16230–16235. https://doi.org/10.1073/pnas.0808830105
Tan Y, Zhang B, Wu T, Skogerbo G, Zhu X, Guo X, He S, Chen R (2009) Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol 10:12. https://doi.org/10.1186/1471-2199-10-12
Younger ST, Corey DR (2011) Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res 39(13):5682–5691. https://doi.org/10.1093/nar/gkr155
Adilakshmi T, Sudol I, Tapinos N (2012) Combinatorial action of miRNAs regulates transcriptional and post-transcriptional gene silencing following in vivo PNS injury. PLoS One 7(7):e39674. https://doi.org/10.1371/journal.pone.0039674
Benhamed M, Herbig U, Ye T, Dejean A, Bischof O (2012) Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol 14(3):266–275. https://doi.org/10.1038/ncb2443
Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N, Mancini M, Nanni M, Cimino G, Lo-Coco F, Grignani F, Nervi C (2012) Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood 119(17):4034–4046. https://doi.org/10.1182/blood-2011-08-371344
Roberts TC (2014) The MicroRNA biology of the mammalian nucleus. Mol Ther Nucleic Acids 3:e188. https://doi.org/10.1038/mtna.2014.40
Miao L, Yao H, Li C, Pu M, Yao X, Yang H, Qi X, Ren J, Wang Y (2016) A dual inhibition: microRNA-552 suppresses both transcription and translation of cytochrome P450 2E1. Biochim Biophys Acta 1859(4):650–662. https://doi.org/10.1016/j.bbagrm.2016.02.016
Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105. https://doi.org/10.1101/gr.082701.108
Seok H, Ham J, Jang ES, Chi SW (2016) MicroRNA target recognition: insights from transcriptome-wide non-canonical interactions. Mol Cells 39(5):375–381. https://doi.org/10.14348/molcells.2016.0013
Pu M, Li C, Qi X, Chen J, Wang Y, Gao L, Miao L, Ren J (2017) MiR-1254 suppresses HO-1 expression through seed region-dependent silencing and non-seed interaction with TFAP2A transcript to attenuate NSCLC growth. PLoS Genet 13(7):e1006896. https://doi.org/10.1371/journal.pgen.1006896
Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27(1):91–105. https://doi.org/10.1016/j.molcel.2007.06.017
Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O (2009) Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11(9):1143–1149. https://doi.org/10.1038/ncb1929
Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, Singh R (2012) Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. PLoS One 7(9):e44873. https://doi.org/10.1371/journal.pone.0044873
Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, Fu XD (2014) MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158(3):607–619. https://doi.org/10.1016/j.cell.2014.05.047
Leung AKL (2015) The whereabouts of microRNA actions: cytoplasm and beyond. Trends Cell Biol 25(10):601–610. https://doi.org/10.1016/j.tcb.2015.07.005
Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15(2):185–197. https://doi.org/10.1016/j.molcel.2004.07.007
Hwang HW, Wentzel EA, Mendell JT (2007) A hexanucleotide element directs microRNA nuclear import. Science 315(5808):97–100. https://doi.org/10.1126/science.1136235
Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR (2014) RNAi factors are present and active in human cell nuclei. Cell Rep 6(1):211–221. https://doi.org/10.1016/j.celrep.2013.12.013
Daneholt B (1997) A look at messenger RNP moving through the nuclear pore. Cell 88(5):585–588
Wei Y, Li L, Wang D, Zhang CY, Zen K (2014) Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J Biol Chem 289(15):10270–10275. https://doi.org/10.1074/jbc.C113.541417
Jeffries CD, Fried HM, Perkins DO (2011) Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA 17(4):675–686. https://doi.org/10.1261/rna.2006511
Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J (2011) Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res 39(2):675–686. https://doi.org/10.1093/nar/gkq776
Politz JC, Hogan EM, Pederson T (2009) MicroRNAs with a nucleolar location. RNA 15(9):1705–1715. https://doi.org/10.1261/rna.1470409
Pitchiaya S, Heinicke LA, Park JI, Cameron EL, Walter NG (2017) Resolving subcellular miRNA trafficking and turnover at single-molecule resolution. Cell Rep 19(3):630–642. https://doi.org/10.1016/j.celrep.2017.03.075
Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y, Zhang L, Ding C, Luo H, Li Y, Peng L, Zhao L, Peng S, Xiao Y, Dong S, Cao J, Yu W (2017) MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol 14(10):1326–1334. https://doi.org/10.1080/15476286.2015.1112487
Gonzalez S, Pisano DG, Serrano M (2008) Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle 7(16):2601–2608. https://doi.org/10.4161/cc.7.16.6541
Younger ST, Corey DR (2011) Transcriptional regulation by miRNA mimics that target sequences downstream of gene termini. Mol BioSyst 7(8):2383–2388. https://doi.org/10.1039/c1mb05090g
Catalanotto C, Cogoni C, Zardo G (2016) MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci 17(10):1712. https://doi.org/10.3390/ijms17101712
Leucci E, Patella F, Waage J, Holmstrom K, Lindow M, Porse B, Kauppinen S, Lund AH (2013) microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep 3:2535. https://doi.org/10.1038/srep02535
Tang R, Li L, Zhu D, Hou D, Cao T, Gu H, Zhang J, Chen J, Zhang CY, Zen K (2012) Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res 22(3):504–515. https://doi.org/10.1038/cr.2011.137
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA 105(5):1608–1613. https://doi.org/10.1073/pnas.0707594105
Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC (2012) Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res 40(4):1695–1707. https://doi.org/10.1093/nar/gkr934
Williams T, Fried M (1986) A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3′ ends. Nature 322(6076):275–279. https://doi.org/10.1038/322275a0
Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, Group RGER, Genome Science G, Consortium F (2005) Antisense transcription in the mammalian transcriptome. Science 309(5740):1564–1566. https://doi.org/10.1126/science.1112009
Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, Consortium F, Group RGER, Genome Science G (2005) The transcriptional landscape of the mammalian genome. Science 309(5740):1559–1563. https://doi.org/10.1126/science.1112014
Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA (2008) Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol 15(8):842–848. https://doi.org/10.1038/nsmb.1444
Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG (2008) Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 4(11):e1000258. https://doi.org/10.1371/journal.pgen.1000258
Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C (2012) Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 30(5):453–459. https://doi.org/10.1038/nbt.2158
Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA (2013) Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res 41(22):10086–10109. https://doi.org/10.1093/nar/gkt777
Liu M, Roth A, Yu M, Morris R, Bersani F, Rivera MN, Lu J, Shioda T, Vasudevan S, Ramaswamy S, Maheswaran S, Diederichs S, Haber DA (2013) The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev 27(23):2543–2548. https://doi.org/10.1101/gad.224170.113
Huang S, Wu S, Ding J, Lin J, Wei L, Gu J, He X (2010) MicroRNA-181a modulates gene expression of zinc finger family members by directly targeting their coding regions. Nucleic Acids Res 38(20):7211–7218. https://doi.org/10.1093/nar/gkq564
Melton C, Judson RL, Blelloch R (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463(7281):621–626. https://doi.org/10.1038/nature08725
Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, Jin Y (2010) miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One 5(2):e9429. https://doi.org/10.1371/journal.pone.0009429
He XH, Zhu W, Yuan P, Jiang S, Li D, Zhang HW, Liu MF (2016) miR-155 downregulates ErbB2 and suppresses ErbB2-induced malignant transformation of breast epithelial cells. Oncogene 35(46):6015–6025. https://doi.org/10.1038/onc.2016.132
Panda AC, Sahu I, Kulkarni SD, Martindale JL, Abdelmohsen K, Vindu A, Joseph J, Gorospe M, Seshadri V (2014) miR-196b-mediated translation regulation of mouse insulin2 via the 5′UTR. PLoS One 9(7):e101084. https://doi.org/10.1371/journal.pone.0101084
Wang S, Pan Y, Zhang R, Xu T, Wu W, Zhang R, Wang C, Huang H, Calin CA, Yang H, Claret FX (2016) Hsa-miR-24-3p increases nasopharyngeal carcinoma radiosensitivity by targeting both the 3′ UTR and 5′ UTR of Jab1/CSN5. Oncogene 35(47):6096–6108. https://doi.org/10.1038/onc.2016.147
Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA (2010) A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′ UTRs. PLoS Pathog 6(6):e1000967. https://doi.org/10.1371/journal.ppat.1000967
Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM (2012) Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA 109(3):941–946. https://doi.org/10.1073/pnas.1112263109
Orom UA, Nielsen FC, Lund AH (2008) MicroRNA-10a binds the 5′ UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30(4):460–471. https://doi.org/10.1016/j.molcel.2008.05.001
Ackerman WE, Buhimschi IA, Brubaker D, Maxwell S, Rood KM, Chance MR, Jing H, Mesiano S, Buhimschi CS (2018) Integrated microRNA and mRNA network analysis of the human myometrial transcriptome in the transition from quiescence to labor. Biol Reprod 98(6):834–845. https://doi.org/10.1093/biolre/ioy040
M’Baya-Moutoula E, Louvet L, Molinie R, Guerrera IC, Cerutti C, Fourdinier O, Nourry V, Gutierrez L, Morliere P, Mesnard F, Massy ZA, Metzinger-Le Meuth V, Metzinger L (2018) A multi-omics analysis of the regulatory changes induced by miR-223 in a monocyte/macrophage cell line. Biochim Biophys Acta 1864(8):2664–2678. https://doi.org/10.1016/j.bbadis.2018.05.010
Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ (2004) The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ UTR. Genes Dev 18(2):132–137. https://doi.org/10.1101/gad.1165404
Yekta S, Shih IH, Bartel DP (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304(5670):594–596. https://doi.org/10.1126/science.1097434
Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of microRNA-target recognition. PLoS Biol 3(3):e85. https://doi.org/10.1371/journal.pbio.0030085
Chi SW, Zang JB, Mele A, Darnell RB (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460(7254):479–486. https://doi.org/10.1038/nature08170
Chi SW, Hannon GJ, Darnell RB (2012) An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19(3):321–327. https://doi.org/10.1038/nsmb.2230
Wang C, Chen Q, Li S, Li S, Zhao Z, Gao H, Wang X, Li B, Zhang W, Yuan Y, Ming L, He H, Tao B, Zhong J (2017) Dual inhibition of PCDH9 expression by miR-215-5p up-regulation in gliomas. Oncotarget 8(6):10287–10297. https://doi.org/10.18632/oncotarget.14396
Bracken CP, Scott HS, Goodall GJ (2016) A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet 17(12):719–732. https://doi.org/10.1038/nrg.2016.134
Zhao Y, Qi X, Chen J, Wei W, Yu C, Yan H, Pu M, Li Y, Miao L, Li C, Ren J (2017) The miR-491-3p/Sp3/ABCB1 axis attenuates multidrug resistance of hepatocellular carcinoma. Cancer Lett 408:102–111. https://doi.org/10.1016/j.canlet.2017.08.027
Sokolova V, Fiorino A, Zoni E, Crippa E, Reid JF, Gariboldi M, Pierotti MA (2015) The effects of miR-20a on p21: two mechanisms blocking growth arrest in TGF-beta-responsive colon carcinoma. J Cell Physiol 230(12):3105–3114. https://doi.org/10.1002/jcp.25051
Zhao Y, Chen J, Wei W, Qi X, Li C, Ren J (2018) The dual-inhibitory effect of miR-338-5p on the multidrug resistance and cell growth of hepatocellular carcinoma. Signal Transduct Target Ther 3:3. https://doi.org/10.1038/s41392-017-0003-4
Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD (2009) New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 19(7):1175–1183. https://doi.org/10.1101/gr.089367.108
Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM, Yang PC (2013) MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun 4:1877. https://doi.org/10.1038/ncomms2876
Zongaro S, Hukema R, D’Antoni S, Davidovic L, Barbry P, Catania MV, Willemsen R, Mari B, Bardoni B (2013) The 3′ UTR of FMR1 mRNA is a target of miR-101, miR-129-5p and miR-221: implications for the molecular pathology of FXTAS at the synapse. Hum Mol Genet 22(10):1971–1982. https://doi.org/10.1093/hmg/ddt044
Lai X, Wolkenhauer O, Vera J (2016) Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic Acids Res 44(13):6019–6035. https://doi.org/10.1093/nar/gkw550
Funding
No funding was received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interest exists.
Rights and permissions
About this article
Cite this article
Pu, M., Chen, J., Tao, Z. et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 76, 441–451 (2019). https://doi.org/10.1007/s00018-018-2940-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2940-7