Abstract
Telomere maintenance is required for chromosome stability, and telomeres are typically elongated by telomerase following DNA replication. In both tumor and yeast cells that lack telomerase, telomeres are maintained via an alternative recombination mechanism. Previous studies have indicated that yeast Sgs1 and Top3 may work together to remove highly negative supercoils that are generated from recombination. However, the mechanism by which cells eradicate highly positive supercoils during recombination remains unclear. In the present study, we demonstrate that TOP2 is involved in telomere–telomere recombination. Disturbance of telomeric structure by RIF1 or RIF2 deletion alleviates the requirement for TOP2 in telomere–telomere recombination. In human telomerase-negative alternative lengthening of telomere (ALT) cells, TOP2α or TOP2β knockdown decreases ALT-associated PML bodies, increases telomere dysfunction-induced foci and triggers telomere shortening. Similar results were observed when ALT cells were treated with ICRF-193, a TOP2 inhibitor. Importantly, ICRF-193 treatment blocks ALT-associated phenotypes in vitro, causes telomere shortening, and inhibits ALT cell proliferation in mice. Taken together, these findings imply that TOP2 is involved in the ALT pathway, perhaps by resolving the highly positive supercoil structure at the front of the helicase. Inhibition of topoisomerase II may be a promising therapeutic approach that can be used to prevent cell proliferation in ALT-type cancer cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Telomeres are dynamic DNA–protein complexes that protect the ends of linear chromosomes, prevent detrimental chromosome rearrangements, and defend against genomic instability and the associated risk of cancer [1]. Telomeres are composed of telomeric DNA, which consists of tandem repeats of short G-rich sequences that are synthesized by the enzyme telomerase [2, 3]. The reverse transcriptase utilizes the RNA component as a template to add the G-rich repeats onto the 3′ ends of the chromosomes [2–4]. In certain human cells, telomerase activity is absent and telomeres are gradually shortened with successive cell divisions due to incomplete replication, which eventually causes replicative senescence. Once telomeres become sufficiently short, they lose the ability to protect the ends of the chromosomes from being recognized as broken ends and being subjected to nuclease digestion, homologous recombination, and nonhomologous end-joining repair. Continuous telomere shortening in human fibroblasts leads to chromosome fusions, crisis, and apoptosis [5]. A limited number of human cells can bypass crisis either through telomerase reactivation [5] or through the alternative ALT pathway for telomere lengthening [6–8].
Even in organisms that normally rely on telomerase, co-existing of telomerase-independent mechanisms allows cells to pass through crisis. Although the majority of Saccharomyces cerevisiae cells that lack the genes for telomerase components eventually enter cell cycle arrest, survivors arise relatively frequently [9]. The generation of survivors requires RAD52-dependent homologous recombination. In S. cerevisiae, the majority of cells that survive in the absence of telomerase activity contain multiple tandem copies of the subtelomeric Y′ element and very short terminal tracts [10] (type I survivors). In a minor fraction of the survivors (type II), the lengths of the telomere sequences are increased heterogeneously from several hundred base pairs to 10 kb or longer [10]. The generation of type II survivors is dependent on the presence of Rad50, Rad59, Rap1, Sgs1, and Top3 [11–15]. The structure of type II telomeres in Saccharomyces resembles that of 15 % of human cell lines and tumors that maintain telomeric DNA via the ALT pathway [6–8]. These ALT cells display unique nuclear foci, termed ALT-associated PML bodies (APBs), that contain RAD52, RAD51, RAD50, RPA, TRF1, TRF2, telomeric DNA, and NBS1 [16].
Telomeres are protected by the Rap1–Rif1–Rif2 complex. Not only does the Rap1–Rif1–Rif2 complex provide a sheltering function for nucleases and telomerase, but it also exhibits a counting ability that balances the length of telomeres in telomerase-positive cells [17]. For the type II telomerase-negative cells that contain long telomeres, the extensive Rap1–Rif1–Rif2 complexes theoretically also form a barrier, preventing the recombinational helicase to move forward easily and creating excessive local supercoiling during telomere recombination [14]. Thus, the cell must carefully control DNA supercoiling to ensure that telomere recombination proceeds smoothly.
Two widely conserved topoisomerases, TOP1 and TOP2, have been demonstrated to be required for DNA replication and to encode functional activities to relieve positive supercoils [18]. In the present study, we examined whether TOP1 and TOP2 are involved in telomere–telomere recombination. We demonstrated that TOP2, but not TOP1, is involved in telomere–telomere recombination in yeast. In addition, the Rap1–Rif1–Rif2 complex is involved in the establishment of TOP2-dependent telomere–telomere recombination. TOP2α or TOP2β knockdown in human ALT cells decreases APB formation and increases telomere dysfunction-induced foci (TIFs). Additionally, ALT cells are more sensitive to the TOP2 inhibitor, ICRF-193 [19], than telomerase-positive cells, and treatment of ALT cells with ICRF-193 causes telomere shortening. Our results demonstrate that hTOP2 is involved in telomere recombination. Therefore, TOP2 inhibitors can be used to block telomere recombination in ALT cells and may provide a novel remedy for telomerase-negative cancer cells.
Materials and methods
Yeast strain and plasmid constructions
All yeast operations were performed by standard methods. In this study yeast strains used were the derivatives of YPH501 (MATa/MATα ura3-52/ura3-52 lys2-801 amber/lys2-801 amber ade2-101 ochre/ade2-101 ochre trp1Δ63/trp1Δ 63 his3Δ 200/his3Δ200 leu2-Δ1/leu2-Δ1). The yeast strains carrying tlc1, tlc1 rad51, tlc1 rif1 and tlc1 rif2 were previously described [10, 14]. The top2 temperature-sensitive mutant is a gift from Dr. Leroy Liu [20]. tlc1 top2-ts and tlc1 rad51 top2-ts mutant strains were generated by mating the deletion mutants, followed by sporulating the diploid and selecting for temperature-sensitive and auxotrophic markers (Supplemental Table 1). To obtain type I and type II survivors, spore cells were serially diluted into or restreaked onto YEPD medium as described [10, 14]. The pRS424-yTOP2, pRS424-hTOP2α and pRS424-hTOP2β were constructed by cloning the PCR-amplified fragment of the yeast TOP2, human TOP2α and TOP2β gene into the vector (Supplemental Table 2).
Yeast culture condition, DNA preparation, enzyme digestion, gel electrophoresis, and Southern analysis
Each spore colony was inoculated into YEPD and grown at 30 °C. top2-ts cells were cultured at semi-permissive temperature (30–32 °C) to observe the phenotype. Several independent isolates for each genotype were analyzed. Genomic DNA preparation and Southern analysis were performed as previously described [15]. To examine the DNA from individual colonies, each colony was expanded in 2 ml of liquid medium to obtain DNA for Southern analysis. The DNA was digested with XhoI to observe the type I pattern or with a mixture of HaeIII, HinfI, HinpII, and MspI four-base cutters to observe the type II pattern. A 270-bp C1–3A fragment was randomly labeled with the random prime labeling system (Invitrogen) and used in Southern hybridization. Data shown here are representative of two or more experiments from independent spores.
Mammalian cell culture, transfection, RNA interference, synthesis of the TOP2 inhibitor and western blot analysis
Cells were cultured in Dulbecco’s modified Eagle’s medium (HeLa, HCT116, U2OS, and VA13-WI38) or RPMI medium (Saos2 and SKLU1) containing fetal bovine serum, penicillin, streptomycin, glutamine, nonessential amino acids (Hyclone), and puromycin (1 μg/ml; Invitrogen). Uninfected cells were cultured in the same media without puromycin. Transfection was conducted using Lipofectamine 2000 (Invitrogen). Helper plasmids and lentiviral vectors expressing shRNAs specifically targeting hTOP2α and hTOP2β were obtained from the National RNAi Core Facility, Taiwan (http://rnai.genmed.sinica.edu.tw). Nonreplicative viral particles were prepared as recommended and their knockdown efficiencies were determined by Western blot analysis. Using (R, S)-2, 3-butanediol as starting material, ICRF-193 was synthesized in five steps with a total yield of 19 % according to the literature with modification [19]. Cells were treated with 150 nM ICRF-193 for 10-50 PD passages.
Cell extracts were loaded on an SDS–polyacrylamide gel. After electrophoresis, the proteins were transferred onto a PVDF membrane. The membrane was then blocked with 5 % non-fat milk for 1 h at room temperature. Primary antibodies used to detect hTOP2α (BD Transduction Laboratories), hTOP2β (Bethyl Laboratories) and actin (GeneTex) were diluted 1:1000 in the blocking buffer and incubated for 1 h at room temperature. After one wash with PBS containing 0.1 % Tween, secondary antibodies were applied for 1 h at room temperature. A conventional chemiluminescence system was used to visualize proteins following the manufacturer’s instructions (PerkinElmer Life Sciences). Data shown here are representative of three or more independent experiments.
Telomere restriction fragment (TRF) analysis
Genomic DNA was isolated from cells by a genomic DNA purification kit (Promega), and 1.5 μg of DNA was digested by RsaI and HinplI. The digested DNA was resolved by pulsed-field gel electrophoresis (BioRad), transferred onto a Hybond N+ nylon membrane and hybridized overnight to a 32P-labeled 800-bp telomere-specific probe.
Immunofluorescence staining and telomere fluorescence in situ hybridization (FISH) staining
Cells were grown on glass coverslips, growth-arrested by withdrawal of methionine for 4 days, fixed in 4 % paraformaldehyde at room temperature for 10 min, followed by permeabilization in 0.05 % Triton X buffer at room temperature for 10 min. For dual immunostaining, slides were blocked with 1 % bovine serum albumin and incubated at 4 °C for overnight with mouse anti-TRF2 (Millipore), rabbit anti-PML (Santa Cruz Biotechnology) or γ-H2AX (Millipore) antibodies. The primary antibodies were detected using Rhodamine Red-conjugated goat anti-rabbit and fluorescein isothiocyanate-conjugated goat anti-mouse antibodies (1:500). The slides were fixed for 10 min in 1× PBS with 4 % (v/v) formaldehyde at room temperature. For telomere FISH staining, slides were rinsed with graded ethanol series (70 % for 2 min, 90 % for 2 min and 100 % for 2 min) and air-dried. Dehydrated slides were soaked with telomeric PNA probe (Panagene) in hybridization solution (30 % formamide, 0.1 % Triton X-100, 50 mM NaCl, 5 mM sodium citrate). Slides were incubated at 80 °C for 3 min, hybridized at room temperature for 2 h, and then washed in PNA wash A (1× PBS, 0.1 % Tween-20), and PNA wash B (30 mM sodium citrate, 300 mM NaCl and 0.05 % Tween-20). DNA was stained with DAPI at room temperature. Immunofluorescence was analyzed with a Zeiss Axioplan fluorescence microscope.
In vivo pharmacology with xenografted mouse tumors
All animal experiments were performed according to regulations approved by the Animal Ethical Committee of National Taiwan University and National Taiwan University College of Medicine and College of Public Health Institutional Animal Care and Use Committee (IACUC) [20120085]. Animals were killed using CO2 asphyxiation and the appropriate tissue harvested. In the pilot toxicity experiments to optimize the working concentration of ICRF-193, doses of 0.3125, 0.625, 1.25, 2.5, and 5 mg/kg body weight were administered every 3 days and the dosage of ICRF-193 in this study was determined by the highest non-lethal dose. Human Saos2 ALT cancer cells and HeLa cells were trypsinized, resuspended as single cell suspensions and injected into left and right flanks of NOD/SCID mice. Mice were injected with 1.25 mg/kg ICRF-193 4–7 days after xenograft via intraperitoneal administration. Tumor size was monitored daily until tumor volume had reached 1,000 mm3.
Results
TOP2 contributes to telomere recombination in telomerase-minus (tlc1Δ) yeast
In the tlc1Δ strain that lacks telomerase activity, yeast cells gradually lose viability after 40–60 generations of propagation. However, a subset of cells in the culture eventually bypasses the senescence and survives. Two classes of survivors, type I and type II, have been identified and can be distinguished by two different Southern blot analyses [10] (Fig. 1a, b; Supplemental Figure 1). The type I telomere pattern is more easily visualized by XhoI digestion Southern blot analysis. XhoI cleaves 0.9 kb from the 3′ end of the Y′ element. In type I survivors, three major XhoI fragments (~1.0, 6.7, and 5.2 kb) hybridized to the telomere probe. The ~1.0-kb terminal fragment from Y′ telomeres consisted mainly of Y′ DNA plus a short stretch of TG1–3/C1–3A repeat. The strong hybridization at 6.7 and 5.2 kb contained the tandemly repeated Y′ long and Y′ short elements, respectively (Fig. 1a; Supplemental Figure 1). In the other Southern blot analysis (Fig. 1b), the genomic DNA is digested with a mixture of four restriction enzymes to very small fragments. The telomere sequences, which are not cut by these enzymes, remain relatively large. Using this technique, DNA from wild-type YPH499 cells yields C1–3A-hybridizing fragments of ~400 ± 75 bp from Y′ telomeres, ~500-bp fragments from tandem Y′ DNA, and fragments of up to 1.1 kb from X telomeres [10] (Fig. 1b, wild type). DNA from type I survivors yields fragments of less than 300 bp from Y′ telomeres (asterisk, terminal Y′, Fig. 1b), in addition to the ~500-bp fragment from tandem Y′ elements (arrow, Fig. 1b). In contrast, type II telomeric DNA digested with these enzymes yields many differently sized fragments of up to 10 kb or even larger (Fig. 1b). In a tlc1 mutant liquid culture, although ~90 % of the survivors are type I, they grow slowly and are quickly overtaken by the type II survivors, which have a growth rate similar to that of wild-type cells [10]. This is also exacerbated by the ability to gradually convert from the type I to the type II pattern of telomere structure [10]. In the liquid culture assay [14], when cultures starting from freshly dissected spores were repeatedly diluted 1:10,000 at 48-h intervals, dramatic telomere lengthening could be observed after several dilutions (tlc1, Fig. 1b).
TOP2 contributes to telomere length maintenance in a tlc1 strain. a Southern blot of XhoI-digested genomic DNA that was isolated from serial liquid dilutions of the tlc1, top2-ts, and tlc1 top2-ts strains, which were probed to detect telomeric sequences. The 5.2 kb short Y′ and 6.7 kb long Y′ are shown on the right. WT wild type. Size markers (kilobases) are shown on the left. b Detection of TRFs in the same set of strains used in a was performed by hybridization of four-base cutter-digested genomic DNA using a telomeric probe. An asterisk marks the position of critically short telomeres, and the arrow indicates the TG1–3/C1–3A fragments between Y′–Y′ tandem repeats. c The diploid strain YPH501 tlc1/TLC1 top2-ts/TOP2 was sporulated, and tetrads were dissected. Individual survivors were isolated from the tlc1 and tlc1 top2-ts strains by restreaking the strains onto YEPD medium, and their DNA was analyzed as in a and b. tlc1 rad51 spores were used as a control. d The diploid strain YPH501 tlc1/TLC1 rad51/RAD51 top2-ts/TOP2 was sporulated and dissected, and freshly isolated spore colonies were restreaked for single colonies. Each strain was repeatedly streaked on solid YEPD plates and grown for 3 days at 30 °C. It should be noted that tlc1 rad51 top2-ts, but not tlc1 rad51 or tlc1 top2-ts, spores senesced extremely fast
TOP2 is an essential gene in yeast and cannot be deleted in haploid cells. Therefore, to investigate whether TOP2 contributes to telomere replication, we used the top2-4 (TOP2 temperature sensitive; ts), which carries a single mutation that replaces proline 821 with glutamine [21], for our study. The top2-4 mutation inhibits the double-strand passage activity, leading chromosomes to topologically interlinked [22, 23]. tlc1 top2-ts yeast strains were cultured at permissive (23 °C) and semi-permissive temperatures (30 °C). Their telomere lengths were then assessed using Southern blot analysis with telomeric DNA probes. Compared to the tlc1 control, the top2-ts strain showed no signs of telomere recombination or cellular senescence at both temperatures (Fig. 1a, b and data not shown). Thus, we determined that the TOP2 defect alone does not cause cells to switch from utilizing telomerase to the telomere recombination pathway in telomerase-positive cells. However, the tlc1 top2-ts cells exhibited a cellular senescence phenotype (see below). Examination of the telomeres of the tlc1 top2-ts strain in liquid revealed that tlc1 top2-ts cells displayed a type II telomeric pattern at the permissive temperature. However, at the semi-permissive temperature, increases in the 5.2 kb short Y′ and 6.7 kb long Y′ signals and the loss of elongated telomere tracks in tlc1 top2-ts cells indicated that the cells mainly use type I recombination for telomere elongation (Fig. 1a, b). In contrast, this kind of switch to type I survivors was not detected in tlc1 top1 cells (Supplemental Figure 2). These data suggest that TOP2 is important for generating type II survivors. To further examine this hypothesis, we used solid plate analysis. On solid plates, we isolated otherwise isogenic spore products by dissecting heterozygous diploids and restreaking them multiple times on plates at the semi-permissive temperature until the survivors appeared. DNA was prepared from the individual survivors, and the telomere patterns of the individual survivors were determined using Southern blot analysis. Significantly, all of the tlc1 top2-ts survivors (100 out of 100) on solid plates displayed the type I pattern, whereas both type I and type II survivors were recovered from the tlc1 strain at a similar ratio (93:7) as previously reported (Fig. 1c). These data suggest that type II recombination is inhibited in tlc1 top2-ts cells at the semi-permissive temperature. Together, these results indicate that TOP2 participates in telomere recombination in telomerase-minus yeast cells.
Cellular senescence was defined as a progressive reduction in the growth rate and an increase in the frequency of cell death in telomerase-minus yeast [24]. Since Rad51 is essential for type I recombination [11, 14], to test whether tlc1, rad51, and top2 mutations could abolish all three telomere maintenance pathways (telomerase, type I, and type II) and accelerate cell death, strains generated from freshly dissected spores with different mutation backgrounds were examined using serial single colony restreaking on solid plates at the semi-permissive temperature (Fig. 1d). In contrast to the tlc1, tlc1 rad51, tlc1 top2-ts strains, which displayed cellular senescence phenotypes on streaking plates, the tlc1 rad51 top2-ts spores were inviable after the second streak (Fig. 1d). On the other hand, the cell viability of rad51 top2-ts cells was not affected after several restreaks (Supplemental Figure 3A), demonstrating that the double mutation of TOP2 and RAD51 does not lead to a synthetic sick phenotype. Rapid telomere shortening was also observed in tlc1 rad51 top2-ts spores (Supplemental Figure 3B). These findings suggest that the triple mutation completely eliminates all three pathways that participate in telomere maintenance, thereby causing accelerated cellular senescence and preventing survivors from forming, and again supporting the important role of TOP2 in type II telomere–telomere recombination.
Mutations in RIF1 and RIF2 restore telomere–telomere recombination in tlc1 top2-ts cells
Rap1, together with Rap1 interacting proteins, Rif1 and Rif2, forms a higher-order telomere structure that protects telomeres and regulates telomere silencing [25]. While Rif1 supports the function of the Cdc13–Stn1–Ten1 complex in telomere capping [26], Rif2 and Rap1 inhibit both nuclease access and nonhomologous end-joining at telomeres [27], indicating that Rif1 and Rif2 play slightly different functions in end protection. Previous studies showed that by interacting with Rap1, Sir4 initiates the sequential assembly of the Rap1–Sir complex at telomeres to form silenced chromatin [28]. The Rap1–Rif complex contributes to anchoring of yeast telomeric DNA, which imposes sufficient topological constraint to accumulate telomeric supercoils [29, 30]. We surmised that the supercoiled DNA structure might also be generated during recombination/helicase movement at Rap1–Rif1–Rif2 protected telomeres. Highly negative supercoiled DNA downstream of the helicase may be resolved by Top3–Sgs1 [15, 31]. However, the biochemical properties of Top3 prevent it from resolving the highly positive supercoiled DNA upstream of the helicase [31]. As previously reported [32, 33], deletion of RIF1 and/or RIF2 led to telomere lengthening (Supplemental Figure 4A). Hence, we next tested whether TOP2 deletion-dependent type I formation is altered in the absence of the Rif1 and/or Rif2 complex. The survivor patterns of tlc1 top2-ts, tlc1 rif1 rif2 and tlc1 rif1 rif2 top2-ts cells were determined by solid plate analysis. The tlc1 rif1 rif2 top2-ts strain displayed higher ratio of type II pattern (14 out of 84, 16.7 %) than the tlc1 top2-ts strain (0 out of 100, 0 %) (Supplemental Figure 4B). rif1 rif2 mutations restored type II telomere–telomere recombination in tlc1 top2-ts mutant cells, such that tlc1 rif1 rif2 top2-ts mutants generated the type II pattern in liquid culture (Supplemental Figure 4C). tlc1 rif1 rif2 top2-ts at 23 and 30 °C and tlc1 rif1 rif2 rad51 top2-ts cells at 23 °C still exhibited cellular senescence phenotypes on streaking plates, whereas the tlc1 rif1 rif2 rad51 top2-ts spores at 30 °C were inviable after the second streak (Supplemental Figure 4D), suggesting that all three pathways for telomere replication (telomerase, type I and type II recombination) are inhibited in the tlc1 rif1 rif2 rad51 top2-ts cells. Together, these data demonstrate that mutations in RIF1 and RIF2 restore telomere–telomere recombination in tlc1 top2-ts cells.
Expression of yeast TOP2 and human TOP2α and TOP2β restores type II survivors in tlc1 top2-ts cells
Yeast TOP2 has two human orthologs, topoisomerase II α and β, which have distinct expression patterns and cellular roles. Human TOP2α is mainly involved in DNA replication and chromosome segregation, whereas hTOP2β plays roles in transcriptional regulation and neuronal development [34]. To further confirm the role that TOP2 may play in the generation of type II survivors, we constructed high copy plasmids containing yeast TOP2, human TOP2α, or human TOP2β. tlc1 top2-ts strains expressing yeast TOP2, human TOP2α, and TOP2β grew at the non-permissive temperature (37 °C), but strains containing the empty plasmid did not (Fig. 2a), suggesting that expressed proteins complement TOP2 activity in yeast. It is worth noting that overexpression of yeast TOP2 resulted in a slow growth phenotype (Fig. 2a), as previously reported [35]. Analysis of the telomere patterns of these strains revealed that, unlike the tlc1 top2-ts strains, which produced the type I pattern at the semi-permissive temperature (32 °C), these complements can at least partially recover the type II pattern (Fig. 2b). Thus, yTOP2, hTOP2α and hTOP2β can functionally complement some defects in telomere–telomere recombination in tlc1 top2-ts cells. Together, these data demonstrate that TOP2 is essential for the generation of type II survivors and that hTOP2α and hTOP2β may replace the yeast TOP2 in type II survivor formation.
Yeast and human TOP2 rescue the type I phenotype in a tlc1 top2 strain. a top2-ts strains were transformed with either the empty vector, a plasmid containing the yeast TOP2, or a plasmid containing the human TOP2α or TOP2β gene under the control of the yeast GAL1 promoter. Each strain was streaked on a complete medium plate lacking tryptophan and grown at 23 or 37 °C. b Spores from the tlc1 top2-ts strains carrying a plasmid expressing wild-type yeast TOP2, human TOP2α, or TOP2β were isolated on complete medium lacking tryptophan and were grown at 32 °C. Southern blot analysis of four-base cutter-digested genomic DNA that was isolated from serial liquid dilutions was performed and probed to detect telomeric sequences. An asterisk marks the position of critically short telomeres, and the arrow indicates the TG1–3/C1–3A fragments between Y′–Y′ tandem repeats. Size markers (kilobases) are shown on the left
Knockdown of TOP2α or TOP2β inhibits telomere maintenance in ALT cells
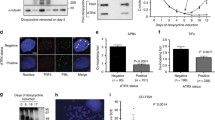
Because we observed that hTOP2α and hTOP2β can replace yTOP2 in type II survivor formation and that the telomeric pattern in yeast type II survivors is similar to that in human ALT cancer cells, we investigated the roles of TOP2α and TOP2β in the ALT pathway. To examine the importance of TOP2α and TOP2β in ALT cells, TOP2α or TOP2β expression was down-regulated using lentiviral short hairpin (sh) RNA interference in telomerase-positive and ALT cells. Western blot analysis demonstrated that the TOP2α and TOP2β protein levels were decreased in cells that had been infected with the shTOP2α and shTOP2β viruses, respectively (Fig. 3a; Supplemental Figure 5). The telomere lengths following population doublings (PDs) were analyzed using Southern blot analysis with a telomeric probe (Fig. 3b) to examine the role of TOP2α or TOP2β in telomere maintenance in ALT cells. TOP2α or TOP2β down-regulated U2OS (Fig. 3b) cells displayed telomere shortening phenotypes compared to control cells, while TOP2 down-regulated HeLa cells did not display any alteration in telomere lengths. These results indicate that TOP2 plays a role in telomere maintenance in ALT cells.
TOP2α and TOP2β are involved in telomere maintenance and APB formation in ALT cells. a Depletion of TOP2α and TOP2β in ALT cells (U2OS) and telomerase-positive cells (HeLa) 3 PD after infection was determined using western blot analysis. b Telomere length was analyzed via the TRF assay in HeLa and U2OS cells. TRF was conducted with cells from the indicated PDs, and the extracted DNA was digested and detected using a telomeric DNA probe. The maximum integrity of the signal was quantified using ImageQuant software and is indicated by a black line. c ALT cells (U2OS, VA13, and Saos2) and telomerase-positive cells (HeLa) were subjected to shRNA knockdown. APBs were analyzed by examining colocalization of TRF2 (red) and PML (green). Representative photos from U2OS cells are shown. d Quantification of the results from the cells shown in c. A total of 200 cells were observed. At least four co-localized PML/TRF2 signals with size larger than 0.9 μm in an individual cell nucleus were scored. e Telomere dysfunction induced a DNA damage response after TOP2α and TOP2β depletion in ALT cells. The cells were infected with either the control lentiviral vector or shTOP2α/β. Telomere (green) and γ-H2AX (red) foci were detected using FISH and immunostaining, respectively. f Quantification of the results from the cells shown in e. More than 200 cells were counted, and cells with greater than four γ-H2AX and telomere colocalized foci were scored. Each bar represents mean ± SD
ALT cells exhibit unique nuclear APB foci, which is a signature of ALT cells [36]. To determine whether the APB status is altered in TOP2-depleted ALT cells, TOP2α or TOP2β expression was down-regulated using lentiviral shRNA interference in one telomerase-positive (HeLa) and three ALT cells (U2OS, VA13 and Saos2) (Fig. 3a; Supplemental Figure 6), APBs were examined using immunofluorescence analysis. APBs-positive cells were scored by examining colocalized foci of PML and the telomeric probe. The majority of the APB foci were decreased in TOP2α- or TOP2β-depleted cells compared to the control virus-infected cells (Fig. 3c). These decreases were observed in all ALT cell lines (Fig. 3d), suggesting that APBs were detected less frequently in the absence of TOP2. Previous studies have shown that TIFs are readily detectable when telomere stability is compromised [37, 38]. To determine whether TOP2α or TOP2β repression affects telomere integrity, we further examined telomere dysfunction in these cells. TIFs can be detected due to the presence of γ-H2AX, a phosphorylated variant of histone 2A that associates with DNA double-strand breaks. Cells with γ-H2AX and telomere colocalized foci were scored. TOP2α- or TOP2β-depleted U2OS cells were analyzed for colocalization of γ-H2AX and telomeric foci (Fig. 3e). Increases in TIFs were observed in shTOP2α and shTOP2β ALT cancer cells but not in telomerase-positive cells (Fig. 3f). These results indicate that TOP2 suppresses TIFs formation.
Topoisomerase II inhibition decreases ALT cell viability, causes ALT telomere shortening, and inhibits ALT cancer progression in mice
Because TOP2 and TOP3 are involved in the ALT pathway, inhibition of these enzymes should hinder telomere maintenance in ALT cells. No anti-TOP3 inhibitors are currently available, but several well-developed TOP2 inhibitors are used clinically [39]. Thus, we examined the abilities of TOP2 inhibitors to block the ALT pathway. Topoisomerase catalytic inhibitors are much less cytotoxic, because, unlike topoisomerase poisons, no DNA lesion is formed. Among many topoisomerase catalytic inhibitors, low concentration of ICRF-193 is able to inhibit TOP2 activity by a unique mechanism in which TOP2 is locked in a ‘closed clamp’ [40]. This conformation occurs after strand passage and DNA religation but before the hydrolysis of ATP [40]. ICRF-193 is a well-known anti-tumor drug for treating acute leukemia and lymphosarcoma [41, 42]. We used ICRF-193 to treat two telomerase-positive (HCT116 and HeLa) and two ALT (U2OS and Saos2) cell lines, and their cell proliferation and ALT features were analyzed. While the telomerase-positive cells only displayed marginal effects and were less sensitive to ICRF-193, ICRF-193-treated ALT cells displayed an inhibition of proliferation in a dose-dependent manner (Fig. 4a; Supplemental Figure 7). Telomere shortening was also observed in ICRF-193-treated ALT cells, but not in telomerase-positive cells (Fig. 4b). We next measured the numbers of APBs and TIFs to determine whether the ALT pathway is affected in drug-treated ALT cells. ICRF-193 treatment decreased APBs (Fig. 5a, b) and increased TIFs (Fig. 5c, d) in ALT cells. These findings suggest that TOP2s participate in telomere–telomere recombination and that ALT cells are more sensitive to the TOP2 inhibitor than telomerase-positive cancer cells.
Treatment with a TOP2 inhibitor results in cell cycle delay and telomere shortening. a The cells were equally seeded in ICRF-193-containing medium, and long-term growth curves for ALT (U2OS and Soas2) and telomerase-positive cells (HCT116 and HeLa) were calculated. b Southern blot hybridization of a telomeric probe 32P (TTAGGG) to equal amounts of genomic DNA from the cells. TRF analysis was conducted for cells treated with ICRF-193 for the indicated PDs. The maximum integrity of the signal was quantified using ImageQuant software and is indicated by a black line
Inhibition of TOP2 activity reduces APBs and promotes TIF formation in ALT cells. a ALT (U2OS, Saos2 and VA13) and telomerase-positive (HeLa) cells were treated with a TOP2 inhibitor ICRF-193. APBs were analyzed by examining the colocalization of PML (red) and the telomeric probe (green). b Quantification of the results from the cells shown in a. c Telomere dysfunction-induced DNA damage response after TOP2 inhibition in ALT (U2OS, Saos2 and VA13) and telomerase-positive (HeLa) cells. The cells were treated with ICRF-193. TIFs were analyzed by examining γ-H2AX foci (red) that colocalized with the telomeric probe (green). d Quantification of the results from the cells shown in c. Cells with greater than four γ-H2AX and telomere colocalized foci were scored. Each bar represents mean ± SD
To validate the efficacy of ICRF-193 in vivo, we generated subcutaneous xenografted tumors in NOD/SCID mice using HeLa and Saos2 ALT cells (Fig. 6a, c). Palpable tumors were evident within 1 week. Mice were injected with ICRF-193 on days 4–7 after xenograft. None of the mice displayed decreases in body weight or displayed any signs of toxicity after the injection of the selected dose of ICRF-193 (Fig. 6b, d). ALT tumor volume and weight regressions were observed within 2 weeks to 1 month after treatment, while telomerase-positive HeLa cell tumor volume and weight did not significantly differ following DMSO and ICRF-193 treatment (Fig. 6e, f). These findings demonstrate that ICRF-193 inhibits TOP2-dependent ALT tumor propagation in vivo.
The topoisomerase II inhibitor ICRF-193 inhibits ALT cancer progression in a mouse model. a NOD-SCID mice bearing HeLa human tumor xenografts were intraperitoneally injected with ICRF-193 at 1.25 mg/kg (red) or DMSO (blue). The mean ± SD from five independent measurements are shown. b Body weight curve. c NOD-SCID mice bearing Saos2 ALT human tumor xenografts were intraperitoneally injected with ICRF-193 at 1.25 mg/kg (red) or DMSO (blue). The mean ± SD from ten independent measurements are shown. d Body weight curve. e Tumor photos. Scale bar 1 cm. f Tumor weight. The mean ± SD from five independent measurements are shown
Discussion
Telomere recombination pathway can occur by homologous recombination-mediated inter-telomeric copying of the telomere. This process requires many recombinational and telomere-binding proteins, and proteins specifically play positive and negative roles in the telomere recombination pathway have been identified in yeast and human cells [43, 44]. While the recombinational proteins execute the homology searching, strand transfer, branch migration and DNA polymerase elongation steps (Supplemental Figure 8), telomere-binding proteins can modulate the efficiency of telomere recombination [14, 45]. Besides, evidence for altered epigenetic state and chromatin structure in human ALT telomeres were revealed by mass spectrometric analysis of the protein composition of telomeric chromatin [46]. A recent study demonstrated that chromatin remodeler ATRX is lost in 90 % of immortalized ALT cell lines [47]. Loss of ATRX has been speculated to increase stalled replication forks at telomeric site and leads to the initiation of aberrant telomeric recombination and ALT pathway activation.
We previously discovered that Top3 is required for telomere recombination [15]. However, the role of Top3 in this process was not clearly defined. Two possible functions of Top3 during telomere recombination have been proposed [15]. It may either resolve the Holliday junction or relieve topological stress. Top3 was demonstrated to resolve a double Holliday junction both in vivo and in vitro [48, 49]. Nevertheless, telomere maintenance in the absence of telomerase has been shown to employ a break-induced replication (BIR) mechanism [50, 51]. The BIR only performs strand transfer once and therefore generates a single Holliday junction, which has not been shown to be an adequate substrate for Top3. This effect led us to speculate that Top3 may remove the highly negative supercoils [31] that are generated during telomere recombination downstream of the helicase. If that is the case, another enzyme should exist to remove the highly positive supercoils that are generated upstream of the helicase during BIR.
Our data reveal that TOP2 is involved in telomere maintenance in yeast and human cells. TOP2 might directly participate in type II formation in telomerase-minus cells. The Gasser lab first demonstrates cytologically that telomeres anchor in clusters at the nuclear membrane [52]. Rif1 was then found to be required for anchorage [30]. This anchorage might impose topological constraints on DNA that might require topoisomerases for resolution [29]. Thus, if ALT repair introduced supercoils cannot escape telomeres by DNA swiveling, TOP2 might be utilized for supercoil relaxation. Our results demonstrated that a TOP2 enzymatic inhibitor, ICRF-193, can inhibit telomere lengthening in ALT cancer cells. Together, these data indicate that the telomeric structure may be a barrier during the movement of the helicase and that TOP2 and Top3 may cooperatively relieve the tension on supercoils (Supplemental Figure 8). Previous studies discovered that TRF2 and Apollo cooperate with TOP2α to maintain telomere integrity in telomerase-positive cells during S phase [50], and TOP2 might also play a role in protecting long telomere structure in telomerase-negative and/or Type II cells. Another findings revealed that TOP2 associates with BLM [53], a telomere-binding helicase in ALT cells [54], which further implied that TOP2 might participate in telomere–telomere recombination. BLM may be a helicase that opens double stranded DNA during BIR and the enzymatic activity of TOP2 may be used to resolve the topological tension generated during helicase movement (Supplemental Figure 8). Of course, our studies did not exclude other possibilities such that TOP2 may regulate the expression of some telomere-binding proteins that are involved in telomere–telomere recombination in ALT cells, although that might not be an explanation in yeast, because deletion of yeast TOP2 does not alter the expression of proteins required in type II recombination [55].
Telomeres are a rational target for anticancer therapeutics [56]. To date, inhibitors that modulate telomere replication have only been described for telomerase-positive cells. The importance of understanding the molecular mechanisms and factor requirements of the ALT pathway goes far beyond solving a crucial and interesting problem in telomere biology, especially because no ALT-specific inhibitor currently exists and the alternative ALT pathway may lead to therapeutic failures and/or acquired resistance during telomerase inhibition-based anticancer therapy [57–60]. More than 20 proteins have been identified in the ALT pathway [44]. However, many of these are structural proteins and there is no easy approach to only inhibit their functions in the ALT pathway. We previously demonstrated that TOP3 is required for human ALT cancer cells. Here, we showed that TOP2 is involved in telomere–telomere recombination. Therefore, TOP2 and TOP3 became two rational enzymatic targets for ALT treatment. While TOP3 suppressors have not been discovered so far, abundant TOP2 suppressors, including poisons and inhibitors, have been developed and applied in cancer therapeutics, which makes TOP2 currently the best drug target for ALT-type cancer cells.
Eukaryotic topoisomerase II catalyzes topological genomic changes that are essential for chromosome segregation, chromatin reorganization, DNA replication, and transcription. Mammalian topoisomerase II exists as two isoforms: TOP2α and TOP2β. Human TOP2α is an important cancer drug target and an established determinant of drug sensitivity. Human TOP2β is also the target of anticancer drugs, but its role in drug sensitivity is less well defined [61]. Here, we show that both TOP2α and TOP2β participate in telomere–telomere recombination. Many TOP2 drugs are presently used as cancer therapeutic agents, have passed preclinical and clinical tests, and can be used against ALT cancer cells. Moreover, because they have been used to treat cancer, TOP2 inhibitors may produce synergistic anti-tumor efficacy in ALT cells without increasing toxicity to non-cancer cells. Since the ALT pathway is one of the main causes of current therapeutic failures and/or acquired resistance in telomerase-inhibition-based anticancer approaches [57–60], combination of a telomerase inhibitor and an ALT pathway inhibitor may represent a valuable anticancer therapeutic strategy that can be used to treat all types of cancers.
Abbreviations
- ALT:
-
Alternative lengthening of telomeres
- APBs:
-
ALT-associated PML bodies
- TIFs:
-
Telomere dysfunction-induced foci
- BIR:
-
Break-induced replication
- SD:
-
Standard deviation
References
Wellinger RJ, Zakian VA (2012) Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191:1073–1105
de Lange T (2002) Protection of mammalian telomeres. Oncogene 21:532–540
Greider CW, Blackburn EH (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405–413
Vega LR, Mateyak MK, Zakian VA (2003) Getting to the end: telomerase access in yeast and humans. Nat Rev Mol Cell Biol 4:948–959
Artandi SE, DePinho RA (2000) A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr Opin Genet Dev 10:39–46
Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR (1997) Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med 3:1271–1274
Dunham MA, Neumann AA, Fasching CL, Reddel RR (2000) Telomere maintenance by recombination in human cells. Nat Genet 26:447–450
Reddel RR, Bryan TM, Colgin LM, Perrem KT, Yeager TR (2001) Alternative lengthening of telomeres in human cells. Radiat Res 155:194–200
Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404–409
Teng SC, Zakian VA (1999) Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19:8083–8093
Chen Q, Ijpma A, Greider CW (2001) Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol 21:1819–1827
Huang P, Pryde FE, Lester D et al (2001) SGS1 is required for telomere elongation in the absence of telomerase. Curr Biol 11:125–129
Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L (2001) The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J 20:905–913
Teng SC, Chang J, McCowan B, Zakian VA (2000) Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell 6:947–952
Tsai HJ, Huang WH, Li TK et al (2006) Involvement of topoisomerase III in telomere-telomere recombination. J Biol Chem 281:13717–13723
Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR (1999) Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res 59:4175–4179
Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275:986–990
Li TK, Liu LF (2001) Tumor cell death induced by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol 41:53–77
Snapka RM, Woo SH, Blokhin AV, Witiak DT (1996) Inhibition of topoisomerase II by ICRF-193, the meso isomer of 2,3-bis(2,6-dioxopiperazin-4-yl)butane. Critical dependence on 2,3-butanediyl linker absolute configuration. Biochem Pharmacol 52:543–549
Wasserman RA, Austin CA, Fisher LM, Wang JC (1993) Use of yeast in the study of anticancer drugs targeting DNA topoisomerases: expression of a functional recombinant human DNA topoisomerase II alpha in yeast. Cancer Res 53:3591–3596
Wasserman RA, Wang JC (1994) Analysis of yeast DNA topoisomerase II mutants resistant to the antitumor drug amsacrine. Cancer Res 54:1795–1800
Koshland D, Hartwell LH (1987) The structure of sister minichromosome DNA before anaphase in Saccharomyces cerevisiae. Science 238:1713–1716
Holm C, Stearns T, Botstein D (1989) DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol 9:159–168
Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633–643
Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73:177–208
Anbalagan S, Bonetti D, Lucchini G, Longhese MP (2011) Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet, e1002024
Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I (2008) Multiple pathways inhibit NHEJ at telomeres. Genes Dev 22:1153–1158
Luo K, Vega-Palas MA, Grunstein M (2002) Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16:1528–1539
Mirabella A, Gartenberg MR (1997) Yeast telomeric sequences function as chromosomal anchorage points in vivo. EMBO J 16:523–533
Park S, Patterson EE, Cobb J, Audhya A, Gartenberg MR, Fox CA (2011) Palmitoylation controls the dynamics of budding-yeast heterochromatin via the telomere-binding protein Rif1. Proc Natl Acad Sci USA 108:14572–14577
Wilson-Sali T, Hsieh TS (2002) Generation of double-stranded breaks in hypernegatively supercoiled DNA by Drosophila topoisomerase IIIbeta, a type IA enzyme. J Biol Chem 277:26865–26871
Hardy CF, Sussel L, Shore D (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6:801–814
Wotton D, Shore D (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev 11:748–760
Corbett KD, Berger JM (2004) Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct 33:95–118
Worland ST, Wang JC (1989) Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J Biol Chem 264:4412–4416
Grobelny JV, Godwin AK, Broccoli D (2000) ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J Cell Sci 113(Pt 24):4577–4585
Takai H, Smogorzewska A, de Lange T (2003) DNA damage foci at dysfunctional telomeres. Curr Biol 13:1549–1556
Saharia A, Stewart SA (2009) FEN1 contributes to telomere stability in ALT-positive tumor cells. Oncogene 28:1162–1167
Nitiss JL (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer 9:338–350
Roca J, Ishida R, Berger JM, Andoh T, Wang JC (1994) Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc Natl Acad Sci USA 91:1781–1785
Creighton AM, Hellmann K, Whitecross S (1969) Antitumour activity in a series of bisdiketopiperazines. Nature 222:384–385
Hellmann K, Newton KA, Whitmore DN, Hanham IW, Bond JV (1969) Preliminary clinical assessment of I.C.R.F. 159 in acute leukaemia and lymphosarcoma. Br Med J 1:822–824
Cesare AJ, Reddel RR (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11:319–330
Nabetani A, Ishikawa F (2011) Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J Biochem 149:5–14
Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119:355–368
Dejardin J, Kingston RE (2009) Purification of proteins associated with specific genomic Loci. Cell 136:175–186
Lovejoy CA, Li W, Reisenweber S et al (2012) Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 8:e1002772
Ira G, Malkova A, Liberi G, Foiani M, Haber JE (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401–411
Plank JL, Wu J, Hsieh TS (2006) Topoisomerase IIIalpha and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci USA 103:11118–11123
Ye J, Lenain C, Bauwens S et al (2010) TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142:230–242
Signon L, Malkova A, Naylor ML, Klein H, Haber JE (2001) Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol 21:2048–2056
Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM (1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol 134:1349–1363
Bhattacharyya S, Keirsey J, Russell B et al (2009) Telomerase-associated protein 1, HSP90, and topoisomerase IIalpha associate directly with the BLM helicase in immortalized cells using ALT and modulate its helicase activity using telomeric DNA substrates. J Biol Chem 284:14966–14977
Stavropoulos DJ, Bradshaw PS, Li X et al (2002) The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet 11:3135–3144
Nikolaou C, Bermudez I, Manichanh C et al (2013) Topoisomerase II regulates yeast genes with singular chromatin architectures. Nucleic Acids Res 41:9243–9256
Buseman CM, Wright WE, Shay JW (2012) Is telomerase a viable target in cancer? Mutat Res 730:90–97
Henson JD, Neumann AA, Yeager TR, Reddel RR (2002) Alternative lengthening of telomeres in mammalian cells. Oncogene 21:598–610
Bechter OE, Zou Y, Walker W, Wright WE, Shay JW (2004) Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res 64:3444–3451
Hu J, Hwang SS, Liesa M et al (2012) Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 148:651–663
Shay JW, Reddel RR, Wright WE (2012) Cancer. Cancer and telomeres—an ALTernative to telomerase. Science 336:1388–1390
Wang JC (1996) DNA topoisomerases. Annu Rev Biochem 65:635–692
Acknowledgments
We thank Drs. Leroy F. Liu and Ginger Zakian for providing plasmids and strains. We also thank Drs. Jing-Jer Lin and Marc Gartenberg for their critical comments on the manuscript. This work was supported by grants from the National Science Council (NRPGM-98-3112-B-002-039); National Taiwan University (NTU-CESRP-102R7602A1); and the National Health Research Institute of Taiwan (NHRI-EX102-9727BI) to SCT.
Author information
Authors and Affiliations
Corresponding author
Additional information
C.-H. Tsai, C.-C. Lin and T.-K. Li contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hsieh, MH., Tsai, CH., Lin, CC. et al. Topoisomerase II inhibition suppresses the proliferation of telomerase-negative cancers. Cell. Mol. Life Sci. 72, 1825–1837 (2015). https://doi.org/10.1007/s00018-014-1783-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-014-1783-0